Abstract
Lateral gene transfer (LGT) is now accepted as an important factor in the evolution of prokaryotes. Establishment of the occurrence of LGT is typically attempted by a variety of methods that includes the comparison of reconstructed phylogenetic trees, the search for unusual GC composition or codon usage within a genome, and identification of similarities between distant species as determined by best blast hits. We explore quantitative assessments of these strategies to study the prokaryotic trait of nitrogen fixation, the enzyme-catalyzed reduction of N2 to ammonia. Phylogenies constructed on nitrogen fixation genes are not in agreement with the tree-of-life based on 16S rRNA but do not conclusively distinguish between gene loss and LGT hypotheses. Using a series of analyses on a set of complete genomes, our results distinguish two structurally distinct classes of MoFe nitrogenases whose distribution cuts across lines of vertical inheritance and makes us believe that a conclusive case for LGT has been made.
Keywords: blast, codon usage, horizontal gene transfer, phylogeny
Lateral gene transfer (LGT) is the process by which genetic material is transferred between distinct evolutionary lineages. This mechanism contrasts with the Darwinian model of vertical descent, where genetic material is inherited from the preceding generation (1). LGT and integration of the transferred DNA into the recipient organism’s chromosome occurs by various extensively studied mechanisms (2). LGT is relatively common among prokaryotes, but less common between prokaryotes and eukaryotes. The spread of the acquired gene(s) in the recipient species population depends on natural selection and/or neutral genetic drift. For example, a gene that has been laterally transferred may confer antibiotic resistance and therefore provide a selective advantage to the organism in the presence of the antibiotic. It is evident that LGT may occur frequently at the cellular level, but it is more difficult for a transferred gene to be sustained in the population and subsequent generations (3, 4). It is commonly accepted that LGT is a source of genetic diversity and has important evolutionary consequences, but opinions vary on the degree of its influence on microbial evolution (4). This topic is of great current interest among biologists and many methods for detecting probable LGT occurrences have been developed (for reviews, see refs. 1, 3, and 5).
The increasing abundance of prokaryotic sequences and completed genomes offers new opportunities to study prokaryotic evolution and LGT. If reconstructed phylogenies from different genes conflict with each other and/or with well studied evolutionary relationships among the species, then this is commonly taken as a sign of LGT (6, 7). Other methods rely on blast searches (8) to detect similar gene sequences between divergent organisms (9, 10) or look for genes with deviations from genome-wide GC or codon composition (11).
The criticism of many of the approaches for identifying cases of LGT is that the observations can also be explained by a variety of other reasons, such as inaccurate phylogenetic reconstruction methods, gene loss in multiple lineages, novel sequences arising from the divergence of gene duplications, and varying mutation rates for different proteins (3, 5). Some strategies for identifying LGT are observational in nature, and evidence obtained for LGT may be prone to investigators’ biases or can be explained by statistical error. Each method has its own strengths and weaknesses, but the most convincing arguments depend on multiple lines of evidence from different methods. We try to address these weaknesses by exploring the use of quantitative measures for LGT and we apply our methods to the trait of nitrogen fixation (NIF)‖ as a case study.
All organisms depend on utilizable nitrogen, but only a few prokaryotes can obtain it from atmospheric nitrogen (N2) through conversion of N2 to ammonia, catalyzed by the heterodimeric enzyme nitrogenase. NIF serves as a good subject for a case study. The process has been exhaustively studied biochemically, and, in certain environments, organisms able to fix nitrogen have a strong selective advantage. Furthermore, the nif genes encoding the core proteins required for NIF are generally closely linked on prokaryotic chromosomes, making the probability of their joint transfer much more likely. Evidence, either supporting or opposing LGT in its evolutionary history, must be in agreement with NIF being a modular and complex trait.
Phylogenies constructed on nif genes are not consistent with the organismal phylogeny (12). There is disagreement on the cause of these contradictory results. The genes have a patchy distribution on the tree-of-life, which some have attributed to LGT (13, 14). Others have argued that the uneven distribution also could be explained by loss of function in certain lineages (12).
We performed a series of evaluations on the completely sequenced genomes of >100 prokaryotic species, of which 14 are nitrogen fixers. First, we performed genome-wide blast searches by using a subset of nif genes as queries and compared the distribution of the blast scores with scores obtained in the same manner using positive and negative control gene sets for LGT. Second, we applied machine learning methods to the 14 species to determine whether codon usage in the nif genes contradicts the discrimination of the species using general codon usage in the genome. Finally, we found that the nitrogen fixers are split into two equally sized groups based on blast scores, distinctive patterns of conserved covariant amino acid residues, and, in NifD, different “signature sequences” around the two invariant amino acids residues that serve as ligands to the MoFe cofactor. However, the placement of these groups is inconsistent with the 16S rRNA-based phylogenetic tree. To explore whether or not the two groups are clades descending from a common ancestor, we performed a statistical test by using bootstrap methodology. In summary, our results support the occurrence of LGT event(s) for NIF across prokaryotic lines.
Results
Our analysis of NIF genes is separated below into three different strategies that rely on the use of control sets for blast similarity searches, genome-wide codon usage, and phylogenetic reconstruction based on 16S rRNA. These methods are applied to complete genomes and to a set of critical NIF proteins. In our analysis, we use a set of 137 complete prokaryotic genomes that currently includes 14 nitrogen fixers (see Table 1). There are a small number of known NIF genes found both in bacterial and archaeal genomes. Our methods are applied to four of these genes (nifD, nifK, nifE, and nifN), which are essential for the function of nitrogenase (13). NifD and nifK encode the α- and β-subunits of nitrogenase, respectively, whereas nifE and nifN are required for the synthesis of the MoFe cofactor. Because the functions of these genes are indispensable for NIF, our conclusions require that there are consistent results for all four genes.
Table 1.
Confusion matrix for the training set of 750 randomly selected genes and four nif genes (values in parentheses)
| br_jap | cl_ace | ch_tep | de_eth | de_vul | ge_sul | me_ace | magnet | me_lot | me_maz | me_the | nostoc | si_mel | wo_suc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| br_jap | 497 (1) | 0 | 18 | 2 | 3 | 2 | 0 | 2 | 77 | 0 | 0 | 0 | 54 | 0 |
| cl_ace | 0 | 737 (4) | 0 | 6 | 1 | 2 | 13 | 1 | 0 | 26 | 3 | 11 | 0 | 0 |
| ch_tep | 49 (3) | 0 | 616 (4) | 11 | 19 | 41 | 1 | 9 | 55 (1) | 1 | 0 | 1 | 55 (1) | 1 |
| de_eth | 0 | 0 | 4 | 684 (3) | 1 | 5 | 12 | 12 | 1 | 10 | 1 | 2 | 2 | 0 |
| de_vul | 15 | 0 | 12 | 4 | 661 (4) | 50 | 1 | 2 | 27 | 0 | 0 | 0 | 27 | 0 |
| ge_sul | 20 | 0 | 30 | 1 | 29 | 614 (4) | 3 | 4 | 19 | 0 | 1 | 0 | 22 | 0 |
| me_ace | 0 | 2 | 11 | 11 | 6 | 6 | 484 (4) | 5 | 3 | 193 (4) | 12 | 6 | 5 | 4 |
| magnet | 2 | 0 | 17 | 7 | 5 | 7 | 5 | 693 (4) | 7 | 1 | 0 | 10 | 5 | 5 |
| me_lot | 78 | 0 | 9 | 0 | 5 | 3 | 0 | 1 | 452 | 0 | 0 | 0 | 71 (1) | 0 |
| me_maz | 0 | 1 | 3 | 9 | 3 | 2 | 205 | 1 | 0 | 496 | 23 | 5 | 1 | 2 |
| me_the | 0 | 0 | 0 | 0 | 0 | 3 | 9 | 0 | 0 | 13 | 708 (4) | 0 | 0 | 1 |
| nostoc | 0 | 10 | 2 | 13 | 0 | 1 | 15 | 14 | 0 | 8 | 1 | 713 (4) | 0 | 7 |
| si_mel | 87 | 0 | 24 | 1 | 15 | 8 | 0 | 0 | 108 (3) | 0 | 0 | 0 | 506 (2) | 0 |
| wo_suc | 2 | 0 | 4 | 1 | 2 | 6 | 2 | 6 | 1 | 2 | 1 | 2 | 2 | 730 (4) |
The nitrogen-fixing species are abbreviated as follows: br_jap, Bradyrhizobium japonicum; cl_ace, Clostridium acetobutylicum ATCC824; ch_tep, Chlorobium tepidum; de_eth, D. ethenogenes; de_vul, D. vulgaris; ge_sul, Geobacter sulfurreducens; me_ace, Methanosarcina acetivorans C2A; magnet, Magnetococcus MC-1; me_lot, Mesorhizobium loti; me_maz, M. mazei Goe1; me_the, Methanobacterium thermoautotrophicum delta H; nostoc, Nostoc sp. PCC7120; si_mel, Sinorhizobium meliloti; wo_suc, Wolinella succinogenes. Correct predictions are indicated in bold.
Control Set Comparisons.
Identification of similarities between distant species as determined by best blast hits is a common method for inferring LGT. Although very simple and fast, the method has been shown to generate incorrect conclusions due to other factors, such as gene loss and rate variation, that may also result in the detection of similar sequences from otherwise distant species (15, 16). As an attempt to account for the weaknesses in using blast, we provide a baseline for the distribution of best blast hits for our case-study genes by making comparisons between the four nif genes and sets of positive and negative control genes for LGT.
Although in general LGT events in the evolutionary history of a gene cannot be determined unequivocally, there are recent cases of LGT events that have been documented. In particular, LGT is known to play an important role in the emergence and spread of antibiotic-resistance genes (17). An example of an antibiotic-resistant gene shown to be transferred across pathogenic bacteria in recent years is CTX-M β-lactamase, which hydrolyzes the β-lactam ring of bacteriostatic penicillin-related antibiotics (18). CTX-M enzymes are cefotaximases, a subclass of plasmid-mediated, extended-spectrum β-lactamases, that were first reported in the second half of the 1980s, a few years after the introduction of the antibiotic cefotaxime. Within 20 years, the genes encoding these enzymes had spread to bacteria belonging to seven genera in three different orders of γ-proteobacteria. The precursors to these genes are chromosomal bla genes in different strains of Kluyvera spp., also γ-proteobacteria, that are opportunistic human pathogens. The CTX-M genes therefore represent a current well understood stage in one form of LGT, in which gene spread is mediated by CTX-M gene containing plasmids. For use as controls in our study, the key point is that, in each of the organisms examined, it is known that the CTX-M gene was acquired by LGT very recently. The outcome of the analysis will be the same whether the gene is currently on a plasmid or on the chromosome. For what we call our positive control set, we use the 40 examples from strains reviewed in ref. 18.
The absence of LGT events in the evolutionary history of a gene cannot be proven unambiguously, but for some genes there is evidence that would strongly argue against the occurrence of LGT. It has been shown that the lateral transfer of informational genes is less likely. This finding has given rise to the so-called complexity hypothesis (19) related to the ideas in ref. 20. With these considerations in mind, for what we call our negative control set of genes, we use three of the larger ribosomal (rib) proteins: S2, S3, and S4. Because of the complexity of ribosomal structure, which involves many protein–protein and protein–RNA interactions, it is likely that LGT did not play a major role in the evolution of ribosomal proteins.
We extracted the sequence for each of the control (rib S2, S3, S4, and CTX-M) and test-set (nifD, nifE, nifK, and nifN) proteins from a query species: Nostoc sp. PCC 7120 for the rib and nif proteins and Kluyvera cryocrescens for CTX-M. The blast software was then used to compare each query protein sequence to all sequences in each of the 137 complete genomes separately. blast returns a score, called the E value, between two sequences that reflects their similarity as determined by the optimal local alignment of the two sequences. In Figs. 1–3, each point symbolizes one of the complete genomes in our data (see ref. 21 for scatter-plot-based methods for evaluating evolutionary relationships). The y axis is a transformation of the E value for the score of the best alignment between the control or test-set sequence and each genome. Increasing values correspond to stronger similarity between the sequences.
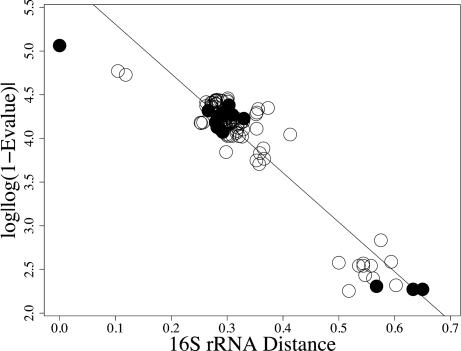
Fig. 1.
Plot of best blast hits E value for rib S2. NIF organisms are indicated by filled circles.
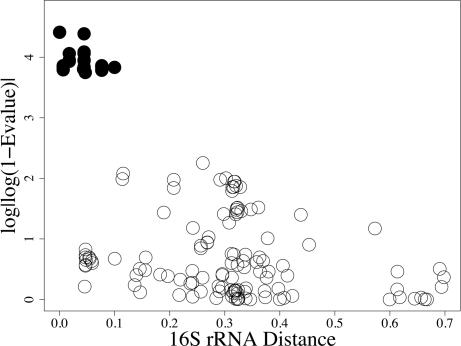
Fig. 2.
Plot of best blast hits E value for CTX-M. E values between the query sequence and the CTX-M enzymes given in ref. 18 are indicated by filled circles.
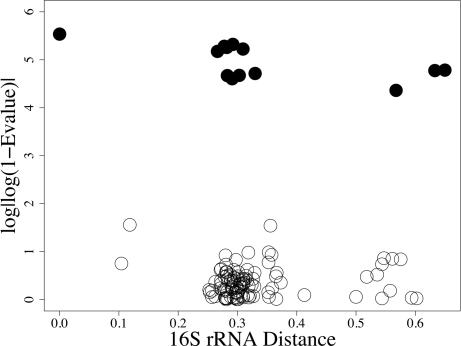
Fig. 3.
Plot of best blast hits E value for nifD. NIF organisms are indicated by filled circles.
As a baseline, we also plotted the evolutionary distance between the query species and the complete genome species as measured by 16S rRNA on the x axis (see Materials and Methods). This molecule was originally used to construct a universal species tree referred to as the “tree-of-life” (22). 16S rRNA is a good candidate for a baseline molecule because it is an integral component of the ribosome where it interacts with several different proteins (23, 24). Although there is coevolution between ribosomal proteins and 16S rRNA, some phylogenies based on the two types of molecules are known to differ (25). The strong selective pressures on 16S rRNA based on the structural constraints and numerous interactions make it an unlikely candidate for LGT, although some countervailing evidence is emerging (26).
In the negative control set (ribosomal proteins), we see a negative linear relationship (Fig. 1). The proteins are less similar as evolutionary distance increases. For the positive control (CTX-M), we see no relationship between evolutionary distance and protein similarity (Fig. 2). Finally, the NIF test set does not show a linear relationship, making these results more similar to those from the positive control for LGT (Fig. 3). These results also are consistent with the remaining nif and ribosomal proteins (see Figs. 6–10, which are published as supporting information on the PNAS web site). For both the positive control and NIF results, some species are very close relatives evolutionarily but are not similar with respect to these proteins. The comparisons with the control sets provide some qualitative support against an evolutionary history of strict vertical inheritance for the nif proteins.
Codon Usage.
Identifying unusual nucleotide or codon usage in a gene also is used to infer LGT. Codon usage varies from genome to genome, so it is possible that transferred sequences from another species may be identified by comparing the codon usage for all genes in a genome (3, 5). We apply machine learning methods to the genomes of nitrogen fixers to determine whether codon usage in the nif genes contradicts the discrimination of the species using general codon usage in the genome.
By using the software random forests (27), we constructed a classifier for the 14 NIF species genomes. The classifier is an algorithm that takes a data vector and assigns a genome label (see Materials and Methods). To construct the classifier, we trained it on 61-dimensional vectors of codon usage (removing stop codons) from 750 randomly selected non-nif genes for each of the 14 NIF species genomes. After the classifier is constructed, we test whether the classifier correctly identifies the genome if it is given the codon usage of a nif gene.
One way to display the performance of the classifier is through a confusion matrix (Table 1), where the column labels are the true genome classes and the row labels are the predicted genome classes. The diagonal entries are the number of correct predictions. For the training set of a random set of 750 genes, the overall error rate is 18.2% (Table 1). Given the four nif genes, the overall error rate is 23.6% (Table 1). Assuming that the nif genes are independent, these error rates are not significantly different at the 0.05 significance level using a normal approximation. However, the nif genes are not a random sample of genes and likely to be dependent. As an alternative classification scheme to account for their interdependencies, we can also define a correct classification if the majority of nif genes (at least two) for a NIF species classify that species. According to this scheme, the error rate is 21.4% and closer to that of the random set of genes.
In summary, we find that the nif genes have similar codon usage to typical genes. Moreover, most of the off-diagonal elements in the table, the incorrect predictions, appear for both gene sets among the same related species: rhizobia (br_jap, me_lot, and si_mel) and methanobacteria (me_ace and me_maz). The few incorrect predictions that do not appear for related species (e.g., ch_tep and rhizobia) are likely due to the background error rate. Our conclusion is that codon usage is not as useful a discriminator in the case of the NIF genes as we might expect if the occurrence of LGT was very ancient.
Structural vs. Phylogenetic Grouping.
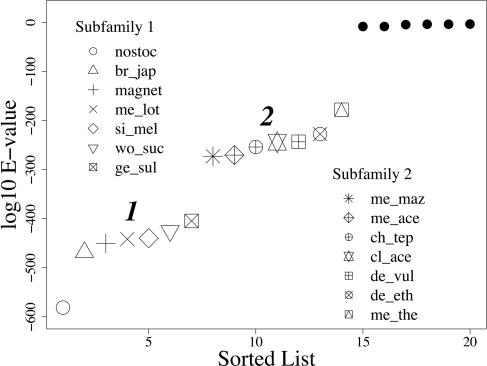
Fig. 4 shows the top 20 scores from the genome-wide blast experiments discussed above using Nostoc sp. PCC 7120 nifD as a query sequence. The best hits are to nifD sequences from the 13 other nitrogen fixers. In Fig. 4, we see that the NIF species are split into two distinct subfamilies. This separation also is observed in the corresponding plot for the other nif genes (see Figs. 11–13, which are published as supporting information on the PNAS web site). The blast E values in these plots measure similarity based on an average across long stretches of aligned positions. To further explore the placement of these two subfamilies, we also examine patterns of conservation of short nonadjacent positions, called motifs, that are covariant within subfamilies using the strong motif algorithm (28). Over 100 motifs are detected for each of the nif protein alignments (Tables 2–5, which are published as supporting information on the PNAS web site). Of these motifs, 62% on average are conserved within one of these two subfamilies, which is significantly more than expected by chance (see Table 6, which is published as supporting information on the PNAS web site). Therefore, these subfamilies also are consistent with patterns of conserved covariant amino acid patterns.
Fig. 4.
Plot of sorted list of top 20 best blast hits E value to Nostoc sp. PCC 7120 nifD. Non-NIF organisms are indicated by filled circles. Labels for the NIF organisms are listed in the key (see Table 1 for full names).
All diazotrophs that have been investigated contain a nitrogenase system based on Mo and Fe (29). In Azotobacter vinelandii nitrogenase, only two residues, Hisα-442 and Cysα-275, serve as ligands to the MoFe cofactors (30). These residues are conserved in all nifD sequences. In addition to the universal MoFe-dependent nitrogenase, some organisms have alternative nitrogenases dependent on V and Fe or Fe alone. Each of these three classes has a distinctive sequence surrounding Hisα-442 (29). Evidently there is a strong interdependence between the primary structure of this region of nifD and the metal composition of the cofactor. We explored the possibility that the two MoFe–nitrogenase subfamilies discovered in our study may differ in the primary structure of the regions surrounding Hisα-442 and Cysα-275 in nifD (by using A. vinelandii residue numbering). Indeed the differences between these regions in the two protein subfamilies are striking (Table 7, which is published as supporting information on the PNAS web site). In all members of subfamily 1, the sequence that includes Hisα-442, FRQMHSWDY, is identical to that in A. vinelandii nifD. In subfamily 2, there are variations at multiple positions in the corresponding sequence, (S,C,L)(R,K,L,V)(Q,L)(L,I)HSY(D,E)(Y,N), where different residues found at each position are shown in brackets. Note that the invariant tryptophan residue in subfamily 1 is replaced by an invariant tyrosine is subfamily 2. In subfamily 1, the sequence about Cysα-275 is L(N,V)(L,I)(L,Y,I)HCYRSMNY, identical in 9 of 12 positions to the corresponding sequence LNLVHCYRSMNY in A. vinelandii nifD. The consensus sequence L(N,S)(L,V,I)(L,V,I)(M,L,Q,R)C(H,Q)RS(A,I)(N,T)Y that includes Cysα-275 in subfamily 2 is much less conserved and identical in only 5 of 12 positions to the corresponding sequence in A. vinelandii nifD. The available information on the ecology, physiology, and biochemistry of the organisms belonging to subfamily 1 indicates that they may perform NIF in the presence of low levels of oxygen, whereas those in subfamily 2 are strict anaerobes (see Table 8, which is published as supporting information on the PNAS web site). It may be that the differences between the sequences around Hisα-442 and Cysα-275 are important in influencing the rate of inactivation of the enzyme by oxygen.
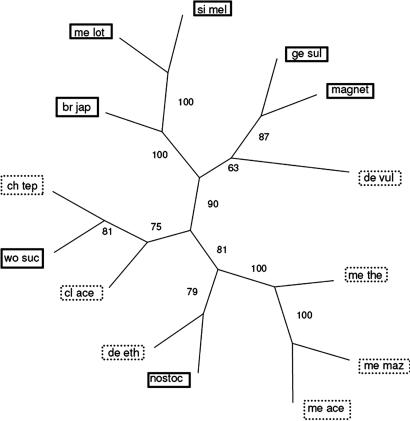
Although the two subfamilies separate by multiple different factors and are consistent with other groupings (see Fig. 14, which is published as supporting information on the PNAS web site) (31), they show a patchy distribution on a phylogeny based on 16S rRNA (Fig. 5; see Materials and Methods). This has been observed by others and attributed to LGT (31). An alternative explanation is that a common evolutionary ancestor exists for the two subfamilies and that the observed patchiness is due to random noise in the 16S rRNA sequences. Using the 16S rRNA sequences of the NIF organisms, we test the hypothesis that the observed patchiness of the two subfamilies on the 16S rRNA tree is still consistent with there being two clades originating from a common ancestor in the 16S rRNA tree. Specifically, we hypothesize that the observed large size of 16S rRNA distances between members of the same subfamilies is due merely to chance. If instead the two subfamilies are clades originating from a single ancestor, then the evolutionary process that led to the observed distances should frequently yield trees in which the two clades originating from the tree root have intraclade distance patterns consistent with those observed within the two subfamilies. If this scenario is false, then it is appropriate to reject the hypothesis that the families arose from a single ancestor without benefit of LGT.
Fig. 5.
Phylogeny of 14 NIF organisms based on 16S rRNA. The two subfamilies are labeled with solid (subfamily 1) and dotted boxes (subfamily 2). Branch lengths are the bootstrap frequencies.
We evaluate this single ancestor hypothesis using a bootstrap procedure. First, we sample 16S rRNA positions randomly for these 14 species with replacement to obtain a bootstrap 16S rRNA sequence sample. We then calculate the distance between all species and construct a neighbor-joining tree. The sum of the average distance (d) within the two clades that split from the root (averaged over all possible roots) is then calculated. This distribution of this statistic, d, in the bootstrap samples is compared to the sum of the average distances within the two observed NIF subfamilies in the original 16S rRNA sequences (dNIF). Using this procedure we calculate a P value for the observed value of dNIF under the null hypothesis that there is a common evolutionary ancestor for the two subfamilies. From 10,000 bootstrap simulations, we obtain P < 10−6. This result indicates that the two NIF populations are unlikely to be descended from a single ancestor.
Discussion
We have developed several methods used for exploring the evolutionary history of the nif genes. These methods are based on adapting existing strategies for identifying LGT: best blast hits, codon usage, and phylogenetic reconstruction and comparison. We have used controls for best blast hits, applied the machine learning methodology random forests to codon usage, and taken advantage of subfamily separation in the phylogeny for hypothesis testing. Furthermore, our conclusions have been put in a structural and functional context by requiring that there are consistent results among all four nif proteins that are essential for NIF. Although we use NIF as a case study, several of these approaches also are applicable to other complex traits.
Of the 137 complete prokaryotic genomes that we analyzed, NIF appears in a small subset of 14 organisms that are widely distributed across two kingdoms. This distribution also is observed by examining the occurrence of nif proteins from organisms with incomplete genomes (14, 31). The history of NIF in prokaryotes can be explained by three possible scenarios. First, this trait arose independently in more than one lineage. Second, the last common ancestor to all 14 organisms had the ability to fix nitrogen, but this trait was lost in multiple lineages. Third, LGT event(s) have spread this trait across prokaryotic families.
The possibility of LGT arising in multiple lineages through gene duplication and mutation is very unlikely (12). In general, nif proteins tend to be highly conserved and although there are four core proteins, there are up to 20 other nif proteins that also are important for proper NIF function (32). Furthermore, within the 14 NIF species that we examined, the gene order for the four nif genes is highly conserved (nifDKEN). These factors make it difficult to argue for independent origins, and there is consensus among the community that this is not a plausible history (31).
The occurrence of numerous gene loss events is an alternative hypothesis to LGT for explaining a gene’s patchy distribution on established phylogenetic relationships. Discriminating between gene loss and LGT unambiguously is very difficult because our data on extant organisms is a single static image of a history of prokaryotic evolution that is dynamic and goes back billions of years (33). Unfortunately, the information on the ancestors of existing organisms that would help to solve the debate is not available. Nevertheless, we will discuss the results of our three methods applied to the nif genes in the context of these two alternative hypotheses.
Unusual codon usage of a gene may indicate the presence of a foreign or laterally transferred gene in a genome. The weakness with this approach is that it cannot recognize the transfer of genes between genomes with similar codon usage or a transfer event that is very old (5). The nif genes do not have unusual codon usage, and they are very similar to “typical” genes in the genome for discriminating between species. However, NIF is believed to be an ancient trait (33, 34). Therefore, the lack of unusual codon usage does not refute the occurrence of LGT. If these genes were transferred, their codon usage is likely to have adapted to the background genomic codon usage over the long history of NIF organisms.
Our analysis of best blast hits with positive and negative control sets show that the history of nif genes shows similarities to recent LGT events like the transfer of CTX-M genes conferring antibiotic resistance, which occurred within the last 20 years. However, we can only make qualitative statements because the sensitivity of this type of test is compromised by the scale in our control sets. That is, CTX-M genes occur in species, particularly pathogens, more closely related to each other than the other NIF species. The nonlinearity observed in Fig. 2 for CTX-M at short distances cannot be extrapolated to the larger distances in Fig. 3 for NIF species. Despite this limitation, this analysis addresses the ability of sequences to discriminate between two different alternative histories as seen in the CTX-M and the ribosomal proteins.
The inconclusive results based on codon usage bias and the control set comparisons illustrate how the age of LGT events obscures the problem of determining LGT. Our analysis of phylogenetic and structural grouping provided more lucid explanations of the NIF history. Regardless of whether there has been widespread gene loss of the NIF trait that would result in its absence in 123 of our 137 species, we can still explore the occurrence of LGT for the nif genes within the 14 species. In particular, we find that these 14 species split into two groups based on the global and covariant sequence similarity of the nif proteins and consistent with ecological niches. The results of our bootstrap test indicate that a common ancestor for the two subfamilies is highly unlikely and support the presence of LGT event(s) in the history of NIF in these species.
Assuming that 16S rRNA provides a reasonable representation of evolutionary relationships, then, these results are incompatible with a history solely consisting of vertical inheritance patterns and gene loss. Our results are in agreement with the following evolutionary history for the nifDKEN genes. A common ancestral gene gave rise to these gene families in the bacterial and archaeal lineages (35). Early in the history of one of these lineages, adaptation to environmental change led to divergence between the nifDKEN sequences in the two lineages leading (at a minimum) to the two subfamilies observed in our analysis. Lateral gene transfer between members of the two subfamilies led to the present nif gene distribution, inconsistent with the 16S RNA phylogeny. There also may have been multiple LGT events within each subfamily, but potential detection of such events would require a finer-grained analysis of many more microorganisms. Our results are consistent with a “mixed scenario” of a combination of LGT event(s) across the subfamilies, vertical inheritance patterns within a subfamily and gene loss in non-NIF species. The possibility of LGT in the history of the nif genes has been discussed by other groups (14, 31), but we hope to have substantiated this claim by multiple sources of evidence and a series of quantitative measures.
Sequencing projects are motivated by many factors and the current inventory of complete genomes may not be entirely reflective of prokaryotic diversity; some taxonomic groupings may be better represented than others. Conclusions based on existing data are limited to the current sampling of species. Nonetheless, we believe that our results, which are based on a large sample of 137 species spanning both archaeal and bacterial kingdoms, are robust. As the number of sequenced genomes increases, we will have a better understanding of prokaryotic diversity and more opportunities to study prokaryotic evolution.
Materials and Methods
Sequence Data.
A library of 135 organisms with complete genomes (as of March 2004) was obtained from The Institute for Genomic Research Comprehensive Microbial Resource database (TIGR CMR, available at www.tigr.org/tigr-scripts/CMR2/CMRHomePage.cgi). The addition of two genomes from nitrogen fixers, Desulfovibrio vulgaris Hildenborough (December 2004) and Dehalococcoides ethenogenes 195 (March 2005), brought the total count of genomes to 137, including 14 nitrogen fixers (see Table 9, which is published as supporting information on the PNAS web site). The entire set of protein sequences for each organism in the library was downloaded from TIGR CMR.
The nif protein sequences were obtained from TIGR CMR or GenBank, and their accession numbers are listed in the Table 10, which is published as supporting information on the PNAS web site. There is no annotated nifN protein for D. ethenogenes 195. The ribosomal protein sequences (S2, S3, and S4) were obtained from TIGR CMR through a batch download for each organism within our library. From the batch download, we found that sometimes a genome contained two copies of one of the ribosomal proteins, labeled either A or B. Only one sequence is taken for each genome by selecting the longer sequence or the A strand if both sequences were identical. The CTX-M protein sequences were obtained from GenBank under the accession numbers given in ref. 18.
For the organisms with complete genomes, we were able to obtain their 16S rRNA sequences from the Ribosomal Database Project-II Release 9 (http://rdp.cme.msu.edu). However, the Ribosomal Database Project did not contain the 16S rRNA for the seven genera containing the CTX-M sequences listed in ref. 18. Therefore, 16S rRNA sequences from five or six pathogenic strains of the species for each of the seven genera were obtained, an alignment was constructed in clustalw (36), and a consensus 16S rRNA sequence was determined (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Algorithms and Software.
blast 2.0 (8) was downloaded from the National Center for Biotechnology Information ftp site. E values were obtained by running blast between the control (e.g., nifD from Nostoc sp. PCC 7120) and a database (e.g., all protein sequences from one genome). We used default parameters except for the filter option, which was turned off, resulting in longer alignments. In our reported results, we selected Nostoc sp. PCC 7120 as the control organism, but do not expect the conclusions to change due to this selection because blast results using different query organisms (e.g., Methanosarcina mazei Goe1) were similar (data not shown). blast E values between some of the 16S rRNA consensus sequences for the CTX-M species were unusable because of their close similarity. We extrapolated the E value from their blast score using the method described in Supporting Materials and Methods.
The fortran source code for random forests, version 5.1 (27), was obtained from A. Cutler (Utah State University, Logan). random forests was run with default parameters, except the options mtry (the number of variables to split at each node) and jbt (the number of trees to be grown in the forest), which were set to 10 and 100, respectively. For each protein sequence, we used codonw (37) with default parameters to calculate the Relative Synonymous Codon Usage (RSCU), which is defined as the observed codon frequency divided by the expected frequency if all synonymous codons for an amino acid were equally likely (38). Proteins annotated as “unknown” or “hypothetical” were excluded from the analysis. All protein and 16S rRNA alignments were generated in clustalw (36).
phylip (Phylogeny Inference Package), version 3.6 (39), was used to create the phylogeny in Fig. 5, which was based on 100 bootstrap samples with default parameters for the following phylip packages: seqboot to generate the bootstrap data; dnadist to calculate distances; neighbor to construct the trees for each bootstrap sample; and consense to construct the final consensus tree. All calculations in the bootstrap procedure are made with default parameters for the following phylip packages: seqboot, dnadist, and neighbor. We ran 10,000 bootstrap runs in increments of 100.
The strong motif algorithm was obtained from the authors of ref. 28. For each of the four nif protein alignments, the algorithm lists a set of motifs that are associated with a subfamily in the data set. For each subfamily size, we tallied the number of motifs and the number of motifs that were associated with a subfamily that was entirely contained in one of the two NIF subfamilies. We calculated the expected number of motifs that would be contained within one of the two NIF subfamilies if the motif subfamilies were chosen at random and then performed a hypergeometric test for each subfamily size.
Supplementary Material
Acknowledgments
We thank Charles Kurland and Rob Knight for helpful comments and suggestions. This research was supported by National Institutes of Health Grant R01 GM075312 (to P.J.B.), a Sloan Foundation/Department of Energy Postdoctoral Fellowship (to K.J.K.), and a National Science Foundation Vertical Integration of Research and Education in the Mathematical Sciences Undergraduate Research Assistantship (to J.C.L.).
Abbreviations
- LGT
lateral gene transfer
- NIF
nitrogen fixation.
Footnotes
Conflict of interest statement: No conflicts declared.
‖We will use the abbreviation nif for the nitrogen fixation process and the designations nif: and nif for nitrogen fixation genes and proteins, respectively.
References
- 1.Koonin E. V., Makarova K. S., Aravind L. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Syvanen M., Kado C. I., editors. Horizontal Gene Transfer. 2nd Ed. London: Academic; 2002. [Google Scholar]
- 3.Kurland C. G., Canback B., Berg O. G. Proc. Natl. Acad. Sci. USA. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurland C. G. BioEssays. 2005;27:741–747. doi: 10.1002/bies.20258. [DOI] [PubMed] [Google Scholar]
- 5.Eisen J. A. Curr. Opin. Genet. Dev. 2000;10:606–611. doi: 10.1016/s0959-437x(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 6.Raymond J., Zhaxybayeva O., Gogarten J. P., Gerdes S., Blankenship R. E. Science. 2002;298:1616–1620. doi: 10.1126/science.1075558. [DOI] [PubMed] [Google Scholar]
- 7.Archibald J. M., Rogers M., Toop M., Ishida K., Keeling P. J. Proc. Natl. Acad. Sci. USA. 2003;100:7678–7683. doi: 10.1073/pnas.1230951100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Rivera M. C., Jain R., Moore J. E., Lake J. A. Proc. Natl. Acad. Sci. USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson K. E., Clayton R. A., Gill S. R., Gwinn M. L., Dodson R. J., Haft D. H., Hickey E. K., Peterson J. D., Nelson W. C., Ketchum K. A., et al. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 11.Mrazek J., Karlin S. Ann. N.Y. Acad. Sci. 1999;18:314–329. doi: 10.1111/j.1749-6632.1999.tb08893.x. [DOI] [PubMed] [Google Scholar]
- 12.Young J. P. W. In: Nitrogen Fixation: From Molecules to Crop Productivity. Pedrosa F. O., Hungria M., Yates M. G., Newton W. E., editors. London: Kluwer–Academic; 1999. pp. 161–164. [Google Scholar]
- 13.Leigh J. A. Curr. Issues. Mol. Biol. 2000;2:125–131. [PubMed] [Google Scholar]
- 14.Boucher Y., Christophe J., Douady R., Papke T., Walsh D. A., Boudreau M. R., Nesbø C. L., Case R. J., Doolittle W. F. Annu. Rev. Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- 15.Genereux D. P., Logsdon J. M. Trends Genet. 2003;19:191–195. doi: 10.1016/S0168-9525(03)00055-6. [DOI] [PubMed] [Google Scholar]
- 16.Koski L. B., Golding G. B. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 17.Dzidic S., Bedekovic V. Acta Pharmacol. Sin. 2003;24:519–526. [PubMed] [Google Scholar]
- 18.Bonnet R. Antimicrob. Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain R., Rivera M. C., Lake J. A. Proc. Natl. Acad. Sci. USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woese C. R. Science. 1990;247:789. doi: 10.1126/science.2305249. [DOI] [PubMed] [Google Scholar]
- 21.Farahi K., Pusch G. D., Overbeek R., Whitman W. B. J. Mol. Evol. 2004;58:615–631. doi: 10.1007/s00239-004-2582-2. [DOI] [PubMed] [Google Scholar]
- 22.Woese C. R., Kandler O., Wheelis M. L. Proc. Natl. Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doolittle W. F. Science. 1999;286:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 24.Gutell R. R., Lee. J. C., Cannone J. J. Curr. Opin. Struct. Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- 25.Wolf Y. I., Rogozin I. B., Grishin N. V., Tatusov R. L., Koonin E. V. BMC Evol. Biol. 2001;1:8. doi: 10.1186/1471-2148-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Berkum P., Terefework Z., Paulin L., Suomalainen S., Lindstrom K., Eardly B. D. J. Bacteriol. 2003;185:2988–2998. doi: 10.1128/JB.185.10.2988-2998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breiman L. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 28.Bickel P. J., Kechris K. J., Spector P. C., Wedemayer G. J., Glazer A. N. Proc. Natl. Acad. Sci. USA. 2002;99:14764–14771. doi: 10.1073/pnas.222508899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eady R. R. Chem. Rev. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 30.Howard J. B., Rees D. C. Chem. Rev. 1996;96:2965–2982. doi: 10.1021/cr9500545. [DOI] [PubMed] [Google Scholar]
- 31.Raymond J., Siefert J., Staples C., Blankenship R. E. Mol. Biol. Evol. 2004;21:541–554. doi: 10.1093/molbev/msh047. [DOI] [PubMed] [Google Scholar]
- 32.Rubio L. M., Ludden P. W. J. Bacteriol. 2005;187:405–414. doi: 10.1128/JB.187.2.405-414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anbar A. D., Knoll A. H. Science. 2002;297:1137–1142. doi: 10.1126/science.1069651. [DOI] [PubMed] [Google Scholar]
- 34.Newton W. In: Prokaryotic Nitrogen Fixation: A Model System for the Analysis of a Biological Process. Triplett W., editor. New York: Springer; 2000. pp. 3–8. [Google Scholar]
- 35.Miller R. V., Day M. J., editors. Microbial Evolution: Gene Establishment, Survival, and Exchange. Washington, DC: Am. Soc. Microbiol.; 2004. [Google Scholar]
- 36.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peden J. Ph.D. thesis. Nottingham, U.K.: Univ. of Nottingham; 1999. [Google Scholar]
- 38.Sharp P. M., Tuohy T. M. F., Mosurski K. R. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.