Abstract
This study compares the relative effects of advancing male age on multiple genomic defects in human sperm [DNA fragmentation index (DFI), chromatin integrity, gene mutations, and numerical chromosomal abnormalities], characterizes the relationships among these defects and with semen quality, and estimates the incidence of susceptible individuals for a well characterized nonclinical nonsmoking group of 97 men (22–80 years). Adjusting for confounders, we found major associations between age and the frequencies of sperm with DFI and fibroblast growth factor receptor 3 gene (FGFR3) mutations associated with achondroplasia (P < 0.01) with no evidence for age thresholds. However, we found no associations between age and the frequencies of sperm with immature chromatin, aneuploidies/diploidies, FGFR2 mutations (Apert syndrome), or sex ratio in this cohort. There were also no consistent correlations among genomic and semen-quality endpoints, except between DFI and sperm motility (r = −0.65, P < 0.001). These findings suggest there are multiple spermatogenic targets for genomically defective sperm with substantially variable susceptibilities to age. Our findings predict that as healthy males age, they have decreased pregnancy success with trends beginning in their early reproductive years, increased risk for producing offspring with achondroplasia mutations, and risk of fathering offspring with Apert syndrome that may vary across cohorts, but with no increased risk for fathering aneuploid offspring (Down, Klinefelter, Turner, triple X, and XYY syndromes) or triploid embryos. Our findings also suggest that the burden of genomic damage in sperm cannot be inferred from semen quality, and that a small fraction of men are at increased risk for transmitting multiple genetic and chromosomal defects.
Keywords: DNA fragmentation, human sperm, achondroplasia, sperm FISH, Apert syndrome
It has become more socially acceptable to delay fatherhood, but the heritable consequences of this trend remain poorly understood. Since 1980, U.S. birth rates have increased up to 40% for men 35–49 years and have decreased up to 20% for men under 30 (1). Although it is well known that as women age, they are at increased risk for infertility, spontaneous abortion, and genetic and chromosomal defects among offspring, the association of male aging with these outcomes has been less well characterized (2).
Advancing paternal age has been implicated in a broad range of abnormal reproductive and genetic outcomes (3, 4), including diminished semen quality (5), reduced fertility (6), increased frequencies of spontaneous abortions (7, 8), ≈20 autosomal dominant diseases including achondroplasia (ACH) and Apert syndrome (AS; see refs. 3 and 9), and several diseases of complex etiology such as schizophrenia (10). Among transmitted chromosomal defects, sex-chromosomal aneuploidy syndromes show substantial paternal contributions with some evidence of paternal-age effects (11–13). However, the mechanisms for age dependency of paternally transmitted genomic defects are poorly understood.
The copy-error hypothesis that continuous stem-cell renewal in males leads to increases in heritable mutations with male age (14) continues to be challenged by increasing lines of evidence: (i) few or ambiguous associations between male age and certain genetic diseases of paternal origin (15, 16) and germ-cell mutations in transgenic mice (17, 18), (ii) nonlinear relationships between specific human sperm mutations and corresponding heritable effects (19, 20), (iii) protein-driven gametic selection mechanisms (21), and (iv) differential susceptibilities of meiotic and postmeiotic cells to heritable damage in mice (22).
Advances in detecting genomic types of defects directly in sperm provide new insights into the spermatogenic targets and mechanisms underlying male age effects. Damage to sperm chromatin, such as DNA fragmentation (DFI) that has been associated with male fertility, successful conception, and sustained pregnancy (23–27), increases with age (28, 29), as do DNA breakage (30–32) and chromosomal aberrations in sperm (33, 34). Sperm mutations associated with ACH (1138G>A mutation in fibroblast growth factor receptor 3 gene, FGFR3; see ref. 20) and with AS (755C>G and 758C>G mutations in the fibroblast growth factor receptor 2 gene, FGFR2; see refs. 19 and 35) increase with age, but less than expected from live-birth data (19, 20). The effects of paternal age at other gene loci (35, 36) and on sperm aneuploidy/diploidy (2, 37) remain inconclusive. It remains unclear whether the inconsistent age results across these studies are due to cohort-specific differences or differing age susceptibilities among the various types of sperm genomic defects.
Our study compares the relative effects of advancing male age on diverse measures of genomic damage in human sperm (chromatin integrity, gene mutations, sex ratio, and numerical chromosomal abnormalities) within a population of well characterized nonsmoking men from a nonclinical setting, assesses intercorrelations among these genomic defects and with semen quality endpoints, and estimates the proportion of men with abnormally high frequencies of sperm with multiple genomic defects. This study also contrasts the effects of age on sperm mutations in two different groups, analyzed in the same laboratory.
Results
Participants were currently employed or active retirees, predominantly white (91%), highly educated (55% postcollege education), and in good to excellent health by self report; they provided a convenience specimen after an average of 5.1 days of sexual abstinence (SD, 3.6; range, 2–20).
Sperm DFI and High DNA Stainability (HDS, Immature Chromatin).
Age was positively associated with all five DFI endpoints (mean, SD, percent, moderate, and high) with significant trends across age decades (P < 0.01) and strong correlations with age (r = 0.64–0.72, P < 0.001; see Table 1 for percent DFI (%DFI); see also Table 2, which is published as supporting information on the PNAS web site, for mean, SD, moderate, and high DFI). Duration of sexual abstinence was positively associated with all DFI endpoints (r = 0.43–0.54, P < 0.001). For percent DFI, the DFI endpoint with the highest correlation to age, the change per year of age was a relative 3.1% after adjusting for abstinence [95% confidence interval (C.I.), 2.3 and 3.9; r2 = 0.54; Fig. 1A]. Approximately 41% of the variance of logarithm %DFI was explained by age (partial r = 0.64, P < 0.001). Regression models of the other four DFI endpoints also had significant increases per year of age (mean DFI, 1.3%; SD DFI, 2.3 channels of fluorescence; moderate DFI, 2.7%; high DFI, 3.7%; P < 0.001). Hockey-stick analyses of the adjusted regression models did not improve the curve fits, suggesting there were no “thresholds” in the age-association curves for the DFI parameters.
Table 1.
Effects of male age on sperm DNA fragmentation, ACH mutations, AS mutations, sex ratio, aneuploidy, and diploidy, with the predicted change per year of age
| Sperm DNA fragmentation* |
ACH mutations* |
AS mutations* | Sex ratio, aneuploidy, and diploidy* |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | %DFI |
HDS |
1138G>A |
755C>G + 758C>G |
n | Sex ratio | Hyper- and hypohaploidy‡ |
Diploidy‡ |
|||||||||||
| n | Mean | (SD) | Abnormal† | Mean | (SD) | Abnormal† | n | Mean | (SD) | n | Mean | (SD) | Mean | (SD) | Mean | (SD) | |||
| Age group, yr | |||||||||||||||||||
| 20–29 | 19 | 12.9 | (7.7) | 2 (10) | 9.1 | (6.1) | 3 (16) | 16 | 0.55 | (0.29) | 18 | 0.18 | (0.26) | 19 | 0.97 | 64.9 | (24.1) | 18.2 | (18.3) |
| 30–39 | 20 | 16.3 | (9.6) | 2 (10) | 8.4 | (6.2) | 2 (10) | 14 | 0.63 | (0.32) | 19 | 0.07 | (0.11) | 20 | 0.98 | 58.4 | (26.8) | 13.9 | (10.3) |
| 40–49 | 16 | 23.2 | (14.9) | 4 (25) | 5.8 | (5.0) | 1 (6) | 12 | 0.92 | (0.61) | 13 | 0.11 | (0.15) | 16 | 0.98 | 49.5 | (19.2) | 8.4 | (4.6) |
| 50–59 | 17 | 35.4 | (18.6) | 8 (47) | 6.6 | (5.2) | 1 (6) | 6 | 1.48 | (0.64) | 13 | 0.04 | (0.07) | 16 | 0.98 | 47.3 | (14.3) | 10.6 | (4.9) |
| 60–80 | 16 | 49.6 | (17.3) | 14 (88) | 5.5 | (2.7) | 0 (0) | 10 | 1.85 | (1.30) | 13 | 0.33 | (0.71) | 19 | 0.98 | 57.5 | (17.9) | 13.4 | (12.2) |
| Total | 88 | 26.6 | (19.1) | 30 (34) | 7.2 | (5.4) | 7 (8) | 58 | 0.97 | (0.83) | 76 | 0.14 | (0.34) | 90 | 0.98 | 56.0 | (21.7) | 13.1 | (11.8) |
| P for trend | <0.01 | 0.03 | <0.01 | 0.68 | 0.41 | 0.20 | 0.17 | ||||||||||||
| Correlation§ | 0.72, <0.001 | −0.22, 0.03 | 0.54, <0.001 | −0.02, 0.8 | 0.14, 0.2 | −0.08, 0.3 | −0.09, 0.2 | ||||||||||||
| Predicted percent change¶ per year of age (95% C.I.) | |||||||||||||||||||
| Unadjusted | 3.6 | (2.8, 4.4) | −1.0 | (−1.9, −0.2) | 3.3 (2.0, 4.6) | 1.9 (−2.1, 6.0) | −0.3 (−0.8, 0.1) | −0.8 (−1.7, 0.2) | |||||||||||
| Adjusted‖ | 3.1 | (2.3, 3.9) | −0.7 | (−1.6, 0.2) | 2.0 (0.5, 3.5) | 0.6 (−3.9, 5.3) | −0.4 (−0.9, 0.1) | −0.4 (−1.4, 0.7) | |||||||||||
*DFI and immature chromatin (HDS) measured by the Sperm Chromatin Structure Assay and (SCSA), ACH mutations measured by PCR per 10,000 genomes, AS mutations by PCR per 50,000 genomes, sex ratio as Y- to X-carrying sperm, and aneuploidy and diploidy by sperm FISH per 10,000 sperm. Detailed results for all subcategoriees of sperm defects measured by each technology are reported in Tables 2–4.
†n (%) above threshold for decreased fertility: %DFI ≥ 30%; HSD ≥ 15% (see Methods).
‡Hyper- and hypohaploidy = sum(X-X-21, Y-Y-21, X-Y-21, X-21-21, Y-21-21, X-0, Y-0, and 21-0); diploidy = sum (X-Y-21-21, X-X-21-21, and Y-Y-21-21).
§Age correlation coefficient, P value. For %DFI and HDS: Pearson correlation. For AS, ACH, aneuploidy, and diploidy: Kendell’s tau.
¶For %DFI and HDS, the change per year of age is a relative percent, as converted from the antilog of the regression coefficient.
‖Adjusted for: %DFI and HDS (abstinence); ACH (radioisotopes exposure based on dosimetry records); AS (abstinence, body mass index, and alcohol use); hyper- and hypohaploidy, abstinence, occupational exposures, scorer); diploidy (occupational exposures, radioisotope exposure).
Fig. 1.
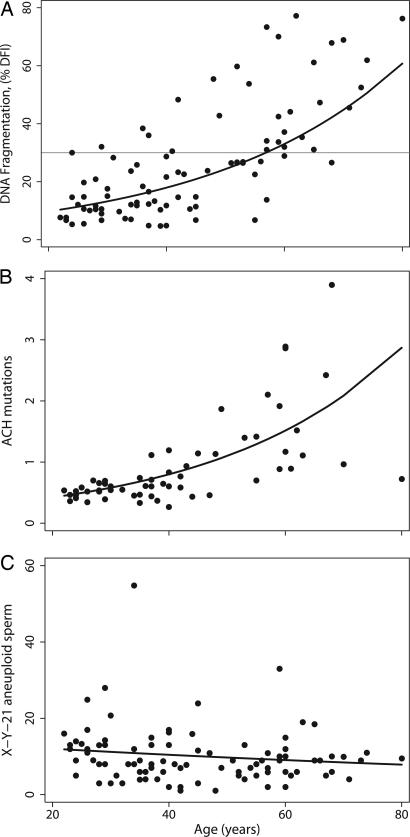
Relationship between male age and selected genomic defects in sperm. Data for each man are plotted with the regression line for age (Table 1; see Fig. 4 for other endpoints). (A) %DFI vs. age. The regression was adjusted for abstinence. The horizontal line is the threshold value (30%) associated with decreased pregnancy rates. (B) ACH mutations (1138G>A) per 10,000 genomes vs. age. The unadjusted negative binomial regression line is shown. Adjusting the model for the history of working with radioisotopes did not significantly change the regression results. (C) X-Y-21 aneuploid sperm per 10,000 sperm vs. age. The unadjusted negative binomial regression line is shown. Adjusting for covariates did not change the relationship (Table 4).
Thirty men had %DFI at or above values previously associated with increased risk of male infertility (Table 1; see refs. 24 and 27), with similar results for mean and SD DFI (Table 2). The proportion of men with abnormal DFI values increased across age categories (P < 0.01), and all five men over 70 had abnormal DFI values (data not shown). Abstinence-adjusted regression models estimated that men reached abnormal DFI values at age 56.9 years for %DFI (Fig. 1A), 55.8 for mean DFI, and 46.1 for SD DFI.
The incidence of sperm with HDS was not correlated with the DFI endpoints after adjusting for age and abstinence (Table 1), providing evidence that HDS is an independent measure of sperm chromatin damage. HDS was slightly negatively associated with age in unadjusted analyses (r = −0.22, P = 0.03) but not after adjusting for abstinence (−0.7% change per year, 95% C.I., −1.6, 0.2; see Fig. 4A, which is published as supporting information on the PNAS web site).
ACH and AS Mutations.
Among 20- to 29-year-old men, the baseline incidence of ACH mutations in sperm was >25-fold higher than the two AS mutations: 0.55 per 10,000 sperm genomes for 1138G>A vs. 0.020 for 755C>G and 0.015 for 758C>G mutations. There was a significant increase with age for the ACH mutation (Table 1; Fig. 1B). The decade-specific incidence of ACH mutations increased from 0.55 per 10,000 genomes for men 20–29 years to 1.85 for men 60+ years (P value for trend <0.01). We found a 3.3% increase in ACH mutation frequency per year of age (unadjusted, 95% C.I., 2.0 and 4.6). Occupational radiation exposure history (film-badge records) was positively associated with the frequency of ACH sperm mutations (1.51 per 10,000 genomes for 18 men with nonzero exposure vs. 0.65 per 10,000 for 40 men with zero exposure; unadjusted P = 0.002; adjusted for age, P = 0.02). After adjusting for radiation history, the frequency of ACH mutations increased 2.0% per year of age (95% C.I., 0.5 and 3.5). Approximately 29% of the variance in ACH mutations was explained by age, after adjusting for radiation history (partial r = 0.54, P < 0.001). Hockey-stick analysis of the adjusted regression model for ACH did not improve the curve fit, suggesting there is no “threshold” in the age association for ACH mutations.
After adjustment for covariates (Table 1), there was no statistically significant increase in AS mutations with age in this cohort for the sum of 755C>G plus 758C>G mutations: 1.9% per year; 95% C.I., −2.1 and 6.0 (Table 1 and Fig. 4B), or for the individual mutations (755C>G: 2.1% change per year; 95% C.I., −3.2 and 7.0; 758C>G: 1.6%, 95% C.I., −4.3 and 7.8; see Table 3, which is published as supporting information on the PNAS web site).
Sex Ratio and Sperm Aneuploidy and Diploidy.
There was no association between male age and sperm sex ratio, aneuploidy, or diploidy in this cohort (Table 1; see Table 4, which is published as supporting information on the PNAS web site, for individual aneuploidy categories), when age was examined by age decades or as a continuous variable. There was a slight decrease in the unadjusted frequency of sex-null sperm with age, which was not significant after adjusting for covariates (−0.52% change per year, 95% C.I., −1.2 and 0.22). As an example, Fig. 1C illustrates the age relationship for XY sperm, with the unadjusted negative binomial regression line (Table 4).
Intercorrelations.
There were significant intercorrelations among the group of five DFI endpoints (0.66 < r < 0.98, P < 0.001) among the 14 aneuploidies and diploidies (see footnote to Table 4) but not among the three gene mutations. The major types of genomic damage were not intercorrelated after adjusting for outliers or age, with the following exceptions: DFI endpoints vs. disomy Y (age adjusted: −0.21 < r < −0.28, P < 0.05) and HDS vs. aneuploidy (disomy 21, sex nullisomy, and total hyper-plus hypohaploidy; age adjusted 0.23 < r < 0.28, P < 0.05). Men with AS mutations in sperm (n = 33) had higher frequencies of sex nullisomic sperm than men with no detectable mutations (n = 43; 20.9 vs. 14.8, P = 0.01) and higher aggregate frequencies of all measured aneuploidies (63.0 vs. 48.4, P = 0.008).
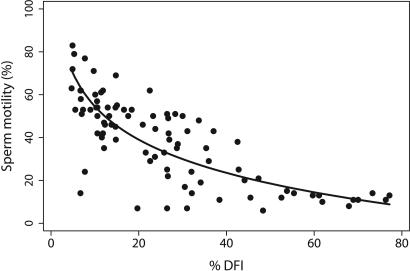
Genomic defects were generally not associated with semen quality (semen volume, sperm concentration, total sperm count, percent motility, percent progressive motility, and total progressive motility; see ref. 5), with the following exceptions. All five measures of DFI were strongly correlated with sperm motility (age adjusted: −0.42 < r < −0.65, P < 0.001); %DFI vs. percent motility had the strongest correlation (Fig. 2), with ≈40% of the variance in motility explained by %DFI. Mean, percent, and moderate DFI were also inversely associated with sperm count (age adjusted: −0.21 < r < −0.28, P < 0.05). HDS was negatively associated with total count, concentration, progressively motile sperm, and total progressively motile sperm (age adjusted: −0.24 < r < −0.30, P < 0.03). Also, men with detectable AS mutations had slightly lower progressive motility compared with men with no detectable mutations (26% vs. 17%, P = 0.04).
Fig. 2.
Relationship between %DFI and sperm motility. The data for each man are plotted with the regression line, adjusted for age and duration of abstinence. The partial correlation coefficient between motility and logarithm (%DFI) is 0.66 (P < 0.001).
Four individuals produced very high frequencies of sperm (>95th percentile) for two or more genomic defects, including defects with no intercorrelations: donor 1, 29 years old, very high for HDS and total hyperhaploidy; donor 2, 60 years old, very high for sex nullisomy, ACH, and AS; donor 3, 68 years old, highest for ACH and disomy 21, high for diploidy (>80th percentile); donor 4, 70 years old, highest for both AS mutations and high for DFI (>90th percentile).
Age-Related Probabilities of Sperm Defects.
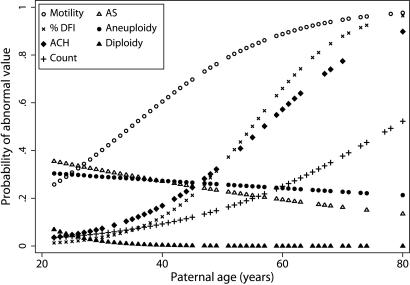
Adjusted logistic regression models were used to predict the age-dependent probabilities for having abnormal values for the individual genomic and semen quality endpoints (Fig. 3and Supporting Text, which is published as supporting information on the PNAS web site). The models predict that age does not increase the probabilities of producing sperm with AS mutations, aneuploidy, or diploidy at higher frequencies than those in 20–29 year olds. For %DFI, the predicted probability of having abnormal values was <5% at age 30, ≈35% at age 50, and 95% by age 80, with similar probabilities for mean and SD DFI (not shown). For ACH, the predicted probability of producing sperm at twice the frequency of 20–29 year olds was 10% at age 30, 34% at age 50, and 85% by age 80. In comparison, men who reach age 50 are predicted to have an 80% probability of poor motility and a 15% probability of total count of <40 million sperm.
Fig. 3.
Probabilities of producing an abnormal semen specimen with increasing age. Maximum-likelihood logit curves were generated to estimate the probabilities at each year of age of producing a semen sample that exceeded the following abnormality values: (○) Motile sperm <50%; ×%DFI > 30%; +total count ≤ 40 × 106; as well as doublings in the frequencies of abnormal sperm compared with the 20- to 29-year-old group for: (♦ ACH mutations (1138G>A in FGFR3); (▵) AS mutations (755C>G + 758C>G in FGFR2); (●) aneuploid sperm (hyper- and hypohaploid); and (▴) diploid sperm (hyper- and hypohaploid). Sperm motility and concentration curves are adapted from Eskenazi et al. (5). The %DFI model was adjusted for abstinence, and the ACH model was adjusted for radiation exposure.
Discussion
Our study identified differential effects of male aging across major categories of sperm genomic defects within a nonclinical cohort of 97 generally healthy nonsmokers. After adjustment for potential confounders, male aging was significantly associated with increases in sperm DFI and the ACH mutation but not with sperm sex ratio, HDS, AS mutations, sperm diploidies, or sperm aneuploidies involving chromosomes X, Y, or 21 within this cohort. The risks for producing sperm with these genomic defects were generally independent of each other and unrelated to conventional semen quality, with a few exceptions discussed below. We also identified four individuals in our cohort who produced unusually high frequencies of sperm in at least two categories of major genomic defects.
DFI.
Among all of the sperm genomic endpoints evaluated in this study, age had the strongest effects on chromatin defects, explaining ≈40% of the variance in DFI endpoints. Our study is the first to model the effect of age on DFI and to estimate the magnitude and shape of the age relationships. We detected no evidence for age thresholds, suggesting a gradual upward trend in the average frequency of sperm with DFI beginning in the early reproductive years. Our DFI findings support observations from previous studies that male age is associated with sperm chromatin defects (28, 29) and DNA damage (30–32). Similar to the findings by Spano et al. (28), we found that average %DFI values more than doubled between 20 and 60 years of age. Because our study included more older men, we also report a 5-fold increase in %DFI between 20 and 80 years of age. Sexual abstinence was positively associated with %DFI, which is consistent with some studies (28, 38) but not others (29, 39). Adjusting for age and abstinence, we found no association between DFI and any environmental, occupational, or medical conditions identified from the questionnaire.
DFI measurements have been shown to be predictive for male fertility and successful pregnancy outcomes (23–25, 31, 40–43). Data from thousands of donors show that samples with %DFI values of <27–30% have higher probabilities of successful pregnancies by natural means (6.5- to 10-fold), intrauterine insemination (7.0- to 8.7-fold), routine in vitro fertilization (≈2-fold), and intracytoplasmic sperm insemination (≈1.5-fold) compared with samples with >30% DFI values. Age-related increases in DFI may explain the clinical experiences of decreased pregnancy and live birth rates after fertility treatments with sperm from older men (44–46), given that abnormal DFI values in sperm were not compensated by using donor eggs from younger females (43). We found significant variation in DFI results within our cohort; e.g., men in their 50s ranged from excellent DFIs (5%) to very poor ones (73%). Even some men in their 20s and 30s had abnormal DFI values, suggesting they too might experience diminished fertility and/or abnormal pregnancy outcomes, consistent with clinical experience.
DFI and Motility.
Previously, we reported strong associations between age and diminished semen quality within this cohort, especially for sperm motility (5). Our current study shows that sperm motility is also inversely correlated with DFI endpoints, independent of age (Fig. 2). This correlation confirms the findings of previous studies (47, 48), whereas other studies found weaker or no associations (25, 27, 39). The finding that DFI and motility were both strongly associated with age and duration of abstinence suggests that a common mechanism related to oxidative stress may be responsible. Sperm residing longer in the male reproductive tract can experience higher exposures to oxidative stress (49), but the implications for male infertility and heritable mutations are likely to be complex. Several studies have suggested that high levels of oxidative stress may render sperm incapable of fertilization, whereas others have suggested that sperm exposed to oxidative stress remain capable (49, 50). Exposure to oxidative stress can induce DNA strand damage and breaks in both somatic and germ cells (49). DNA damage in stem or differentiating cells may yield sperm with various genomic defects. Male postmeiotic cells have impaired repair capacities that put them at special risk for DNA lesions that may be converted to mutations or aberrations in zygotes affecting the development and health of the offspring (22).
HDS.
Our study did not find an association of age and HDS. However, independent of age, HDS was positively correlated with frequency of aneuploidy and negatively correlated with sperm concentration and progressive motility. The association with aneuploidy is consistent with a previous finding of higher frequencies of aneuploidy among subpopulations of immature sperm (51). The inverse association between HDS and progressive motility, in the context of no consistent associations between HDS and the DFI endpoints, suggests that increases in HDS may arise from mechanisms different from those that lead to DFI. HDS sperm may represent late-stage spermatids that were released from the seminiferous epithelium before they were fully differentiated and biochemically mature. Such cells would be expected to have both poor motility and immature chromatin. Evenson et al. (52) reported a transient increase in HDS sperm in a man with fever, with a corresponding increase in the fraction of protamine 2 precursor in his sperm nuclei, suggesting a possible biochemical basis for immature chromatin.
ACH and AS.
It is well established that ACH and AS are strongly associated with paternal age, and that sporadic cases are paternal in origin (15, 53, 54). However, within our cohort, only the frequencies of sperm with the ACH mutations increased with male age. Our ACH findings are consistent with those of Tiemann-Boege et al. (20), which was expected because our cohort comprised a subset of their study population. Because the magnitude of the increase in the ACH mutation frequency in sperm is sensitive to the background of the assay (0.32 of 10,000 genomes), the age effect might actually be greater than reported. We also found that the frequency of ACH mutations significantly increased with prior exposure to ionizing radiation within Occupational Safety and Health Administration regulations; however, the small sample size and the varied types of low-level work-site exposures prevented further analyses for dose–response or effects of radiation quality. Both the ACH (1138G>A in FGFR3) and AS mutations (755C>G in FGFR2) occur in CpG dinucleotides, whereas the AS mutation (758C>G in FRFG2) does not, suggesting that methylation differences are an unlikely explanation of the ACH vs. AS difference in our study.
Cohort-Specific AS Effects.
Our AS findings are in conflict with prior sperm studies (21, 35), including the combination study of our age and genetic effects in sperm (AGES) group and the men recruited at the Johns Hopkins Medical Institutions (JHMI) in Baltimore (19). The AGES and JHMI groups had similar average ages (40 years) and frequencies of 755C>G mutations but differed in average sperm concentrations (P = 0.06, two-tailed t test), frequencies of 758C>G mutations (0.06 vs. 0.16, P = 0.004, two-tailed t test), and proportions of samples with nondetectable mutations at both loci (56% vs. 29%, P = 0.001, χ2 test). Using Poisson regression as described in Glaser et al. (19), the JHMI group showed a significant age association at each locus individually and combined (P < 0.01), but our AGES group showed no association at either locus. Technical explanations are unlikely, because both groups were analyzed in the laboratory of R.L.G. and E.W.J. The AGES group was predominantly white, and the JHMI group had more individuals of African-American and Asian-Pacific descent, suggesting sociodemographic explanations for the disparity in results. In addition to ACH and AS, there are ≈20 autosomal dominant genes with significant paternal-age components and strong evidence of paternal origin of mutation including Crouzon, Pfeiffer, MEN 2A, MEN 2B, progeria, and familial adenomatous polyposis (9, 55–57). Comparative studies of the underlying mutations in sperm may help us to better understand the mechanisms of age effects across loci, selection during spermatogenesis, genetic variation within loci, and group-specific differences.
Aneuploidy and Diploidy.
Our AGES study with ≈90 men of whom 25 were over the age of 60 is the largest investigation to date of the association between age and frequencies of aneuploid and diploid sperm for genotypes linked to Klinefelter, triple X, XYY, Turner, and Down syndromes, and triploid pregnancies. Several earlier studies found small inconsistent associations with age, generally based on fewer than 40 men with few men over 60 and using a variety of assay methods (12, 58–65). The lack of association between age and Klinefelter sperm (X-Y-21) in the AGES group is inconsistent with our earlier positive finding in fathers of boys with Klinefelter syndrome (12), both of which were analyzed by using the same assay in the laboratory of A.J.W. The disparity is suggestive of another cohort-specific difference for the effects of age on XY sperm.
We identified intercorrelations among the subcategories of sperm aneuploidy and diploidy (Table 4) independent of age, suggesting that defects in chromosome number may share mechanisms affecting both meiosis I and meiosis II disjunction. We also identified several confounding factors for numerically defective sperm: abstinence, potential occupational exposures, history of mumps, and working with radioisotopes. Others have suggested that age may be associated with increased risks for sperm defects in chromosomal structure (2, 33, 34), suggesting that aneuploidy and aberrations may arise from different spermatogenic targets with different sensitivities for induced damage (66).
Implications.
Our findings are based on convenience samples of generally healthy nonsmoking workers and retirees in a nonclinical setting and may not be representative of men attending fertility clinics and those with health problems. Furthermore, markers for other gene mutations, translocations, and deletions were not explored. Also, evidence for group-specific age associations raises questions of whether sperm defects were due to age per se or arise from lifestyles or increased opportunities for mutagenic exposures in older men.
Our sperm findings provide further evidence that men choosing to delay fatherhood may have a lower likelihood of a successful pregnancy free of early loss and gene defects. However, unlike women, older men do not appear to be at increased risk for trisomic or triploid pregnancies. The poor correlations among sperm defects suggest that multiple measures of genomic damage are needed to fully assess the reproductive and genetic burden in sperm, and that men with good semen quality may still be at risk for fathering a child with a genomic defect. Our study also identified a small fraction of men who may be at increased risk for transmitting multiple genetic and chromosomal defects, raising further concerns for men who delay fatherhood.
Methods
The AGES study consisted of 97 men, 22–80 years of age (median, 44 years), employed or retired from a government research laboratory. It received Institutional Review Board approval, and each man gave informed consent. Donor recruitment, selection, and conventional semen procedures were described (5). Exclusion criteria were: current cigarette smokers (last 6 months), current fertility or reproductive problems, previous semen analysis with zero sperm count, vasectomy, history of prostate cancer or undescended testicle, or cancer chemo- or radiotherapy. Single specimens from each man were stored at −80°C before genomic analyses (see Supporting Text).
DFI and Immature Chromatin (HDS) were measured for 88 men (median, 41.5 years; range, 20–80) by flow cytometry using the sperm chromatin structure assay (SCSA) (67). Independent replicates (5,000 cells each) were analyzed for mean and standard deviation of DFI (both in channels of fluorescence); percent of cells with detectable DFI, i.e., >250 channels (%DFI, formerly known as COMPαt); and percent of HDS cells. %DFI was further divided into moderate (250 < DFI <650 channels) and high DFI (DFI >650). Values of mean DFI >300, SD DFI >200, %DFI >30, and HDS >15 have been correlated with decreased male fertility (24, 27). Frequencies of sperm with 1138G>A mutations (ACH mutation) in FGFR3 were determined for 58 men (median, 38.5 years; range 20–80) using a PCR-based assay (20). Each sample was analyzed an average of 6.7 times (range, 5–12) in three to six experiments (mean, 3.4) with a background of 0.32 mutants per 10,000 genomes (20). Frequencies of sperm with mutations in FGFR2 (755C>G and 758C>G; AS mutations) were determined in 76 men (median, 40 years; range, 20–80) by PCR (19). Each sample was analyzed 7–16 times, equivalent to 350,000–800,000 sperm. Multicolor FISH (68, 69) was applied to >10,000 sperm from each of 90 men (median, 42 years; range 20–80) to measure disomies X, Y, XY, and 21; sex nullisomy; total diploidy (X-Y-21-21, meiosis I; X-X-21-21 and Y-Y-21-21, meiosis II); total hyperhaploidy, and hyperhaploidy plus hypohaploidy. The group of men analyzed by SCSA and sperm FISH were not significantly different in age or semen quality from the entire AGES group after exclusion of men due to no or very low sperm counts. Men analyzed for ACH were slightly younger (41.6 vs. 46.4 years, P = 0.06) with slightly higher semen volume and total sperm number (P < 0.05) but not different for concentration or motility. Men analyzed for AS had slightly higher semen volume (P = 0.04) but were not significantly different in age and other semen-quality endpoints.
Statistical analyses were performed by using stata 8.0 (Stata, College Station, TX), with age examined as both a continuous and categorical variable (by decade). For each outcome, we constructed multivariate regression models with continuous age controlling for covariates (see Supporting Text). Hockey-stick models were fit to the adjusted data to determine whether there was a change in the relationship between age and sperm outcomes at any age (70). Associations among genomic endpoints and between genomic and semen quality endpoints were determined by using Pearson correlation coefficients and partial correlation coefficients, adjusting for age. We applied maximum-likelihood logit modeling to estimate the probability at each year of age for having abnormal values of %DFI, aneuploidy, diploidy, ACH, and AS mutations, in comparison to abnormal motility and sperm numbers, as determined (5).
Supplementary Material
Acknowledgments
We acknowledge E. Sloter, X. Lowe, F. Marchetti, T. Schmid, L. Jost, K. Kim, C. Garry, L. Tomasik-Cheeseman, A. Alme, S. Kidd, L. Moore, E. Silver, and R. Weldon for assisting in laboratory work, statistics, and preparation of this work. This research was funded by National Institute on Environmental Health Sciences/Environmental Protection Agency Grant P42 ES04705 (to A.J.W. and B.E.), National Institutes of Health Grant GM36745 (to N.A.), the L. H. Gross Foundation, Mrs. J. S. Sutland, and Dr. and Mrs. L. Pakula (to R.L.G. and E.W.J.). This work was performed in part under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract W-7405-Eng-48.
Abbreviations
- ACH
achondroplasia
- AS
Apert syndrome
- DFI
DNA fragmentation index
- HDS
high DNA stainability
- AGES
age and genetic effects in sperm
- C.I.
confidence interval
- %DFI
percent DFI.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Martin J. A., Hamilton B. E., Sutton P. D., Ventura S. J., Menacker F., Munson M. L. Natl. Vital Stat. Rep. 2005;54:1–116. [PubMed] [Google Scholar]
- 2.Sloter E., Nath J., Eskenazi B., Wyrobek A. J. Fertil. Steril. 2004;81:925–943. doi: 10.1016/j.fertnstert.2003.07.043. [DOI] [PubMed] [Google Scholar]
- 3.Crow J. F. Nat. Rev. Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 4.Kuhnert B., Nieschlag E. Hum. Reprod. Update. 2004;10:327–339. doi: 10.1093/humupd/dmh030. [DOI] [PubMed] [Google Scholar]
- 5.Eskenazi B., Wyrobek A. J., Sloter E., Kidd S. A., Moore L., Young S., Moore D. Hum. Reprod. 2003;18:447–454. doi: 10.1093/humrep/deg107. [DOI] [PubMed] [Google Scholar]
- 6.Kidd S. A., Eskenazi B., Wyrobek A. J. Fertil. Steril. 2001;75:237–248. doi: 10.1016/s0015-0282(00)01679-4. [DOI] [PubMed] [Google Scholar]
- 7.McIntosh G. C., Olshan A. F., Baird P. A. Epidemiology. 1995;6:282–288. doi: 10.1097/00001648-199505000-00016. [DOI] [PubMed] [Google Scholar]
- 8.Fisch H., Hyun G., Golden R., Hensle T. W., Olsson C. A., Liberson G. L. J. Urol. 2003;169:2275–2278. doi: 10.1097/01.ju.0000067958.36077.d8. [DOI] [PubMed] [Google Scholar]
- 9.Glaser R. L., Jabs E. W. Sci. Aging Knowledge Environ. 2004 doi: 10.1126/sageke.2004.3.re1. re. 1. [DOI] [PubMed] [Google Scholar]
- 10.Sipos A., Rasmussen F., Harrison G., Tynelius P., Lewis G., Leon D. A., Gunnell D. Biol. Med. J. 2004;329:1070. doi: 10.1136/bmj.38243.672396.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassold T. J. Curr. Top. Dev. Biol. 1998;37:383–406. doi: 10.1016/s0070-2153(08)60181-7. [DOI] [PubMed] [Google Scholar]
- 12.Lowe X., Eskenazi B., Nelson D. O., Kidd S., Alme A., Wyrobek A. J. Am. J. Hum. Genet. 2001;69:1046–1054. doi: 10.1086/323763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnedo N., Templado C., Sanchez-Blanque Y., Rajmil O., Nogues C. Hum. Reprod. 2005;21:524–528. doi: 10.1093/humrep/dei321. [DOI] [PubMed] [Google Scholar]
- 14.Penrose L. S. Lancet. 1955;269:312–313. doi: 10.1016/s0140-6736(55)92305-9. [DOI] [PubMed] [Google Scholar]
- 15.Risch N., Reich E. W., Wishnick M. M., McCarthy J. G. Am. J. Hum. Genet. 1987;41:218–248. [PMC free article] [PubMed] [Google Scholar]
- 16.Ketterling R. P., Vielhaber E., Li X., Drost J., Schaid D. J., Kasper C. K., Phillips J. A., 3rd, Koerper M. A., Kim H., Sexauer C., et al. Hum. Genet. 1999;105:629–640. doi: 10.1007/s004399900158. [DOI] [PubMed] [Google Scholar]
- 17.Hill K. A., Buettner V. L., Halangoda A., Kunishige M., Moore S. R., Longmate J., Scaringe W. A., Sommer S. S. Environ. Mol. Mutagen. 2004;43:110–120. doi: 10.1002/em.20004. [DOI] [PubMed] [Google Scholar]
- 18.Ono T., Ikehata H., Nakamura S., Saito Y., Hosoi Y., Takai Y., Yamada S., Onodera J., Yamamoto K. Mutat. Res. 2000;447:165–177. doi: 10.1016/s0027-5107(99)00200-6. [DOI] [PubMed] [Google Scholar]
- 19.Glaser R. L., Broman K. W., Schulman R. L., Eskenazi B., Wyrobek A. J., Jabs E. W. Am. J. Hum. Genet. 2003;73:939–947. doi: 10.1086/378419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiemann-Boege I., Navidi W., Grewal R., Cohn D., Eskenazi B., Wyrobek A. J., Arnheim N. Proc. Natl. Acad. Sci. USA. 2002;99:14952–14957. doi: 10.1073/pnas.232568699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goriely A., McVean G. A., van Pelt A. M., O’Rourke A. W., Wall S. A., de Rooij D. G., Wilkie A. O. Proc. Natl. Acad. Sci. USA. 2005;102:6051–6056. doi: 10.1073/pnas.0500267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchetti F., Wyrobek A. J. Birth Defects Res. C Embryo Today. 2005;75:112–129. doi: 10.1002/bdrc.20040. [DOI] [PubMed] [Google Scholar]
- 23.Evenson D. P., Jost L. K., Marshall D., Zinaman M. J., Clegg E., Purvis K., de Angelis P., Claussen O. P. Hum. Reprod. 1999;14:1039–1049. doi: 10.1093/humrep/14.4.1039. [DOI] [PubMed] [Google Scholar]
- 24.Larson K. L., DeJonge C. J., Barnes A. M., Jost L. K., Evenson D. P. Hum. Reprod. 2000;15:1717–1722. doi: 10.1093/humrep/15.8.1717. [DOI] [PubMed] [Google Scholar]
- 25.Larson-Cook K. L., Brannian J. D., Hansen K. A., Kasperson K. M., Aamold E. T., Evenson D. P. Fertil. Steril. 2003;80:895–902. doi: 10.1016/s0015-0282(03)01116-6. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal A., Saleh R. A., Bedaiwy M. A. Fertil. Steril. 2003;79:829–843. doi: 10.1016/s0015-0282(02)04948-8. [DOI] [PubMed] [Google Scholar]
- 27.Virro M. R., Larson-Cook K. L., Evenson D. P. Fertil. Steril. 2004;81:1289–1295. doi: 10.1016/j.fertnstert.2003.09.063. [DOI] [PubMed] [Google Scholar]
- 28.Spano M., Kolstad A. H., Larsen S. B., Cordelli E., Leter G., Giwercman A., Bonde J. P. Hum. Reprod. 1998;13:2495–2505. doi: 10.1093/humrep/13.9.2495. [DOI] [PubMed] [Google Scholar]
- 29.Bonde J. P., Joffe M., Apostoli P., Dale A., Kiss P., Spano M., Caruso F., Giwercman A., Bisanti L., Porru S., et al. Occup. Environ. Med. 2002;59:234–242. doi: 10.1136/oem.59.4.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh N. P., Muller C. H., Berger R. E. Fertil. Steril. 2003;80:1420–1430. doi: 10.1016/j.fertnstert.2003.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Morris I. D., Ilott S., Dixon L., Brison D. R. Hum. Reprod. 2002;17:990–998. doi: 10.1093/humrep/17.4.990. [DOI] [PubMed] [Google Scholar]
- 32.Migliore L., Naccarati A., Zanello A., Scarpato R., Bramanti L., Mariani M. Hum. Reprod. 2002;17:2912–2918. doi: 10.1093/humrep/17.11.2912. [DOI] [PubMed] [Google Scholar]
- 33.Martin R. H., Rademaker A. Am. J. Hum. Genet. 1987;41:484–492. [PMC free article] [PubMed] [Google Scholar]
- 34.Bosch M., Rajmil O., Egozcue J., Templado C. Eur. J. Hum. Genet. 2003;11:754–759. doi: 10.1038/sj.ejhg.5201049. [DOI] [PubMed] [Google Scholar]
- 35.Goriely A., McVean G. A., Rojmyr M., Ingemarsson B., Wilkie A. O. Science. 2003;301:643–646. doi: 10.1126/science.1085710. [DOI] [PubMed] [Google Scholar]
- 36.Brohede J., Arnheim N., Ellegren H. Mol. Biol. Evol. 2004;21:58–64. doi: 10.1093/molbev/msg242. [DOI] [PubMed] [Google Scholar]
- 37.Wyrobek A. J., Schmid T. S., Marchetti F. Environ. Mol. Mutagen. 2005;45:271–283. doi: 10.1002/em.20121. [DOI] [PubMed] [Google Scholar]
- 38.Richthoff J., Spano M., Giwercman Y. L., Frohm B., Jepson K., Malm J., Elzanaty S., Stridsberg M., Giwercman A. Hum. Reprod. 2002;17:3162–3169. doi: 10.1093/humrep/17.12.3162. [DOI] [PubMed] [Google Scholar]
- 39.Evenson D. P., Jost L. K., Baer R. K., Turner T. W., Schrader S. M. Reprod. Toxicol. 1991;5:115–125. doi: 10.1016/0890-6238(91)90039-i. [DOI] [PubMed] [Google Scholar]
- 40.Spano M., Bonde J. P., Hjollund H. I., Kolstad H. A., Cordelli E., Leter G. Fertil. Steril. 2000;73:43–50. doi: 10.1016/s0015-0282(99)00462-8. [DOI] [PubMed] [Google Scholar]
- 41.Bungum M., Humaidan P., Spano M., Jepson K., Bungum L., Giwercman A. Hum. Reprod. 2004;19:1401–1408. doi: 10.1093/humrep/deh280. [DOI] [PubMed] [Google Scholar]
- 42.Henkel R., Hajimohammad M., Stalf T., Hoogendijk C., Mehnert C., Menkveld R., Gips H., Schill W. B., Kruger T. F. Fertil. Steril. 2004;81:965–972. doi: 10.1016/j.fertnstert.2003.09.044. [DOI] [PubMed] [Google Scholar]
- 43.Evenson D. P., Wixon R. L. Reprod. Biomed. 2006;12:466–472. doi: 10.1016/s1472-6483(10)62000-7. [DOI] [PubMed] [Google Scholar]
- 44.Mathieu C., Ecochard R., Bied V., Lornage J., Czyba J. Hum. Reprod. 1995;10:1090–1097. doi: 10.1093/oxfordjournals.humrep.a136100. [DOI] [PubMed] [Google Scholar]
- 45.Klonoff-Cohen H. S., Natarajan L. Am. J. Obstet. Gynecol. 2004;191:507–514. doi: 10.1016/j.ajog.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 46.Brzechffa P. R., Buyalos R. P. Hum. Reprod. 1997;12:29–33. doi: 10.1093/humrep/12.1.29. [DOI] [PubMed] [Google Scholar]
- 47.Giwercman A., Richthoff J., Hjøllund H., Bonde J. P., Jepson K., Frohm B., Spano M. Fertil. Steril. 2003;80:1404–1412. doi: 10.1016/s0015-0282(03)02212-x. [DOI] [PubMed] [Google Scholar]
- 48.Sills E. S., Fryman J. T., Perloe M., Michels K. B., Tucker M. J. J. Obstet. Gynaecol. 2004;24:74–77. doi: 10.1080/01443610310001620369. [DOI] [PubMed] [Google Scholar]
- 49.Aitken R. J., Baker M. A., Sawyer D. Reprod. Biomed. Online. 2003;7:65–70. doi: 10.1016/s1472-6483(10)61730-0. [DOI] [PubMed] [Google Scholar]
- 50.Evenson D. P., Jost L. K., Baer R. K. Environ. Mol. Mutagen. 1993;21:144–153. doi: 10.1002/em.2850210208. [DOI] [PubMed] [Google Scholar]
- 51.Kovanci E., Kovacs T., Moretti E., Vigue L., Bray-Ward P., Ward D. C., Huszar G. Hum. Reprod. 2001;16:1209–1217. doi: 10.1093/humrep/16.6.1209. [DOI] [PubMed] [Google Scholar]
- 52.Evenson D. P., Jost L. K., Corzett M., Balhorn R. J. Androl. 2000;21:739–746. [PubMed] [Google Scholar]
- 53.Wilkin D. J., Szabo J. K., Cameron R., Henderson S., Bellus G. A., Mack M. L., Kaitila I., Loughlin J., Munnich A., Sykes B., et al. Am. J. Hum. Genet. 1998;63:711–716. doi: 10.1086/302000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moloney D. M., Slaney S. F., Oldridge M., Wall S. A., Sahlin P., Stenman G., Wilkie A. O. M. Nat. Genet. 1996;13:48–53. doi: 10.1038/ng0596-48. [DOI] [PubMed] [Google Scholar]
- 55.Aretz S., Uhlhaas S., Caspari R., Mangold E., Pagenstecher C., Propping P., Friedl W. Eur. J. Hum. Genet. 2004;12:52–58. doi: 10.1038/sj.ejhg.5201088. [DOI] [PubMed] [Google Scholar]
- 56.Tartaglia M., Cordeddu V., Chang H., Shaw A., Kalidas K., Crosby A., Patton M. A., Sorcini M., van der Burgt I., Jeffery S., Gelb B. D. Am. J. Hum. Genet. 2004;75:492–497. doi: 10.1086/423493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rannan-Eliya S. V., Taylor I. B., De Heer I. M., Van Den Ouweland A. M., Wall S. A., Wilkie A. O. Hum. Genet. 2004;115:200–207. doi: 10.1007/s00439-004-1151-5. [DOI] [PubMed] [Google Scholar]
- 58.Griffin D. K., Abruzzo M. A., Millie E. A., Cheean L. A., Feingold E., Sherman S. L., Hassold T. J. Hum. Mol. Genet. 1995;4:2227–2232. doi: 10.1093/hmg/4.12.2227. [DOI] [PubMed] [Google Scholar]
- 59.Martin R. H., Spriggs E., Ko E., Rademaker A. W. Am. J. Hum. Genet. 1995;57:1395–1399. [PMC free article] [PubMed] [Google Scholar]
- 60.Robbins W. A., Baulch J. E., Moore D., 2nd, Weier H. U., Blakey D., Wyrobek A. J. Reprod. Fertil. Dev. 1995;7:799–809. doi: 10.1071/rd9950799. [DOI] [PubMed] [Google Scholar]
- 61.Kinakin B., Rademaker A., Martin R. Cytogenet. Cell. Genet. 1997;78:116–119. doi: 10.1159/000134641. [DOI] [PubMed] [Google Scholar]
- 62.Asada H., Sueoka K., Hashiba T., Kuroshima M., Kobayashi N., Yoshimura Y. J. Assist. Reprod. Genet. 2000;17:51–59. doi: 10.1023/A:1009454114973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bosch M., Rajmil O., Martinez-Pasarell O., Egozcue J., Templado C. Eur. J. Hum. Genet. 2001;9:533–538. doi: 10.1038/sj.ejhg.5200659. [DOI] [PubMed] [Google Scholar]
- 64.Rives N., Langlois G., Bordes A., Simeon N., Mace B. J. Med. Genet. 2002;39:E63. doi: 10.1136/jmg.39.10.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luetjens C. M., Rolf C., Gassner P., Werny J. E., Nieschlag E. Hum. Reprod. 2002;17:1826–1832. doi: 10.1093/humrep/17.7.1826. [DOI] [PubMed] [Google Scholar]
- 66.Wyrobek A. J., Schmid T. S., Marchetti F. J. Natl. Cancer. Inst. Monogr. 2005:31–35. doi: 10.1093/jncimonographs/lgi001. [DOI] [PubMed] [Google Scholar]
- 67.Evenson D. P., Larson K. L., Jost L. K. J. Androl. 2002;23:25–43. doi: 10.1002/j.1939-4640.2002.tb02599.x. [DOI] [PubMed] [Google Scholar]
- 68.Baumgartner A., Van Hummelen P., Lowe X. R., Adler I. D., Wyrobek A. J. Environ. Mol. Mutagen. 1999;33:49–58. doi: 10.1002/(sici)1098-2280(1999)33:1<49::aid-em6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 69.Frias S., Van Hummelen P., Meistrich M. L., Lowe X. R., Hagemeister F. B., Shelby M. D., Bishop J. B., Wyrobek A. J. Cancer Res. 2003;63:44–51. [PubMed] [Google Scholar]
- 70.Bacon D. W., Watts D. G. Biometrika. 1971;58:525–534. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.