Abstract
Regulatory T cells that express the Foxp3 transcription factor play important roles in preventing autoimmune diseases. Although several studies have demonstrated that the lack of the forkhead DNA-binding domain of Foxp3 caused severe autoimmune disease in scurfy mutant mice, the other functional domains of Foxp3 are less well characterized. Here, we show that the deletion of glutamic acid (ΔE250) in the leucine-zipper domain of Foxp3 causes a loss of hyporesponsiveness when compared with wild-type Foxp3 upon antigenic stimulation. CD4 T cells that ectopically express the glutamic acid mutant show significant losses of suppressor activity both in vitro and in vivo. We also demonstrate that regulation of both Th1- and Th2-type cytokine secretion in CD4 T cells that express wild-type Foxp3 is significantly altered by the deletion of glutamic acid. Defects are also observed in the expression of adhesion molecules, such as l-selectin (CD62L) and CD103, suggesting an important role of glutamic acid in the migratory behavior of regulatory T cells. Finally, this mutation reduces transcriptional repressor activity and impairs the homodimerization of Foxp3. Taken together, our results provide insight into the mechanism that controls autoimmune diseases via the deletion of this single glutamic acid residue in the leucine-zipper domain of Foxp3.
Keywords: autoimmunity
Regulatory T cells are known to play key roles in the maintenance of immunological homeostasis by the suppression of potential self-reactive CD4+CD25− T cells in the periphery (1). Naturally arising CD4+CD25+ T cells isolated from both mouse and human show similar behaviors in terms of potent suppressive functions on their CD25− counterparts via cell–cell contact (2, 3).
Recent studies on CD4+CD25+ T cells have shown that Foxp3, a forkhead (FKH) family transcription factor, has an important role in regulatory T cell development and is currently the best lineage marker in mice (4). In addition, Foxp3-expressing regulatory T cells themselves are hyporesponsive to antigenic stimulation for both proliferation and inhibition of cytokine secretion (4, 5). Transgenic and knockout studies of Foxp3 revealed that Foxp3 plays a critical role for the prevention of autoimmune disease, adoptive transfer of Foxp3-expressing CD4 T cells have also demonstrated that ectopic expression of Foxp3 is sufficient for the mediation of immunological tolerance in vivo (5–7).
In humans, it has been shown that Foxp3 mutations are responsible for ≈70% of patients with severe X-linked autoimmunity and allergic disorders in immune dysfunction polyendocrinopathy enteropathy X-linked (IPEX). The onset of similar autoimmune disease occurs in scurfy mutant mice that lacks the important FKH-binding domain in Foxp3 because of the presence of a 2-bp insertion that results in an early stop codon (8–10). Although members of the Fox family can act as transcriptional activators and repressors, it has been shown experimentally that the FKH domain of Foxp3 is required for the suppression of various cytokine genes (11). A common feature of FKH family proteins is the DNA-binding activity of the FKH domain on the specific DNA sequence A(A/T)TpyTT(G/T)py (py:pyrimidine) (4). Various single amino acid mutations in the FKH domain were found in human patients, suggesting the importance of the DNA-binding domain of Foxp3 (8, 10). Recently, it was also reported that amino acid deletions in either the leucine-zipper or zinc finger domains within Foxp3 occur in IPEX patients cases (12). The functions of these domains have not yet been thoroughly examined.
In this study, we show that incorporation of the glutamic acid deletion in the leucine-zipper domain of wild-type Foxp3 (ΔE250) eliminates its suppressor function in vitro and in vivo. Biochemical analysis shows that wild-type Foxp3 forms homodimers and that the glutamic acid deletion causes impairment of Foxp3 homodimerization. Interestingly, the loss of such suppressive function may be partly explained by the effects of Foxp3 mutant- expressing T cells on cytokine gene transcription. We also examined the influences of the ectopic expression of Foxp3 on both Th1− and Th2− cytokine production. Culture of wild-type Foxp3 transduced CD4 T cells, but not cells expressing the ΔE mutant transduced CD4 T cells, inhibited Th1 cytokine (IFN-γ) as well as Th2 cytokine (IL-4 and IL-5) secretion. The ectopic expression of the ΔE mutant also failed to increase expression of migration-related adhesion molecules, such as CD62L and CD103. The results of our study provide insight into the mechanisms that regulate autoimmune diseases via the deletion of this single glutamic acid residue in the leucine-zipper domain of Foxp3 and the importance of leucine-zipper domain.
Results
Lack of the Glutamic Acid Residue in the Leucine-Zipper Domain of Foxp3 Impairs the Generation of Suppressive Regulatory T Cells in Vivo.
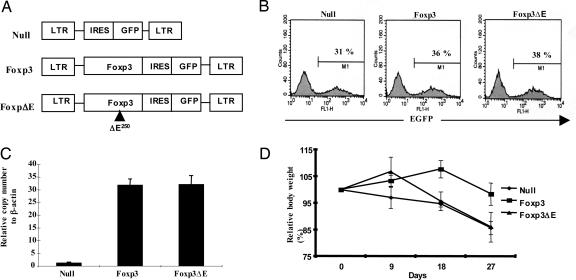
To examine the effects of IPEX mutations outside of the FKH DNA-binding domain of Foxp3, we chose the human JM2 mutation, which is a deletion of the glutamic acid residue in the leucine-zipper domain of Foxp3. In humans, the JM2 mutation corresponds to amino acid 251 of Foxp3, which is the second glutamic acid (E) in the EKEK motif and corresponds to amino acid 250 in murine Foxp3. Based on the very high sequence homology (90%) in the leucine-zipper domains of both mouse and human, we deleted this glutamic acid residue in mouse Foxp3 by site-directed mutagenesis and subsequently generated retroviral constructs (Fig. 1A). The expression of both wild-type Foxp3 and its mutant was confirmed by Western blot (see Fig.7, which is published as supporting information on the PNAS web site). As ectopic expression of Foxp3 in CD4+CD25− T cells has been shown to generate a regulatory T cell-like phenotype, we decided to use a retrovirus-mediated gene transfer system to compare wild-type Foxp3 and its mutant that lacks the glutamic acid (Foxp3ΔE). After 72 h of infection with these vectors, 31–38% of the CD4+ T cells were GFP-positive (Fig. 1B). To measure the expression levels of Foxp3 in the sorted cells, we performed real-time RT-PCR, which resulted in a 30-fold increase in Foxp3 expression (Fig. 1C). Next, we asked whether this mutant has the potential to generate functional regulatory T cells activity comparable to wild-type Foxp3 using an inflammatory bowel disease (IBD) model. IBD was induced by CD4+CD25− cells, which were coinjected with null vector-, wild-type Foxp3-, or ΔE mutant Foxp3-transduced CD4+CD25− T cells into RAG knockout mice. We observed that the null-vector-transduced T cells resulted in consistent weight loss initially, whereas the ΔE mutant transduced T cells showed an initial increase and then a substantial decrease in body weight (Fig. 1D). By contrast, suppression with the wild-type Foxp3-expressing T cells showed little loss of body weight. Wild-type Foxp3-induced regulatory T cells prevented disease, whereas the ΔE mutant does not generate functional regulatory T cells.
Fig. 1.
The ΔE mutation in the leucine-zipper domain of Foxp3 prevents generation of functional regulatory T cells. (A) Each Foxp3 and Foxp3 ΔE cDNA was cloned into MSCV 2.2 retroviral vectors containing the marker GFP. (B) CD4+CD25− T cells from C57BL/6 mice were transduced with retroviruses, and GFP-positive cells were then sorted. (C) GFP-positive cells were analyzed by real-time PCR for Foxp3 expression. (D) The effect of transduced CD4+ T cells on the progression of IBD was monitored by mean body weight. Closed circles, null vector; closed squares, wild-type Foxp3; closed triangles, Foxp3ΔE.
Glutamic Acid Deletion of Foxp3 Induced Loss of Suppressor Function.
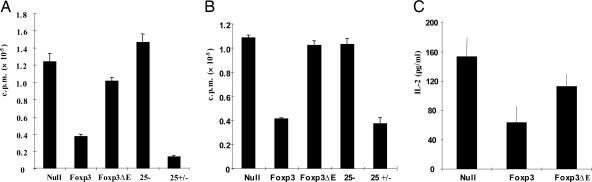
Based on our findings using the IBD model and data from previous reports, we investigated the possible defects in Foxp3 mutant-expressing CD4 T cells. Because one of the key features of Foxp3-expressing T cells is hyporesponsiveness to antigenic stimulation, the effects of wild-type Foxp3 vs. the Foxp3 ΔE mutation on T cell proliferation were examined. The wild-type Foxp3-transduced cells were almost as effective as purified CD4+CD25+ cells in inhibiting proliferation. A significant loss of hyporesponsiveness was found in Foxp3 mutant-expressing T cells, suggesting the importance of this glutamic acid in regulating proliferation (Fig. 2A). It is well known that Foxp3-expressing CD4 T cells can suppress their CD25− T cell counterparts, thereby preventing proliferation of autoreactive T cells. To determine whether the Foxp3 ΔE mutant could affect the suppressor function on CD25− T cells, we cocultured CD25− T cells and Foxp3-transduced cells sorted by FACS that was based on GFP expression in the presence of antigen-presenting cells (APCs) and T cell receptor stimulation. We found that suppressor function was significantly lost after glutamic acid deletion (Fig. 2B). To further investigate whether IL-2 production was affected in these GFP-sorted cells, we stimulated cells with irradiated APCs and anti-CD3 mAb and then assessed IL-2 secretion. The inhibition of IL-2 secretion from Foxp3-transduced CD4+ T cells was largely reversed in Foxp3ΔE-transduced cells (Fig. 2C). Our results demonstrate that the glutamic acid deletion in the leucine-zipper domain of Foxp3 significantly impairs the suppressive function of Foxp3.
Fig. 2.
Effects of ΔE mutation on proliferation, suppressor function, and IL2 secretion. The ΔE mutation in Foxp3 leads to loss of suppressor function of Foxp3-transduced CD4 cells cultured alone (A) or with purified CD4+CD25− T cells (B). (C) IL-2 secretion was measured in transduced CD4 cells that were cultured alone.
Loss of Control in Th1 and Th2 Cytokine Secretion Is Induced by the Foxp3 Mutant.
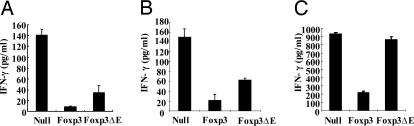
We decided to test the effect of the glutamic acid deletion on production of IFN-γ, which is one of the main characteristics of inflammatory disorders. To assess the ability of CD4+ T cells expressing either the Foxp3ΔE mutant or wild-type Foxp3 to produce IFN-γ, we stimulated transduced GFP- positive cells with irradiated APCs and anti-CD3 mAbs for 72 h. Interestingly, the ability of mutant Foxp3 to suppress IFN-γ production was impaired but not completely abrogated (Fig. 3A). Next, we cocultured CD25− T cells with Foxp3- or the ΔE mutant-expressing T cells to determine the effects on CD4+CD25− T cells. However, this pattern was essentially unchanged, suggesting that the glutamic acid mutant still retains a partially suppressive role on naïve CD25− T cells in terms of IFN-γ production (Fig. 3B).
Fig. 3.
Effects of ΔE mutation on IFN-γ (Th1 cytokine) secretion. The ΔE mutation in Foxp3 impairs Foxp3-induced inhibition of CD4 T cells cultured alone (A) or with either purified CD4+CD25− T cells (B) or Th1 differentiated CD4 T cells (C).
This finding prompted us to further investigate the regulatory effects on the control on Th1-differentiated CD4+ T cells. C57BL/6 (B6) CD4 T cells were cultured for 4 days under conditions to generate Th1 cells (see Materials and Methods) and were later cocultured for 3 days with either wild-type Foxp3- or the Foxp3 ΔE mutant-transduced CD4+ T cells. The suppressive effect of wild-type Foxp3 was maintained, whereas the ΔE mutant did not inhibit IFN-γ secretion (Fig. 3C). The maintenance of this suppressive activity by wild-type Foxp3 in a Th1 environment correlates well with an in vivo study that describes the inhibition of IBD by the transfer of Foxp3-expressing CD4+CD25+ regulatory T cells in that they can cure IBD even after the onset of a Th1 environment (13).
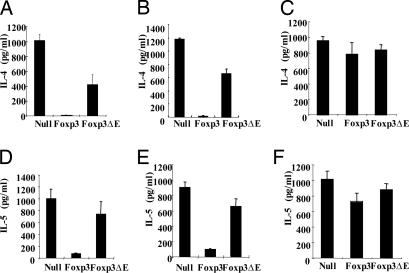
The ΔE mutant patient phenotype has been reported as akin to a Th2 phenotype, and thus the effect of the single glutamic acid residue deletion on IL-4 and IL-5 cytokine production was examined. First, we showed that these two cytokines were potently suppressed by wild-type Foxp3-expressing CD4 T cells, in both single culture of GFP-positive cells (Fig. 4A and D) and coculture with CD25− T cells (Fig. 4 B and E). In coculture experiments with CD4+CD25− T cells, the ΔE mutant showed partial inhibition of IL-4 secretion but little inhibition of IL-5 secretion. These data suggest that the effects of the ΔE mutation may have different effects on regulation of Th1 and Th2 cytokine secretion in the ΔE mutant-expressing T cells. To extend our study from Th1-polarized cells, we also tested coculture with Th2-differentiated cells. Little suppressive activity of either wild-type or ΔE mutant Foxp3 was observed under these conditions (Fig. 4 C and F). This is consistent with previous findings (14), in that freshly isolated Treg cannot suppress polarized Th2 cells in vitro without prior activation of Treg.
Fig. 4.
Effects of ΔE mutation on Th2 cytokines. The ΔE mutation in Foxp3 attenuates Foxp3-induced suppression of IL-4 and IL-5 secretion in CD4 cells (A and D) cultured alone or with CD4+CD25− T cells (B and E). Cytokine expression by coculture of transduced CD4 cells with Th1- and Th2-differentiated T cells (C and F).
Deletion of Glutamic Acid in Foxp3 Inhibits Induction of Cell Surface Markers of Regulatory T Cells, Including Adhesion Molecules.
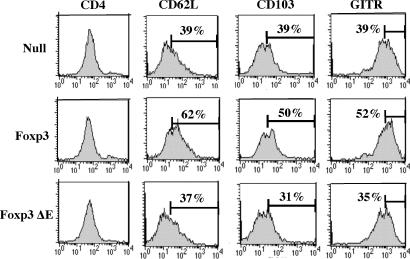
It has been previously shown that the ectopic expression of Foxp3 changed cell surface markers into regulatory T cell-like phenotypes (5). Because it has been also demonstrated that Foxp3+CD62L+ cells prevented autoimmune diseases such as type 1 autoimmune diabetes and myasthenia gravis (15, 16), it was of interest whether the ΔE mutant could inhibit these events. We performed FACS analysis after transducing wild-type Foxp3− or the Foxp3 ΔE mutant in CD4+CD25− T cells. As a control, we showed that CD4 levels after retrovirus transduction were not altered. We found that wild-type Foxp3-transduced T cells showed increase in CD62L expression, whereas the ΔE mutant failed to up-regulate this adhesion molecule (Fig. 5). This suggests that CD4 T cells expressing the ΔE mutant can migrate more like null-vector-transduced cells, thereby losing regulatory T cell-like character. We also tested glucocorticoid-induced tumor necrosis factor-receptor family related (GITR) and CD103, because it has been shown that both of them are increased by wild-type Foxp3 expression. GITR has been known as one of the regulatory T cell surface markers (17), and αEβ7 integrin (CD103) plays a crucial role for localization of T cells in the intestine, thereby preferentially preventing IBD (18, 19). We observed that the ΔE mutant impaired up-regulation of CD103 and GITR, whereas wild-type Foxp3 increased expression of both cell surface markers. Taken together, our data suggest that the ΔE mutation affects not only cytokine production but also cell surface markers of regulatory T cells.
Fig. 5.
Deletion of a functional glutamic acid residue down-regulates cell surface markers including adhesion molecule. Each group of GFP-positive cells was sorted and stained with APC-conjugated anti-CD4, phycoerythrin-conjugated anti-CD62L, anti-CD103, and anti-GITR Abs. A representative result from three independent experiments is shown.
Lack of Glutamic Acid Decreases Transcriptional Repression of Foxp3 with Concomitant Loss of Homodimerization.
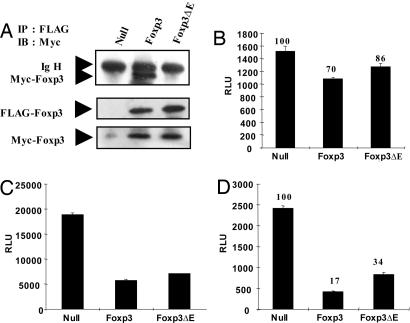
It is thought that Foxp3 may homodimerize through its leucine-zipper domain (12). We have examined this notion directly in transiently transfected HEK293 cells using either wild-type or ΔE Foxp3-encoding plasmids that are epitope-tagged with either a FLAG or Myc tag, respectively. Nuclear extracts were immunoprecipitated with anti-FLAG Ab and then immunoblotted with anti-Myc Ab. Fig. 6A Top shows that the FLAG-tagged wild-type Foxp3 coprecipitated the Myc-tagged wild-type Foxp3. By contrast, the ΔE mutant tagged with Myc was not coprecipitated with the FLAG-tagged ΔE mutant Foxp3. Fig. 6A Middle and Lower show that similar levels of FLAG- and Myc-tagged Foxp3 proteins were present in the transfected cells. These results show that Foxp3 can homodimerize, and more importantly, that this homodimerization is crippled by the ΔE mutation.
Fig. 6.
The glutamic acid residue in the leucine-zipper domain of Foxp3 is required for homodimerization. (A) FLAG- and Myc-tagged Foxp3 or Foxp3ΔE were cotransfected for 48 h. After harvesting of cells, lysates were incubated with FLAG antibody (M2)-conjugated agarose beads, washed and boiled with sample buffer for SDS/PAGE. Immunoprecipitates were analyzed by anti-Myc Ab. Lysates were also analyzed for protein expression by using anti-FLAG and anti-Myc Abs. RLU, relative luciferase unit. (B–D) Each null vector, Foxp3, and Foxp3 ΔE was cotransfected with the FKH luciferase construct in Jurkat T cells with lipid-mediated transfection (B). After 48 h, luciferase activity was measured (C and D). Each NFAT and NF-κB luciferase construct was cotransfected with null vector, Foxp3, or Foxp3ΔE in HEK293 cells. After 48-h transfection, cells were harvested and assessed for luciferase activity. All samples were normalized by cotransfection and analysis of TK-Renilla activity. A representative result from at least three independent experiments is shown.
Based on our findings of the loss of suppressor function of Foxp3ΔE, we determined whether the transcriptional repression activity of Foxp3 was also affected. We examined whether the activity from a FKH reporter construct differed from either the wild-type or the ΔE mutant in transiently transfected Jurkat T cells. The wild-type Foxp3 reduced the FKH reporter activity by 30%, whereas the Foxp3 ΔE reduced its activity by 14%. Thus, ≈50% of repressor activity was consistently lost with the single glutamic acid residue deletion in the leucine-zipper domain (Fig. 6B). We also assessed the effect of wild-type and ΔE mutant Foxp3 on both NF-κB- and NFAT-mediated gene transcription. For NF-κB, we cotransfected p65 with a NF-κB luciferase construct and observed that the glutamic acid deletion did not impair the suppressive activity of wild-type Foxp3 in HEK293 cells (Fig. 6C). For NFAT, we measured NFAT-mediated gene transcription with a constitutively active form of NFAT (NFAT-ca), showing that NFAT-mediated gene transcription was partially impaired when NFAT-ca was overexpressed with the ΔE mutant (Fig. 6D). Overall, our data suggest that the glutamic acid residue deletion in the leucine-zipper domain of Foxp3 contributes to the defect in Foxp3 homodimerization and decreases its transcriptional repressor activity.
Discussion
We have shown that the deletion of the glutamic acid residue in the leucine-zipper domain of murine Foxp3 largely abolishes its ability to confer suppression on transduced CD4+CD25− T cells both in vitro and in vivo. Surprisingly, this mutation has differential effects on cytokine production in both Th1 and Th2 cells, and these effects appear to be particular for inflammatory and allergic events. The lack of dimerization potential, therefore, must be a critical factor that contributes to the loss of the regulatory T cell-like phenotype.
In our study, the suppressor activity of Foxp3-transduced CD4+CD25− T cells in vitro was significantly decreased by the deletion of glutamic acid at position 250. We also observed that IL-2 secretion and proliferation of Foxp3ΔE-expressing T cells were significantly but not completely impaired. Possibly, a single amino acid deletion mutation can cause different types of defects in the repressive properties of Foxp3 when compared with the scurfy mutant, which lacks the DNA-binding domain and nuclear localization signal. It has been demonstrated that both NFAT- and NF-κB-mediated gene transcription was critically impaired in the scurfy mutant. Our data indicate that the glutamic acid residue deletion is not involved in the impairment of the inhibition of p65- mediated gene transcription by Foxp3 and only partially affects NFAT-mediated gene transcription. Alternatively, studies have reported differential gene expression in regulatory T cells (20, 21), which suggest that this mutant can cause altered repressor function on different genes with varying degrees, corresponding to decreased FKH transcriptional activity of the ΔE mutant.
A previous comparison of amino acid sequences between N-Myc and human FOXP3 in three-heptad repeats demonstrated that either homo-or heterodimerization may mediate the proper function of Foxp3 (11). In N-Myc, it was shown that the second heptad is involved in both homodimerization and heterodimerization with Max (22). Our study shows a similar finding for Foxp3. Given our finding that the ΔE mutant affects control of proliferation, we attempted to predict possible molecules involving interactions with proteins that regulate cell cycle. Our computer search based on a motif scan identified both cdc2 and cdk5 kinases as candidates under very strict conditions (http://scansite.mit.edu). It has been also very recently suggested that FOXP dimers can bind cognate DNA sites separated far from each other or located on different DNA strands, possibly suggesting that the function of FOXP3 proteins for certain genes (e.g., cytokines affected in our study) highly depends on promoting the assembly of higher-order protein–DNA complexes with varying degrees (23).
We observed that ectopic expression of wild-type Foxp3 can efficiently suppress IFN-γ production, whereas this function was partially lost in ΔE mutant-transduced cells cocultured with CD25− T cells. The loss of suppression on Th1 polarized cells that secrete IFN-γ by ΔE mutant-expressing CD4 T cells correlates with our IBD model in that control of IFN-γ production is significantly impaired by the ΔE mutant after the differentiation of CD4 T cells into Th1. Hence, it is likely that the effect of Foxp3 mutant protein may be amplified and thereby exert greater effects after the differentiation of CD4 T cells into Th1 cells leading to inflammatory disorders.
Along with the control of autoimmune disorders that are mediated by Th1 responses, it has recently been shown that induction of Th1-like regulatory cells that express Foxp3 can protect airway hyperreactivity (24). Moreover, it was reported that IPEX patients having the glutamic acid deletion developed eczema, increased Ig E levels, and peripheral eosinophilia (11). It is conceivable that the glutamic acid residue deletion can affect both naturally arising and adaptively induced regulatory T cell generation, thereby exacerbating inflammatory disorders. Our results demonstrate that the tight control on IL-4 and IL-5 production by CD4+ T cells was hampered by the ΔE mutant when cocultured with CD25− T cells. Thus wild-type Foxp3-expressing T cells can exert significant inhibition of the development of Th1 or Th2 cells, whereas the Foxp3ΔE-expressing CD4 Treg cells would lose that regulatory ability. Although the suppressive activity of either wild-type or ΔE mutant Foxp3 was minimal under our conditions for Th2-polarized T cells, the effect of the ΔE mutant for Th2 responses would be significant based on the suppressive function of Foxp3-expressing Treg cells in vivo.
Although very little is known about the trafficking behavior of regulatory T cells, CD62L+ regulatory T cells also expresses CCR7 and delayed diabetes in vivo (25), suggesting that these regulatory T cells retain a naïve T cell-like character rather than an effector/memory phenotype. Cells expressing ΔE mutant Foxp3 failed to induce CD62L, suggesting that this deletion might confer different migratory behavior than wild-type Foxp3-expressing CD4 T cells. During the preparation of this manuscript, a paper appeared showing that CD103 expression in regulatory T cells is not essential for their effects on CD25− T cells (26). However, CD103+ regulatory T cells are known to be important for preventing IBD by playing unique roles especially in gut-associated lymphoid tissues (27). The lack of increased CD103 and GITR expression by the ΔE mutant Foxp3 demonstrated that the effects of mutation was not confined to cytokine regulation.
For both mouse and human, it has been thought that Foxp3 can function as a potent transcriptional repressor, either directly or indirectly with its high mRNA levels in Foxp3-expressing regulatory T cells. It is possible that affected human offspring who have the ΔE mutation from early infancy can accumulate pathogenic regulatory T cells. Recently, the process that is required for both the development of regulatory T cells and the induction of Foxp3 has been characterized (28). In the murine model and possibly in human patients bearing the ΔE mutation, mutant Foxp3 can be induced by the same mechanisms. It is possible that these mutant Foxp3-expressing T cells may develop from thymus. Also, it is known that TGF-β can induce Foxp3 in CD25− T cells (29), and this process would also induce mutant Foxp3 expression given that the foxp3 gene is mutated. In both cases, these ΔE mutant-expressing T cells would aggravate Th1-type autoimmune diseases.
A study of a large number of IPEX patients who have mutations in the FKH domain of FOXP3 has been published, showing different patterns of gene expression (30). For example, it has been reported that human CD4+CD25+ regulatory T cells express one isoform of Foxp3 that lacks exon 2, potentially functioning as a dominant-negative form of Foxp3 (31). From our findings in mouse Foxp3, a human IPEX patient who has the ΔE mutation may be critically affected with this Foxp3 isoform. Taken together, the deletion of glutamic acid residues in the Foxp3 protein provides a valuable approach to explain how this deletion event can contribute to molecular defects in severe autoimmune and allergic diseases.
Materials and Methods
Mice.
C57BL/6 mice (B6) and RAG2 knockout mice (C57BL/6 background) at 6–8 weeks of age were used. Mice were housed in a specific pathogen-free facility at Yale University. All mouse experiments were approved by the Institutional Animal Care and Use Committee of Yale University.
Plasmids and Antibodies.
Anti-Myc antibody and an anti-FLAG M2 affinity gel were purchased from Sigma. Murine and human Foxp3 were kind gifts from Shimon Sakaguchi (Kyoto University, Kyoto). The FLAG epitope tag was inserted into the N-terminal region of mouse Foxp3 between positions 1 and 2, and the Myc epitope was inserted immediately before the translation termination codon using standard methods. The constitutively active form of NFAT was provided by Mohammed Oukka (Harvard Medical School, Boston). Murine anti-CD3 (145.2C-11), anti-CD28 (37.51), phycoerythrin-conjugated anti-CD62L, anti-CD4, anti-GITR, and anti-CD103 mAbs were purchased from eBioscience (San Diego). Anti-p38 antibody was purchased from Cell Signaling Technology, Beverly, MA.
Luciferase Assays and Transfection.
Jurkat cells were costransfected with NFAT-, NF-κB-, or FKH (Forkhead/winged helix)-luciferase reporter constructs using the XtremeGene Q2 reagent (Roche Molecular Biochemicals). Mouse FLAG and Myc-tagged Foxp3 and Foxp3 ΔE were cloned into the pcDNA3.1(−)myc-his A expression vector. Briefly, 1.5 × 105 cells were transfected with control null vector and vectors that encoded mouse Foxp3 or Foxp3 ΔE, and each assay was normalized with relative normalization for Renilla using TK-Renilla. After a 48-h transfection, cells were harvested and analyzed using dual luciferase assay reagents (Promega). For HEK293 cells, lipofectamine 2000 (Invitrogen) was used to cotransfect p65 and the constitutively active form of NFAT. For each condition for transient transfection, expression of Foxp3 was confirmed by Western blot in Jurkat and 293 cells, respectively, as shown in Fig. 8, which is published as supporting information on the PNAS web site.
Ectopic Expression of Foxp3 with Retroviral Infection.
Murine wild-type Foxp3 and Foxp3ΔE cDNAs were cloned into the MSCV2.2 GFP retroviral plasmid. The protein expression level of each wild-type Foxp3 and Foxp3ΔE cDNA was tested as shown in Fig. 7. For retrovirus packaging, the Phoenix-Eco cell line was used. Retrovirus-containing supernatant was harvested after a 48-h incubation from transfected Phoenix-Eco cells and filtered before infection. Freshly isolated magnetic-activated cell sorting purified CD4+CD25− T cells were activated by irradiated APCs, anti-CD3, and anti-CD28 for 24 h before infection. At the time of infection, 50 units/ml of IL-2 and 8 μg/ml of polybrene were used, and cells were infected by centrifugation at for 1 h at 583 × g. At 72 h postinfection, GFP-positive cells were sorted by using MoFlo (DAKO).
Proliferation and Suppression Assay.
To measure proliferation of sorted GFP-positive cells, 0.5 × 105 GFP-positive Foxp3 and Foxp3 ΔE-expressing CD4 T cells were incubated with 1.0 × 105 cells of APCs that had been irradiated (2,000 rads) APCs. Cells were plated in round-bottom 96-well plates with anti-CD3 mAbs. Proliferation was assessed after 72 h, after the addition of [3H]thymidine [1 μCi per well (1 Ci = 37 GBq); NEN]. For analysis of suppressive capacity of GFP-positive sorted cells, 0.5 × 105 transduced CD4 T cells were added to 1.0 × 105 cells of CD4+CD25− T cells in the presence of anti-CD3 mAbs and 1.0 × 105 irradiated APCs. Subsequently, suppression was measured by determining the [3H] thymidine incorporation. To obtain freshly isolated CD4+ CD25− and CD25+ cells as controls, APC-conjugated murine anti-CD4 and phycoerythrin-conjugated murine anti-CD25 Abs were used and cells were sorted by using MoFlo (DAKO).
Th1/2 Differentiation and Cytokine Bead Array.
For measuring cytokines in GFP-sorted cells, 0.5 × 105 GFP-positive T cells were incubated with 1.0 × 105 cells irradiated APCs. Cells were plated in round-bottom 96-well plates with anti-CD3 mAbs. After 72 h, supernatants were harvested and analyzed by the Bioplex cytokine bead array system (Bio-Rad). To assess cytokine secretion in coculture with CD25− T cells, 0.75 × 105 GFP-positive T cells were cocultured with an equal number of CD25− T cells under identical conditions. To generate differentiated CD4 T cell cultures, magnetic-activated cell sorting-purified CD4 T cells were incubated either with Th1 (25 units/ml IL-2/5 ng/ml IL-12/1 μg/ml anti-IL-4 Ab) or Th2 conditions (25 units/ml IL-2/10 ng/ml IL-4/1 μg/ml anti-IFN-γ Ab) for 4 days with anti-CD3 mAb. After 3 additional days of resting after two times of wash with 5% FCS containing Bruff’s media, 0.5 × 105 Th1- and Th2-differentiated cells were cocultured with an equal number of GFP-sorted cells for 72 h in the presence of irradiated APCs and anti-CD3 mAb.
Immunoprecipitation and Western Blot.
To prepare nuclear extracts, cells were washed twice with PBS and lysed for 10 min on ice in buffer A (10 mM Hepes, pH 7.9/10 mM KCl/0.1 mM EDTA/0.5 mM PMSF/1.5 μg/ml aprotinin/1 μg/ml leupeptin/1 μg/ml pepstatin) in the presence of 0.2% Nonidet P-40. The lysates were clarified by centrifugation for 5 min at 16,000 × g. Pellets were washed with buffer A and incubated with buffer C (20 mM Hepes, pH 7.9/0.4M NaCl/1 mM EDTA/1 mM PMSF/1.5 μg/ml aprotinin/1 μg/ml leupeptin/1 μg/ml pepstatin) to extract nuclear proteins for 30 min in an agitator (Eppendorf). Lysates were recovered by centrifugation for 20 min at 4°C. For immunoprecipitation, anti-FLAG (M2)-conjugated agarose beads (Sigma) were added and rotated for 2 h at 4°C. Immunoprecipitates were washed with TBS, pH 7.4, three times followed by the addition of 2× sample buffer and heated at 95°C for 5 min. Immunoprecipitated proteins were separated by SDS/PAGE and transferred to a poly(vinylidene difluoride) membrane. Immunoblotting was performed by probing with anti-Myc Ab (9E10, Sigma), followed by horseradish peroxidase-conjugated anti-rabbit IgG Ab and detected by enhanced chemiluminescence (Pierce).
Real-Time PCR.
RNA was prepared from GFP-sorted Foxp3-expressing CD4 T cells with TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed by using oligo(dT)15 primers with BD Sprint PowerScript (BD Biosciences), and cDNA was generated according to standard procedures. Samples were analyzed by real-time PCR analysis on Stratagene MX3000 under standard conditions. Relative copy numbers of mRNA abundance were normalized against β-actin. Foxp3 primers were as follows: Foxp3 forward, 5′-CTTTCACCTATGCCACCCTTATCC-3′; Foxp3 reverse, 5′-ATTCATCTACGGTCCACACTGCTC-3′; β-actin forward, 5′-CCTCTCAGCTGTGGTGGTGAAGC-3′; and β-actin reverse, 5′-GCGTGCTGTCCCTGTATGCCTCT-3′.
Generation of Tetracycline-Inducible Jurkat.
The stability of the FoxP3 and FoxP3 ΔE proteins was also evaluated by using a tetracycline-inducible Jurkat system. A time-course study after induction with doxycycline is shown in Fig. 9 which is published as supporting information on the PNAS web site. FLAG-tagged Foxp3 and FoxP3 ΔE was cloned into the pREV-TRE2 expression vector with BamHI and HindIII restriction enzyme sites. The Jurkat Tet-on cell line was purchased from Clontech (BD Biosciences) and infected with the pREV-TRE2 Foxp3 expression vector. FoxP3 and FoxP3 ΔE Jurkat clones were pulsed with 1 μg/ml of doxycycline (Clontech) for 24 h. After 24-h induction, cells were washed with PBS extensively to remove residual doxycycline and incubated 24, 48, 72, and 96 h in the absence of doxycycline. Subsequently, nuclear lysates were prepared for each time point to monitor protein degradation. Western blot was performed with anti-FLAG mAb and p38 was used as a control for protein quantitation.
IBD.
To induce IBD, each 2.5 × 105-sorted GFP-positive T cells of MSCV2.2 (Null vector), wild-type Foxp3, or glutamic acid residue deletion (ΔE) Foxp3 were cotransferred with 5.0 × 105 CD25− T cells, and animal body weight was measured over time.
Supplementary Material
Acknowledgments
We are thankful to Frederick Rodriguez for animal handling and care and to Gouzel Tokmulina for cell sorting. This work was supported by Korean Research Foundation Grant M07-2003-000-20134-0 and National Institutes of Health Grant P01 AI036529 and in part by research grants to S.-K.L. from Korea Institute of Industrial Technology Evaluation and Planning (M1-0310-40-0000), Korea Health Industry Development Institute (0412-DB00-0101-0011), Korea Science and Engineering Foundation (2005-00117), and Korea Rural Economic Institute (204081-3).
Abbreviations
- IBD
inflammatory bowel disease
- APC
antigen-presenting cell
- FKH
forkhead
- IPEX
immune dysfunction polyendocrinopathy enteropathy X-linked
- GITR
glucocorticoid-induced tumor necrosis factor-receptor family related.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sakaguchi S. Annu. Rev. Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Gavin M., Rudensky A. Curr. Opin. Immunol. 2003;15:690–696. doi: 10.1016/j.coi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Baecher-Allan C., Viglietta V., Hafler D. A. Semin. Immunol. 2004;16:89–98. doi: 10.1016/j.smim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Schubert L. A., Jeffery E., Zhang Y., Ramsdell F., Ziegler S. F. J. Biol. Chem. 2001;276:37672–37679. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- 5.Hori S., Nomura T., Sakaguchi S. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 6.Fontenot J. D., Gavin M. A., Rudensky A. Y. Nat. Immunol. 2003;4:330–336. [PubMed] [Google Scholar]
- 7.Wildin R. S., Ramsdell F., Peake J., Faravelli F., Casanova J. L., Buist N., Levy-Lahad E., Mazzella M., Goulet O., Perroni L., et al. Nat. Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 8.Brunkow M. E., Jeffery E. W., Hjerrild K. A., Paeper B., Clark L. B., Yasayko S. A., Wilkinson J. E., Galas D., Ziegler S. F., Ramsdell F. Nat. Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 9.Bennett C. L., Christie J., Ramsdell F., Brunkow M. E., Ferguson P. J., Whitesell L., Kelly T. E., Saulsbury F. T., Chance P. F., Ochs H. D. Nat. Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 10.Bettelli E., Dastrange M., Oukka M. Proc. Natl. Acad. Sci. USA. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatila T. A., Blaeser F., Ho N., Lederman H. M., Voulgaropoulos C., Helms C., Bowcock A. M. J. Clin. Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mottet C., Uhlig H. H., Powrie F. J. Immunol. 2003;170:3939–3943. doi: 10.4049/jimmunol.170.8.3939. [DOI] [PubMed] [Google Scholar]
- 13.Gallatin W. M., Weissman I. L., Butcher E. C. Nature. 1983;304:30–34. doi: 10.1038/304030a0. [DOI] [PubMed] [Google Scholar]
- 14.Stassen M., Jonuleit H., Muller C., Klein M., Richter C., Bopp T., Schmitt S., Schmitt E. J. Immunol. 2004;173:267–274. doi: 10.4049/jimmunol.173.1.267. [DOI] [PubMed] [Google Scholar]
- 15.Pop S. M., Wong C. P., Culton D. A., Clarke S. H., Tisch R. J. Exp. Med. 2005;201:1333–1346. doi: 10.1084/jem.20042398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aruna B. V., Sela M., Mozes E. Proc. Natl. Acad. Sci. USA. 2005;102:10285–10290. doi: 10.1073/pnas.0504578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHugh R. S., Whitters M. J., Piccirillo C. A., Young D. A., Shevach E. M., Collins M., Byrne M. C. Immunity. 2002;16:311–323. doi: 10.1016/s1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 18.Banz A., Peixoto A., Pontoux C., Cordier C., Rocha B., Papiernik M. Eur. J. Immunol. 2003;33:2419–2428. doi: 10.1002/eji.200324205. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann J., Huehn J., de la Rosa M., Maszyna F., Kretschmer U., Krenn V., Brunner M., Scheffold A., Hamann A. Proc. Natl. Acad. Sci. USA. 2002;99:13031–13036. doi: 10.1073/pnas.192162899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khattri R., Cox T., Yasayko S. A., Ramsdell F. Nat. Immunol. 2003;4:337–342. [PubMed] [Google Scholar]
- 21.Gavin M. A., Clarke S. R., Negrou E., Gallegos A., Rudensky A. Nat. Immunol. 2002;3:33–41. doi: 10.1038/ni743. [DOI] [PubMed] [Google Scholar]
- 22.Wenzel A., Cziepluch C., Hamann U., Schurmann J., Schwab M. EMBO J. 1991;10:3703–3712. doi: 10.1002/j.1460-2075.1991.tb04938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stroud J. C., Wu Y., Bates D. L., Han A., Nowick K., Paabo S., Tong H., Chen L. Structure (London) 2006;14:159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Stock P., Akbari O., Berry G., Freeman G. J., Dekruyff R. H., Umetsu D. T. Nat. Immunol. 2004;5:1149–1156. doi: 10.1038/ni1122. [DOI] [PubMed] [Google Scholar]
- 25.Szanya V., Ermann J., Taylor C., Holness C., Fathman C. G. J. Immunol. 2002;169:2461–2465. doi: 10.4049/jimmunol.169.5.2461. [DOI] [PubMed] [Google Scholar]
- 26.Annacker O., Coombes J. L., Malmstrom V., Uhlig H. H., Bourne T., Johansson-Lindbom B., Agace W. W., Parker C. M., Powrie F. J. Exp. Med. 2005;202:1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huehn J., Siegmund K., Lehmann J. C., Siewert C., Haubold U., Feuerer M., Debes G. F., Lauber J., Frey O., Przybylski G. K., et al. J. Exp. Med. 2004;199:303–313. doi: 10.1084/jem.20031562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tai X., Cowan M., Feigenbaum L., Singer A. Nat. Immunol. 2005;6:152–162. doi: 10.1038/ni1160. [DOI] [PubMed] [Google Scholar]
- 29.Chen W., Jin W., Hardegen N., Lei K. J., Li L., Marinos N., McGrady G., Wahl S. M. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coffer P. J., Burgering B. M. Nat. Rev. Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- 31.Allan S. E., Passerini L., Bacchetta R., Crellin N., Dai M., Orban P. C., Ziegler S. F., Roncarolo M. G., Levings M. K. J. Clin. Invest. 2005;115:3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.