Abstract
Cockayne syndrome (CS) is an inherited neurodevelopmental disorder with progeroid features. Although the genes responsible for CS have been implicated in a variety of DNA repair- and transcription-related pathways, the nature of the molecular defect in CS remains mysterious. Using expression microarrays and a unique method for comparative expression analysis called L2L, we sought to define this defect in cells lacking a functional CS group B (CSB) protein, the SWI/SNF-like ATPase responsible for most cases of CS. Remarkably, many of the genes regulated by CSB are also affected by inhibitors of histone deacetylase and DNA methylation, as well as by defects in poly(ADP-ribose)-polymerase function and RNA polymerase II elongation. Moreover, consistent with these microarray expression data, CSB-null cells are sensitive to inhibitors of histone deacetylase or poly(ADP-ribose)-polymerase. Our data indicate a general role for CSB protein in maintenance and remodeling of chromatin structure and suggest that CS is a disease of transcriptional deregulation caused by misexpression of growth-suppressive, inflammatory, and proapoptotic pathways.
Keywords: DNA repair, L2L, microarray, transcription
Cockayne syndrome (CS) is a devastating inherited disease characterized by severe postnatal growth failure and progressive neurological dysfunction, along with a variety of symptoms reminiscent of aging, including retinal degeneration, sensorineural hearing loss, cataracts, and loss of subcutaneous fat (1). The nature of the molecular defect that causes CS remains elusive, although CS has been intensively studied for many years, and much is known about the cellular functions of the five genes responsible for the disease. Most cases of CS are caused by defects in two genes: CS groups A (CSA) and B (CSB). The CSB protein is a SWI/SNF-like DNA-dependent ATPase (2–4) that can wind DNA (5) and remodel chromatin in vitro (6). CSB is required for translocation of the CSA protein to the nuclear matrix after DNA damage (7). Rare alleles of three xeroderma pigmentosum (XP) genes (XPB, XPD, and XPG) are responsible for the remaining cases of CS; these patients usually lack the severe predisposition to skin cancer typical of XP (8). CS genes have been implicated, alone or in various combinations, in transcription-coupled repair (TCR) of UV-induced DNA lesions (9, 10), repair of oxidative DNA damage (11), transcription initiation by RNA polymerase I (pol I; refs. 12 and 13) and RNA polymerase II (pol II; ref. 14), elongation by pol II (15, 16), transcription of induced genes (17), protein ubiquitination (18), and metaphase chromosome condensation (19).
To understand how CSB defects cause CS, we characterized the array of genes regulated by CSB. Previous expression array studies, although encouraging, had not been conclusive (20) or had focused on the role of CS genes in the transcriptional response to oxidative (17) or ultraviolet (21) DNA damage. Our hope was to work backwards from effect to cause, deducing the biochemical functions of CSB from genes it regulates. We found that CSB causes significant changes in gene expression, even in the absence of external stress. We then identified patterns among CSB-regulated genes using L2L, a unique tool for comparative gene expression analysis we had developed (22). Remarkably, the strongest patterns in the data demonstrate a general role for CSB in chromatin maintenance and remodeling.
Results
hTERT-Immortalized CSB Cell Lines.
All previous studies of CSB function in immortal cells have used SV40-transformed lines derived from primary fibroblasts (23); however, SV40 large T antigen interacts with p53 (24), which in turn interacts with CSB in vitro and probably in vivo (19, 25). To avoid confounding functional interactions between large T antigen and CSB and to provide a well controlled isogenic pair of cell lines for analyzing CSB-dependent gene expression, we created telomerase reverse transcriptase (hTERT)-immortalized CSB lines derived from the same primary CS1AN fibroblast line used previously for immortalization by SV40. We then derived two daughter lines, CSB-wt, expressing wild-type CSB cDNA, and CSB-null, expressing EGFP cDNA as a control. We confirmed functional CSB rescue experimentally by Western blotting, chromosome fragility assay, and UV sensitivity (Fig. 5, which is published as supporting information on the PNAS web site).
CSB Induces Significant Changes in Gene Expression.
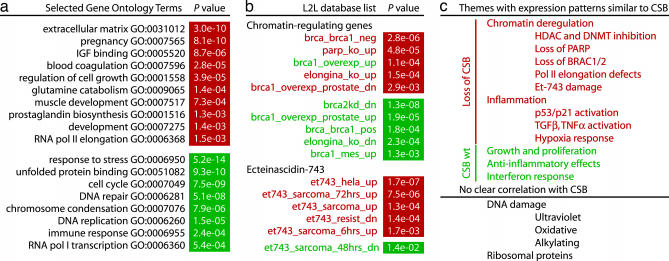
We found significant changes in gene expression after rescue of immortalized CSB cells by expression of wild-type CSB cDNA. Of the 44,928 probe sets on the Affymetrix Human Genome U133 Set microarrays, ≈1,000 were reliably regulated by CSB: 551 probe sets were down-regulated, and 428 were up-regulated (Table 1, which is published as supporting information on the PNAS web site). These “reliable” changes were defined as the same change call in seven of nine comparisons regardless of average fold change, criteria for which we calculated a false-discovery rate of ≈1% (Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site). Many of the most “robust” changes, which we defined as probe sets with the same change call in all nine pairwise comparisons and an average fold change greater than ±2.0, have potentially relevant biological roles. The 141 probes sets (representing 90 genes) that were robustly down-regulated by CSB include a number of tumor suppressors, growth inhibitors, and inflammatory mediators, along with several pregnancy- and lactation-related genes and many genes related to the extracellular matrix (Table 2, which is published as supporting information on the PNAS web site). The 67 probe sets (representing 52 genes) that were robustly up-regulated by CSB include a more varied assortment of functions: oxidative metabolism, proliferation, cell cycle progression, neuronal survival, the immune response, DNA repair, RNA processing, and drug resistance (Table 3, which is published as supporting information on the PNAS web site). We found similar patterns when we searched for overabundance of Gene Ontology terms (26) among the lists of reliably up- and down-regulated genes (Fig. 2a; complete results may be found in Table 4, which is published as supporting information on the PNAS web site).
Fig. 2.
Results of analyzing the gene expression patterns of CSB-null and CSB-wt cell lines with the L2L software suite and microarray database. Green and red names of lists denote up- and down-regulation within a particular theme in the database (e.g., all lists of genes up-regulated by Et-743 are red; all lists of genes down-regulated by Et-743 are green). The P values for overlap with genes up- and down-regulated by CSB are indicated in green and red boxes, respectively. (a) Selected Gene Ontology terms found to be significantly overabundant (P < 0.02) among genes regulated by CSB. (b) CSB-regulated genes overlap significantly (P < 0.02) with genes regulated by Et-743, as well as by BRCA1 and -2, PARP, and Elongin A. (c) Summary of themes identified by L2L in the gene expression patterns of CSB-null and CSB-wt cell lines.
We selected 26 of the most robustly regulated genes for validation by real-time quantitative RT-PCR (Q-RT-PCR), using independent samples of RNA isolated by two different methods. Of the 26 genes, 22 exhibited similar expression differences as assayed by Q-RT-PCR or microarray when using the same RNA isolation protocol (RNeasy) and a similar normalization method (overall signal or mass of RNA). The choice of isolation protocol and normalization method produced surprising variability in the Q-RT-PCR results, although no fewer than 21 genes were validated by using any combination of conditions (Fig. 7, which is published as supporting information on the PNAS web site).
CSB, HDAC Inhibitors, and Chromatin Disruption.
Intriguingly, a number of genes robustly regulated by CSB are known to be regulated by factors that modify or disrupt chromatin structure, such as histone deacetylase (HDAC) inhibitors. These genes include the cyclin-dependent kinase inhibitor CDKN1A (p21/WAF1/CIP1; ref. 27), multidrug-resistance gene ABCB1 (MDR1; ref. 28), matrix-metalloprotease inhibitor RECK (29), and cyclooxygenase PTGS1 (COX1; ref. 30). The latter was particularly surprising, because PTGS1 is generally considered to be a housekeeping gene with a TATA-less promoter (31) that few known stimuli, other than HDAC inhibitors, can induce. The apparent derepression of a variety of pregnancy-specific glycoproteins, as well as the imprinted paternally expressed gene PEG3, also suggested that the absence of CSB might phenocopy an agent that disrupts normal chromatin structure.
To examine this idea more rigorously, we sought to compare our list of CSB-regulated genes with lists of genes known to be regulated by HDAC inhibitors. Initially, we performed a literature search of the previous 24 months, manually compiling a list of 24 genes that are up-regulated and 8 that are down-regulated by the HDAC inhibitor trichostatin A (TSA). The overlaps between these lists and our data were so tantalizing that we sought a more systematic approach to confirm and extend the conclusions. We extracted lists of differentially expressed genes from published articles studying the effects of HDAC inhibitors on gene expression, converted these lists and our own to a common gene identifier, and analyzed the significance of overlap among lists, if any. The idea proved so fruitful in concept, yet tedious in execution, that we gradually developed, refined, and expanded it into a software suite we named L2L for “list-to-list” comparisons (22). We initially demonstrated the utility of L2L by reanalyzing published data on diabetic nephropathy (22); a similar but more focused approach was used to discover, within disparate microarray data, a common host transcriptional response to pathogens (32). We statistically validated our L2L results for diabetic nephropathy (22) and for CSB (this work) by comparing them with the results of extensive random-data simulations (Table 5, which is published as supporting information on the PNAS web site; see Supporting Text for details).
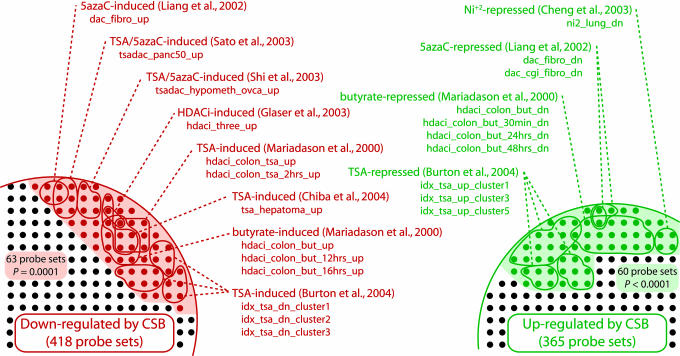
Using the L2L suite, we compared our CSB data with previously published studies of gene expression changes induced by HDAC inhibitors and other agents that disrupt normal chromatin structure, looking for consistent patterns between up- and down-regulated lists. The results were striking (Fig. 1; see also Fig. 8a, which is published as supporting information on the PNAS web site). Our list of genes reliably up-regulated by the absence of CSB overlapped significantly (P < 0.02) with 16 lists of genes up-regulated by the DNA methylation inhibitor 5-aza-2′-deoxycytidine (5azaC); by HDAC inhibitors, including TSA and butyrate; and by combinations of 5azaC and HDAC inhibitors. Significantly, the inverse was also evident. Our list of genes down-regulated in the absence of CSB overlapped with 10 lists of genes down-regulated by the chromatin-disrupting factors 5azaC, TSA, and butyrate, as well as nickel. Because there are many lists in the database, we quantified the significance of these concentrations of overlaps within a single theme (such as chromatin disruption) by determining how frequently they were found in the results of random data. Of the 10,000 random data sets we tested, none overlapped significantly (P < 0.02) with as many lists involving up-regulation by chromatin disruption as our CSB-Down data set (Table 6, which is published as supporting information on the PNAS web site). Inversely, only one of 10,000 overlapped significantly with as many lists involving down-regulation by chromatin disruption as our CSB-Up data set. We next examined the total cast of genes that are regulated by chromatin disruption. In all, 63 probe sets down-regulated by CSB are found on a list involving up-regulation by chromatin disruption, and 60 probe sets up-regulated by CSB are found on a list involving down-regulation by chromatin disruption. The former was matched by only one of the 10,000 random data sets, and the latter by none. Together, these data suggest that loss of CSB phenocopies treatment with a chromatin-disrupting agent like TSA, and that CSB may be required for maintaining repression of silenced genes.
Fig. 1.
Many genes that are regulated by CSB are also regulated by HDAC inhibitors (HDACi), inhibitors of DNA methylation, and other factors that disrupt chromatin structure. Shading indicates all such regulated probe sets; colored circles represent significant overlaps between CSB-regulated genes and specific chromatin-related lists in the L2L Microarray Database. Only probe sets on the arrays that represent named genes are shown.
The role of CSB in several DNA repair pathways has been studied extensively, and several lists of damage-regulated genes overlapped significantly with CSB-regulated genes. However, the direction of overlap was inconsistent (Fig. 8b; see also Table 6). For example, genes down-regulated by CSB correlated with genes that are both up- and down-regulated by oxidative stress. At least two scenarios might explain these puzzling overlaps: (i) damage-regulated genes might be coincidentally regulated by other stress-response pathways in CSB-null cells, or (ii) CSB-null cells might attempt to generate a proper transcriptional response to DNA damage but fail. Previous studies of CSB and XPB/CS cells found that their transcriptional response to DNA damage was deficient (17) or delayed (21), consistent with our evidence for a chromatin remodeling defect. Moreover, the sole strong correlation between CSB-regulated genes and the DNA damage response, a striking overlap between CSB and the anticancer drug Ecteinascidin 743 (Et-743; see Fig. 2b), appears to reflect a common failure in transcriptional regulation rather than a common response to DNA damage (see below). We also found no compelling evidence for differential regulation of ribosomal proteins in the presence or absence of CSB (see Supporting Text) despite CSB’s role in rRNA transcription (12).
CSB, PARP, BRCA1, BRCA2, and Elongin A.
Other L2L results also strongly support a general role for CSB in transcriptional control through chromatin maintenance or remodeling (Fig. 1b). First, we observed significant overlap between genes induced by loss of CSB and knockout of poly(ADP-ribose)-polymerase (PARP)-1 in mice. PARP-1 has a variety of functions including gene induction, heterochromatin maintenance, and chromosome stability (33). Our data are consistent with recent evidence that CSB and PARP-1 work together in DNA repair (34) and may indicate that the two proteins function cooperatively in chromatin remodeling to regulate transcription of DNA repair factors or to repair DNA.
Second, we observed strong bidirectional overlaps between genes regulated by elongation factor Elongin A in mice and genes regulated by CSB. CSB and its budding yeast homolog, Rad26, have been implicated in pol II elongation (16, 35), and CSB is believed to associate primarily with an elongating polymerase complex (36). Our expression data are the first in vivo evidence that CSB plays a general role in regulating elongation-dependent genes.
Third, we observed broad overlaps between genes regulated by BRCA1 or BRCA2 and genes regulated by CSB. Four lists of genes up-regulated by BRCA1 or BRCA2 overlapped with our list of genes up-regulated by CSB, whereas two lists of genes down-regulated by BRCA1 overlapped with the genes down-regulated by CSB. These overlaps indicate that the three proteins participate in a common pathway other than DNA repair, because we found no clear evidence for induction of a general DNA damage response in CSB-null cells (see above). Interestingly, BRCA1 functions together with BARD1 as a ubiquitin ligase responsible for ubiquitination of stalled pol II (37, 38). Moreover, ubiquitination and degradation of stalled pol II are deficient in CSB-null cells after UV damage (39), although the precise nature of the defect is unclear. If BRCA1 and BRCA2 work together with CSB to degrade or mobilize stalled pol II, these three proteins might coregulate genes where pol II frequently stalls. Such genes could be preferentially sensitive to DNA damage, perhaps because of persistent single-strandedness, or could have sequences and/or chromatin structures that challenge pol II elongation. Intriguingly, defects in BRCA1 and BRCA2 cause the same locus-specific metaphase chromosome fragility (A.D.B. and A.M.W., unpublished work), as seen for CSB defects (19).
Treatment with the Antitumor Drug Et-743 Partially Phenocopies Loss of CSB.
Alone among DNA-damaging agents in the L2L database, the novel antitumor drug Et-743, a minor groove-binding alkylator isolated from the Caribbean sea squirt Ecteinascidia turbinate, significantly and with consistent direction regulated many of the same genes as loss of CSB (Fig. 2b). Five lists of genes up-regulated by Et-743 overlap strongly with the list of genes up-regulated by loss of CSB. The shared pattern of gene regulation between Et-743 and CSB is especially intriguing, because the cytotoxicity of Et-743, unlike that of all other antitumor drugs, specifically requires an intact TCR pathway, including CS group A and CSB (40). The current model of Et-743 cytotoxicity suggests that the TCR pathway attempts to repair Et-743 lesions but fails, instead forming a covalent adduct that triggers apoptosis. In the absence of TCR, translesion synthesis and/or homologous recombination uneventfully repair the lesions. Despite the common link with TCR, it is not obvious why gene expression patterns induced by treatment with Et-743 and loss of CSB should overlap. No comparable overlap is seen for other DNA alkylating drugs, or for UV or global oxidative damage, which are both repaired by CSB-dependent pathways. However, repair-mediated apoptosis is not the only mechanism of action of Et-743. The drug also inhibits transcription induced by heat shock (41), the steroid receptor SXR (42), and HDAC inhibitors (43). Intriguingly, Et-743 disrupts induction by HDAC inhibitors of two genes that are also strongly regulated by CSB, ABCB1 and CDKN1A. Therefore, the inability to dynamically regulate transcription is probably responsible for more of the overlap between CSB- and Et-743-related gene expression changes than fatally stalled TCR complexes or sequestration of CSB at sites of DNA damage.
Gene Expression and Survival of CSB-Null Cells Are Sensitive to HDAC and PARP Inhibitors.
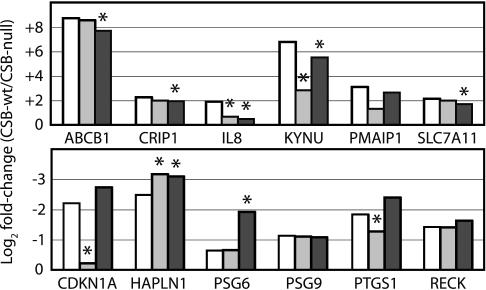
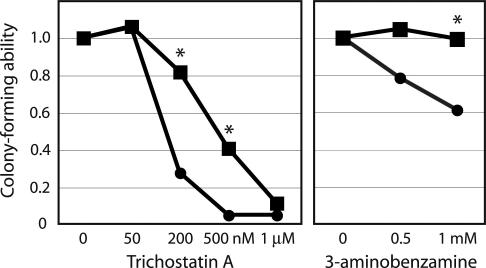
To confirm that loss of CSB partially phenocopies treatment with a HDAC inhibitor, we examined the effect of TSA treatment on expression of 12 genes that are robustly regulated by CSB and are reported to be regulated by HDAC inhibitors. CSB-wt and CSB-null cell lines were treated with 1 μM TSA or ethanol only as a control for 24 h before RNA harvest; relative gene expression levels were then determined by real-time quantitative RT-PCR. TSA treatment significantly reduced the difference in gene expression between CSB-wt and CSB-null lines for four genes, two induced and two repressed by CSB, notably including CDKN1A and PTGS1 (Fig. 3). Interestingly, as our data implicate PARP in CSB function (see above), treatment with 5 mM 3-aminobenzamine (3AB, a PARP inhibitor) also reduced the difference in gene expression between CSB-wt and CSB-null lines for several genes but, in contrast to TSA, only for genes induced by CSB (Fig. 3). In view of the evidence that both HDAC and PARP inhibition influence CSB-regulated gene expression, we asked whether CSB-null cells are sensitive to treatment with either drug. We found that growth in the presence of 3AB significantly reduced colony formation by CSB-null cells, and that CSB-null cells were substantially more sensitive to treatment with TSA for 48 h (Fig. 4). These data suggest that CSB regulates HDAC inhibitor-sensitive genes directly, that PARP may coregulate a subset of CSB-regulated genes, and that these pathways are important for normal cell function. The prominence of growth-inhibitory and inflammatory factors (like CDKN1A and PTGS1) among these putative CSB target genes encouraged us to ask whether loss of CSB induced a broader pattern of inflammatory gene expression.
Fig. 3.
Treatment with TSA and 3AB partially phenocopy lack of CSB as judged by RT-PCR. Bars are the log2 ratio of gene expression levels in CSB-wt vs. CSB-null cell lines when treated with ethanol alone (white), 1 μM TSA (light gray), or 5 mM 3AB (dark gray). Zero represents no difference in expression between the two cell lines. ∗, P < 0.05 for difference between drug treated and control by Student’s t test of triplicate experiments.
Fig. 4.
CSB-null cells are sensitive to treatment with TSA and 3AB. The number of colonies is normalized to the ethanol control (1.0). ∗, P < 0.05 for the difference between CSB-wt (squares) and CSB-null (circles) by Student’s t test of triplicate experiments.
CS May Be an Inflammatory Disease.
The L2L suite discovered several patterns in CSB-regulated genes suggesting that a variety of inflammatory pathways are up-regulated in CS (Fig. 2c; see Fig. 8c for details). First, the expression of genes regulated by the p53/p21, TGFβ, and TNF-α/JNK pathways are altered by loss of CSB. Moreover, when we used OPOSSUM (44) to predict the abundance of transcription-factor-binding sites in the promoters of CSB-regulated genes, the most significant result was an overabundance of NF-κB-related sites (c-REL, p65, and NF-κB) in genes up-regulated by loss of CSB (Table 7, which is published as supporting information on the PNAS web site). Second, CSB up-regulates many genes that are also up-regulated by antiinflammatory drugs. Third, hypoxia-response pathways are up-regulated in the absence of CSB. Fourth, genes up-regulated by CSB overlap with several lists associated with highly proliferative cancers and with the response to VEGF and EGF growth factors, consistent with our initial impressions of robustly regulated genes and our formal Gene Ontology analysis (Fig. 2a and Table 4). Overall, CSB-wt cells exhibit the gene expression profile of rapidly proliferating cells, whereas CSB-null cells appear to be under inflammatory stress.
IFN-induced genes are often up-regulated by inflammatory stimuli. NF-κB, for instance, directly induces the IFN-β promoter (45). However, we paradoxically saw robust expression of IFN-regulated genes in CSB-wt cells, a pattern that may further suggest a chromatin-maintenance role for CSB. Several of the genes most robustly up-regulated by CSB are involved in the immune response, and L2L identified 11 lists of genes associated with IFN induction or the viral response that overlapped significantly with genes up-regulated by CSB (Fig. 8d). The expression of upstream signaling genes, however, was unchanged: IRF-, STAT- and, critically, IFN-family gene expression was either stable or undetectable. The robust up-regulation of IFN-induced genes in CSB-wt is therefore unlikely to be due to induction of IFNs themselves by upstream inflammatory signals but may be related to the unique structure of IFN-sensitive promoters. Most cells, including these CS fibroblasts, express low but detectable levels of IFNs that are thought to be critical for priming robust induction of IFNs in response to viral infection (46). Intriguingly, HDAC activity is required for IFN-induced, but not basal, expression of IFN target genes (47). The relatively low expression of IFN-induced genes in CSB-null cells may indicate that induction by IFNs is both HDAC- and CSB-dependent, consistent with our observation that HDAC inhibitors phenocopy loss of CSB, and that both cause dysfunctional chromatin remodeling.
Finally, many genes regulated by CSB are also regulated in models of human aging (Fig. 8e). These data imply that CS disrupts the regulation of genes involved in normal aging, and that CS is a true progeria, as the clinical symptoms suggest. The link between disruption of DNA methylation patterns and aging is well established (48); moreover, growing evidence indicates that aging causes deregulation of specific genes, especially in the brain (49–51), and that the outward symptoms of aging in various tissues may reflect uncontrolled or chronic inflammatory processes (52). Our data suggest that some of these inflammatory signals may be generated endogenously in affected cells as a result of gene-specific transcriptional deregulation.
Discussion
Our microarray analysis demonstrates that the patterns of gene deregulation in CSB-null cells resemble those induced by agents that disrupt chromatin structure or modifications. CSB is structurally similar to SWI/SNF chromatin remodeling proteins and exhibits ATP-dependent chromatin remodeling activity in vitro (3) but, unlike many other SWI/SNF family members, it is not known to exist in complexes with histone modifying enzymes. Moreover, CSB does not appear to affect the expression of any histone acetyltransferases or methyltransferases and only modestly affects one HDAC (HDAC2) and one histone gene (H2AFO). Given this evidence that CSB does not regulate chromatin modifying factors, we speculate that CSB may play a direct role in facilitating chromatin modification. CSB may make histones more accessible to modifying enzymes, perhaps by transiently moving (3) or even displacing (5) nucleosomes or by remodeling histone–histone or histone–DNA contacts.
Remarkably, complete absence of CSB protein does not cause CS (53), although CS is inherited recessively, suggesting that the CSB alleles associated with CS may generate protein fragments that interfere with otherwise redundant pathways. Indeed functional redundancy may explain why CSB’s biological functions have not been as readily characterized as those of other SWI/SNF-like complexes. These functions could include actions at specific promoters. For example, up- or down-regulation of an entire class of genes (e.g., the IFN or hypoxia response) could indicate that CSB is required for full activity of a class-specific transcription factor (e.g., ISGF3 or HIF-1). Alternatively, CSB could be recruited primarily to polymerases stalled at sites of disrupted chromatin or at DNA lesions where local chromatin structure must be modified before, during, or after repair (54). Interestingly, many stalled polymerases in normal cells are probably caused by structural obstacles to elongation, rather than by DNA damage. Stalling may be especially common for spuriously initiated polymerases that probably explain some of the transcriptional “dark matter” identified by recent tiling microarray experiments (55), or for “pathfinder” polymerases that begin the complex reorganization of chromatin structure downstream from a newly activated promoter (56).
As for most factors involved in chromatin remodeling, including the prototypical SWI/SNF (57), CSB is required only for regulation of particular genes. However, many of the CSB-regulated genes identified by our microarray analysis play critical roles in cell growth and the stress response. As a result, loss of CSB leads to an inflammatory proapoptotic gene expression profile, which may provide the link between CSB function and the CS phenotype. Loss of CSB would cause a general defect in chromatin maintenance and remodeling, initiating a vicious cycle of dysfunction in which the cell is unable to respond appropriately to stimuli, including the derepression of harmful genes. The resulting “inflammatory phenotype” may be responsible for the extraordinary neurodegenerative and wasting symptoms of this progeroid disease and, perhaps, for some of the similar symptoms found in normal human aging.
Methods
Supporting Information.
For further details and additional methods, see Supporting Text, Tables 1–7, and Figs. 5–8.
Cell Lines.
GM00739B primary cells, obtained from Coriell Cell Repositories, are derived from a compound heterozygote CS patient with severe clinical symptoms (CS1AN). The cells were immortalized by PG-13/neo retroviral transduction of telomerase reverse transcriptase (hTERT) cDNA (58). Wild-type CSB cDNA (23) and EGFP cDNA were cloned into pIRESpuro (Clontech), and stable daughter lines expressing each construct were generated by transfection of the linearized plasmid. Immortalized lines were maintained in MEMα (Invitrogen) with 10% FBS and supplements, as well as 1 mg/ml G418 and 0.5 μg/ml puromycin for transgene selection.
Microarray Hybridization and Data Analysis.
Cells were grown to 60–80% confluence, trypsinized, and collected. Total RNA was isolated by using RNeasy midi spin columns (Qiagen, Valencia, CA) and enriched for polyA+ RNA on Oligotex polydT beads (Qiagen). Synthesis of target cRNAs for hybridization to HG-U133 Set GeneChips was carried out as specified by the manufacturer (Affymetrix). Hybridization to and scanning of the arrays was performed by the Center for Expression Arrays core facility at the University of Washington. Intensity data from the scanned arrays was analyzed with Affymetrix microarray suite 5.0 (MAS5.0) using default parameters and normalized to an arbitrary mean value of 800. Data are deposited at National Center for Biotechnology Information Gene Expression Omnibus (GSE3407). Three sets of biological replicates provided for nine pairwise comparisons between the CSB-wt and CSB-null cell lines. A biological replicate represented independent RNA harvests and independent cRNA synthesis using the same stable cell line at a different passage number. The significance of changes in gene expression was defined by the frequency of MAS5.0 change calls among the pairwise comparisons. The change call, or test statistic, is a qualitative measure of the significance of an expression change that is calculated independently from fold change (59) and is more reliable than fold change cutoffs for detecting significant expression changes (60). We considered an identical change call in at least seven of the nine pairwise comparisons to be reliably significant, based on false-discovery rates calculated from randomized data (see Supporting Text and Fig. 6). The L2L Microarray Analysis Tool (22) was used to mine the lists of reliable changes for underlying biological patterns and for overabundance of Gene Ontology terms; a prerelease build of version 2006.2 of the L2L database was used. L2L generates a nominal P value for the significance of overlap or overabundance based on the binomial distribution. We performed random-data simulations to generate adjusted P values and determine false discovery rates (see Supporting Text and Table 5). The results showed that a binomial P value of 0.02 was a conservative cutoff for true significance. The abundance of predicted transcription factor-binding sites in CSB-regulated genes was calculated by opossum (44).
Supplementary Material
Acknowledgments
We thank Hua-Ying Fan for generating the anti-CSB antibody and Tom Pavelitz for valuable advice. We are grateful for the generous support of Peter Rabinovitch and the Nathan Shock Center of Excellence in the Basic Biology of Aging at the University of Washington. Roger Bumgarner of the University of Washington Center for Expression Arrays provided invaluable help throughout this project. J.C.N. is supported by the National Institute of General Medical Sciences Medical Scientist Training Program and by fellowships from the Cora May Poncin Foundation and the Merck Research Laboratories. This work is supported by National Institutes of Health Grant GM41624 (to A.M.W.).
Abbreviations
- CS
Cockayne syndrome
- CSB
CS group B
- HDAC
histone deacetylase
- PARP
poly(ADP-ribose)-polymerase
- pol
RNA polymerase
- TCR
transcription-coupled repair
- TSA
trichostatin A
- Et-743
Ecteinascidin 743
- 3AB
3-aminobenzamine.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE3407).
References
- 1.Nance M. A., Berry S. A. Am. J. Med. Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 2.Selby C. P., Sancar A. J. Biol. Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 3.Citterio E., Rademakers S., van der Horst G. T., van Gool A. J., Hoeijmakers J. H., Vermeulen W. J. Biol. Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen M., Stevnsner T., Modin C., Martensen P. M., Brosh R. M., Jr, Bohr V. A. Nucleic Acids Res. 2003;31:963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beerens N., Hoeijmakers J. H., Kanaar R., Vermeulen W., Wyman C. J. Biol. Chem. 2005;280:4722–4729. doi: 10.1074/jbc.M409147200. [DOI] [PubMed] [Google Scholar]
- 6.Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R. E., Hoeijmakers J. H., Vermeulen W. Mol. Cell. Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamiuchi S., Saijo M., Citterio E., de Jager M., Hoeijmakers J. H., Tanaka K. Proc. Natl. Acad. Sci. USA. 2002;99:201–206. doi: 10.1073/pnas.012473199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapin I., Lindenbaum Y., Dickson D. W., Kraemer K. H., Robbins J. H. Neurology. 2000;55:1442–1449. doi: 10.1212/wnl.55.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleaver J. E., Thompson L. H., Richardson A. S., States J. C. Hum. Mutat. 1999;14:9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Venema J., Mullenders L. H., Natarajan A. T., van Zeeland A. A., Mayne L. V. Proc. Natl. Acad. Sci. USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuo J., Jaruga P., Rodriguez H., Dizdaroglu M., Bohr V. A. J. Biol. Chem. 2002;277:30832–30837. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- 12.Bradsher J., Auriol J., Proietti de Santis L., Iben S., Vonesch J. L., Grummt I., Egly J. M. Mol. Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 13.Iben S., Tschochner H., Bier M., Hoogstraten D., Hozak P., Egly J. M., Grummt I. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 14.Dubaele S., Egly J. M. J. Eur. Acad. Dermatol. Venereol. 2002;16:220–226. doi: 10.1046/j.1468-3083.2002.00453.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee S. K., Yu S. L., Prakash L., Prakash S. Cell. 2002;109:823–834. doi: 10.1016/s0092-8674(02)00795-x. [DOI] [PubMed] [Google Scholar]
- 16.Selby C. P., Sancar A. Proc. Natl. Acad. Sci. USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyng K. J., May A., Brosh R. M., Jr., Cheng W. H., Chen C., Becker K. G., Bohr V. A. Oncogene. 2003;22:1135–1149. doi: 10.1038/sj.onc.1206187. [DOI] [PubMed] [Google Scholar]
- 18.Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A. F., Tanaka K., Nakatani Y. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 19.Yu A., Fan H. Y., Liao D., Bailey A. D., Weiner A. M. Mol. Cell. 2000;5:801–810. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]
- 20.Selzer R. R., Nyaga S., Tuo J., May A., Muftuoglu M., Christiansen M., Citterio E., Brosh R. M., Jr, Bohr V. A. Nucleic Acids Res. 2002;30:782–793. doi: 10.1093/nar/30.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Costa R. M., Riou L., Paquola A., Menck C. F., Sarasin A. Oncogene. 2005;24:1359–1374. doi: 10.1038/sj.onc.1208288. [DOI] [PubMed] [Google Scholar]
- 22.Newman J. C., Weiner A. M. Genome Biol. 2005;6:R81. doi: 10.1186/gb-2005-6-9-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J. H. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 24.Pipas J. M., Levine A. J. Semin. Cancer Biol. 2001;11:23–30. doi: 10.1006/scbi.2000.0343. [DOI] [PubMed] [Google Scholar]
- 25.Wang X. W., Yeh H., Schaeffer L., Roy R., Moncollin V., Egly J. M., Wang Z., Freidberg E. C., Evans M. K., Taffe B. G., et al. Nat. Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 26.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., et al. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang L., Sowa Y., Sakai T., Pardee A. B. Oncogene. 2000;19:5712–5719. doi: 10.1038/sj.onc.1203963. [DOI] [PubMed] [Google Scholar]
- 28.Castro-Galache M. D., Ferragut J. A., Barbera V. M., Martin-Orozco E., Gonzalez-Ros J. M., Garcia-Morales P., Saceda M. Int. J. Cancer. 2003;104:579–586. doi: 10.1002/ijc.10998. [DOI] [PubMed] [Google Scholar]
- 29.Liu L. T., Chang H. C., Chiang L. C., Hung W. C. Cancer Res. 2003;63:3069–3072. [PubMed] [Google Scholar]
- 30.Taniura S., Kamitani H., Watanabe T., Eling T. E. J. Biol. Chem. 2002;277:16823–16830. doi: 10.1074/jbc.M200527200. [DOI] [PubMed] [Google Scholar]
- 31.Chandrasekharan N. V., Simmons D. L. Genome Biol. 2004;5:241. doi: 10.1186/gb-2004-5-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenner R. G., Young R. A. Nat. Rev. Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 33.Kraus W. L., Lis J. T. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 34.Thorslund T., von Kobbe C., Harrigan J. A., Indig F. E., Christiansen M., Stevnsner T., Bohr V. A. Mol. Cell. Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S. K., Yu S. L., Prakash L., Prakash S. Mol. Cell. Biol. 2001;21:8651–8656. doi: 10.1128/MCB.21.24.8651-8656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tantin D., Kansal A., Carey M. Mol. Cell. Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleiman F. E., Wu-Baer F., Fonseca D., Kaneko S., Baer R., Manley J. L. Genes Dev. 2005;19:1227–1237. doi: 10.1101/gad.1309505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starita L. M., Horwitz A. A., Keogh M. C., Ishioka C., Parvin J. D., Chiba N. J. Biol. Chem. 2005;280:24498–24505. doi: 10.1074/jbc.M414020200. [DOI] [PubMed] [Google Scholar]
- 39.Bregman D. B., Halaban R., van Gool A. J., Henning K. A., Friedberg E. C., Warren S. L. Proc. Natl. Acad. Sci. USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takebayashi Y., Pourquier P., Zimonjic D. B., Nakayama K., Emmert S., Ueda T., Urasaki Y., Kanzaki A., Akiyama S. I., Popescu N., et al. Nat. Med. 2001;7:961–966. doi: 10.1038/91008. [DOI] [PubMed] [Google Scholar]
- 41.Minuzzo M., Marchini S., Broggini M., Faircloth G., D’Incalci M., Mantovani R. Proc. Natl. Acad. Sci. USA. 2000;97:6780–6784. doi: 10.1073/pnas.97.12.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Synold T. W., Dussault I., Forman B. M. Nat. Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 43.Friedman D., Hu Z., Kolb E. A., Gorfajn B., Scotto K. W. Cancer Res. 2002;62:3377–3381. [PubMed] [Google Scholar]
- 44.Ho Sui S. J., Mortimer J. R., Arenillas D. J., Brumm J., Walsh C. J., Kennedy B. P., Wasserman W. W. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merika M., Thanos D. Curr. Opin. Genet. Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 46.Taniguchi T., Takaoka A. Nat. Rev. Mol. Cell. Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 47.Chang H. M., Paulson M., Holko M., Rice C. M., Williams B. R., Marie I., Levy D. E. Proc. Natl. Acad. Sci. USA. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson B. Age. Res. Rev. 2003;2:245–261. doi: 10.1016/s1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 49.Lu T., Pan Y., Kao S. Y., Li C., Kohane I., Chan J., Yankner B. A. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 50.Kyng K. J., May A., Kolvraa S., Bohr V. A. Proc. Natl. Acad. Sci. USA. 2003;100:12259–12264. doi: 10.1073/pnas.2130723100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blalock E. M., Geddes J. W., Chen K. C., Porter N. M., Markesbery W. R., Landfield P. W. Proc. Natl. Acad. Sci. USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrucci L., Ble A., Bandinelli S., Lauretani F., Suthers K., Guralnik J. M. Aging Clin. Exp. Res. 2004;16:240–243. doi: 10.1007/BF03327390. [DOI] [PubMed] [Google Scholar]
- 53.Horibata K., Iwamoto Y., Kuraoka I., Jaspers N. G., Kurimasa A., Oshimura M., Ichihashi M., Tanaka K. Proc. Natl. Acad. Sci. USA. 2004;101:15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong F., Kwon Y., Smerdon M. J. DNA Rep. 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 55.Johnson J. M., Edwards S., Shoemaker D., Schadt E. E. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Morillon A., Karabetsou N., Nair A., Mellor J. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Sudarsanam P., Iyer V. R., Brown P. O., Winston F. Proc. Natl. Acad. Sci. USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiyono T., Foster S. A., Koop J. I., McDougall J. K., Galloway D. A., Klingelhutz A. J. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 59.Affymetrix . Microarray Suite User Guide, Version 5. Santa Clara, CA: Affymetrix; 2001. [Google Scholar]
- 60.Irizarry R. A, Bolstad B. M, Collin F, Cope L. M, Hobbs B, Speed T. P. Nucleic Acids Res. 2003;31 doi: 10.1093/nar/gng015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.