Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1), when activated by DNA damage, promotes both cell death and inflammation. Here we report that PARP-1 enzymatic activity is directly inhibited by minocycline and other tetracycline derivatives that have previously been shown to have neuroprotective and anti-inflammatory actions. These agents were evaluated by using cortical neuron cultures in which PARP-1 activation was induced by the genotoxic agents N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) or 3-morpholinosydnonimine (SIN-1). In both conditions, neuronal death was reduced by >80% either by 10 μM 3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone, an established PARP inhibitor, or by 100 nM minocycline. Neuronal NAD+ depletion and poly(ADP-ribose) formation, which are biochemical markers of PARP-1 activation, were also blocked by 100 nM minocycline. A direct, competitive inhibition of PARP-1 by minocycline (Ki = 13.8 ± 1.5 nM) was confirmed by using recombinant PARP-1 in a cell-free assay. Comparison of several tetracycline derivatives showed a strong correlation (r2 = 0.87) between potency as a PARP-1 inhibitor and potency as a neuroprotective agent during MNNG incubations, with the rank order of potency being minocycline > doxycycline > demeclocycline > chlortetracycline. These compounds are known to have other actions that could contribute their neuroprotective effects, but at far higher concentrations than shown here to inhibit PARP-1. The neuroprotective and antiinflammatory effects of minocycline and other tetracycline derivatives may be attributable to PARP-1 inhibition in some settings.
Keywords: death, doxycycline, inflammation, neuron
Minocycline and other tetracycline derivatives have neuroprotective effects unrelated to their antimicrobial properties (1). Minocycline can reduce neuronal death after excitotoxicity and ionizing radiation in culture (2, 3), and in animal models of stroke (1, 3–5), Parkinson's disease (6, 7), Huntington's disease (8), and amyotrophic lateral sclerosis (9). The neuroprotective effects of minocycline have been attributed both to reduced inflammation and a direct effect on neuronal survival (2, 3, 5, 9, 10).
Poly(ADP-ribose) polymerase-1 (PARP-1) plays a key role in neuronal death and survival under stress conditions (11). PARP-1 is the most abundant of several PARP family members, accounting for >85% of nuclear PARP activity (11). When activated by DNA damage, PARP-1 consumes NAD+ to form branched poly(ADP-ribose) on target proteins. Poly(ADP-ribose) formation on histones and enzymes involved in DNA repair appears to facilitate DNA repair by preventing chromatid exchange and by loosening histone wrapping (12, 13). Poly(ADP-ribose) formation also has effects on gene transcription through interactions with transcription factors, notably NF-κB (14–16), and PARP-1 inhibition or gene deletion attenuates the brain microglial response to cytokines and other triggers (17–19). Extensive PARP-1 activation can, in addition, lead to neuronal death through mechanisms linked to NAD+ depletion and release of apoptosis inducing factor from the mitochondria (20–22). PARP-1 activation is a key mediator of neuronal death during excitotoxicity, ischemia, and oxidative stress, such that PARP-1 gene deletion or pharmacological inhibition can markedly improve neuronal survival in these settings (20, 23–25).
The overlap of minocycline and PARP inhibitor effects on cell responses to stress suggests the possibility of a common mode of action. Here we show that minocycline and other tetracycline derivatives block PARP-1-mediated neuronal death in culture by directly inhibiting PARP-1 activity. These results identify a mechanism by which these agents can potently influence neuronal survival.
Results
Minocycline Prevents PARP-1-Mediated Neuron Death.
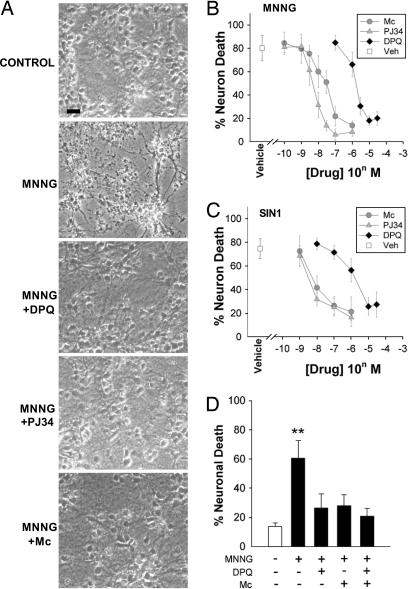
Primary mouse cortical neurons were incubated with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) to induce DNA damage and PARP-1-mediated neuron death. Prior studies have established PARP-1 activation as a key factor mediating neuron death under these conditions (22, 26, 27). This was confirmed here by near-complete inhibition of MNNG-induced neuronal death with the PARP inhibitors 3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone (DPQ) or N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-N,N-dimethylacetamide hydrochloride (PJ34) (Fig. 1 A and B). The IC50 values for DPQ and PJ34 were 2.1 ± 0.2 μM and 6.0 ± 1.6 nM, respectively. Minocycline also reduced MNNG-induced neuronal death, with an IC50 of 42.0 ± 9.5 nM. Thus, minocycline prevents PARP-1-mediated neuronal death with potency exceeding the second generation PARP inhibitor DPQ (28), and approaching that of the third-generation PARP inhibitor PJ34 (29).
Fig. 1.
Minocycline prevents PARP-1-mediated neuronal death. (A) Photomicrographs of neuronal cultures, 24 h after 30-min incubations with MNNG (75 μM) alone or in combination with minocycline (Mino, 100 nM) or the established PARP inhibitors DPQ (25 μM) or PJ34 (100 nM). Control wells received medium exchanges only. Live cells are phase-bright with well demarcated cell bodies. (Scale bar, 20 μm.) (B) Dose–response curves of Mc, DPQ, and PJ34 on MNNG-induced neuronal death. (C) Dose–response curves of Mc, DPQ, and PJ34 on SIN-1-induced neuronal death. Neurons were incubated with 2 mM SIN-1 for 60 min. (D) The neuroprotective effects of minocycline (Mc, 100 nM) are not additive with DPQ (25 μM). ∗∗, P < 0.01 vs. all other conditions; P > 0.1 for comparisons between the DPQ plus Mc condition vs. DPQ alone or vs. Mc alone. Data in B–D are means ± SEM of three independent experiments, each performed in triplicate.
PARP-1 activation also mediates cell death induced by peroxynitrite (20, 21, 25), and we also evaluated the effect of minocycline in cultures treated with the peroxynitrite generator SIN-1 (30, 31). Like MNNG, SIN-1-induced neuronal death was blocked by minocycline with potency intermediate between DPQ and PJ34 (Fig. 1C). Treatment with maximally effective concentrations of DPQ and minocycline together had no additive effect on neuron survival (Fig. 1D), suggesting that minocycline acts along the same cell death pathway as the PARP inhibitors.
Minocycline Inhibits PARP-1 Activity in Neurons.
Effects of minocycline on neuronal PARP-1 activity were evaluated by measuring poly(ADP-ribose) formation and NAD+ consumption. PAR accumulation was evident in almost all neuronal nuclei after incubation with MNNG (Fig. 2A). This accumulation was nearly completely suppressed by minocycline, as well as by DPQ and PJ34. These results were corroborated by Western blots of PAR formation in the neuron cultures, prepared with a different antibody (Fig. 2 B and C). Of note, the Western blots showed PAR formation below basal levels in the presence of 25 μM DPQ or 100 nM minocycline, consistent with prior reports of significant basal PARP activity in neuron cultures (32). In parallel studies, NAD+ consumption by PARP-1 was evaluated by measuring NAD+ in neuronal cultures treated with MNNG. Incubation with 75 μM MNNG for 30 min reduced neuronal NAD+ levels by ≈70% (Fig. 2D). Minocycline attenuated the NAD+ depletion in a dose-dependent manner, with maximal efficacy observed at 100 nM. This effect was comparable to that achieved with a maximally effective concentration of DPQ (25 μM). To exclude the possibility that minocycline was acting by blocking the DNA damage induced by MNNG, DNA-polymerase I-mediated biotin-dATP nick translation (PANT) was used to identify neurons with extensive single-strand DNA breaks. MNNG produced a severalfold increase in PANT-stained cells, and this increase was not attenuated by minocycline or by DPQ (Fig. 2E).
Fig. 2.
Minocycline inhibits of PARP-1 activity in neurons. (A) Photomicrographs show immunostaining for poly(ADP-ribose) (PAR, Left), and nuclei identified in the same fields with propidium iodide (Right). Neurons were fixed for staining 15–30 min after a 10-min incubation with 75 μM MNNG. PAR formation was suppressed by the PARP inhibitors DPQ (25 μM) and PJ34 (100 nM), and by minocycline (Mc, 100 nM). (Scale bar, 20 μm.) Representative of three independent studies. (B) Western blot of PAR formation in neurons harvested 15 min after a 10-min incubation with 75 μM MNNG alone or in combination with the designated concentrations of DPQ or minocycline. Control wells received medium exchanges only. PAR is seen as a discrete, major band at 116 kDa, corresponding to PAR formation on PARP-1 itself, and a smear of other bands resulting from PAR formation on other proteins. β-actin bands indicate relative protein loading in each lane. (C) Quantification of PAR Western blots. Data are means ± SEM of three independent experiments, each performed in triplicate. ∗∗, P < 0.01 compared to MNNG alone. (D) NAD+ content in neurons harvested after 30-min incubation with 75 μM MNNG. Data are means ± SEM of three independent experiments, each performed in triplicate. ∗∗, P < 0.01 compared to MNNG alone. (E) DNA damage in neurons visualized with the PANT method, conditions as in A. The increase in PANT-labeled neurons induced by MNNG was not blocked by DPQ or Mc.
Minocycline Directly Inhibits PARP-1 Enzyme Activity in Cell-Free Assay.
The observed effects of minocycline on poly(ADP-ribose) formation, NAD+ consumption, and PARP-1-mediated cell death could result from either direct or indirect effects on PARP-1 enzymatic activity in neurons. The possibility of direct inhibition was evaluated by using recombinant PARP-1 in a cell-free assay. As expected, PARP-1 activity was markedly reduced by DPQ and PJ34 (Fig. 3A). Minocycline likewise showed a dose-dependent effect on PARP-1 activity, producing a 74.4 ± 8.5% inhibition at the maximally neuroprotective dose (100 nM). Higher concentrations of minocycline produced no further reduction in PARP activity (data not shown). We additionally examined the effect of minocycline on PARP-1 enzyme activity at varying NAD+ concentrations (Fig. 3B). A double-reciprocal (Lineweaver–Burke) plot of these data (Fig. 3C) showed the inhibition to be competitive with respect to NAD+, and a Dixon plot (Fig. 3D) gave a Ki value of 13.8 ± 1.5 nM.
Fig. 3.
Minocycline inhibition of recombinant PARP-1. (A) Activity of isolated, recombinant PARP-1 was inhibited by DPQ, PJ34, and minocycline. Data from each of five independent experiments were normalized to the control (no inhibitor) synthetic rate. ∗∗, P < 0.01 vs. control. (B) Plot of PARP-1 enzyme activity at varying NAD+ concentrations in the presence of minocycline (25, 50, and 100 nM) or vehicle. A Lineweaver–Burke plot of these data (C) show the inhibition to be competitive with respect to NAD+, and a Dixon plot (D) shows the Ki value to be 13.8 ± 1.5 nM.
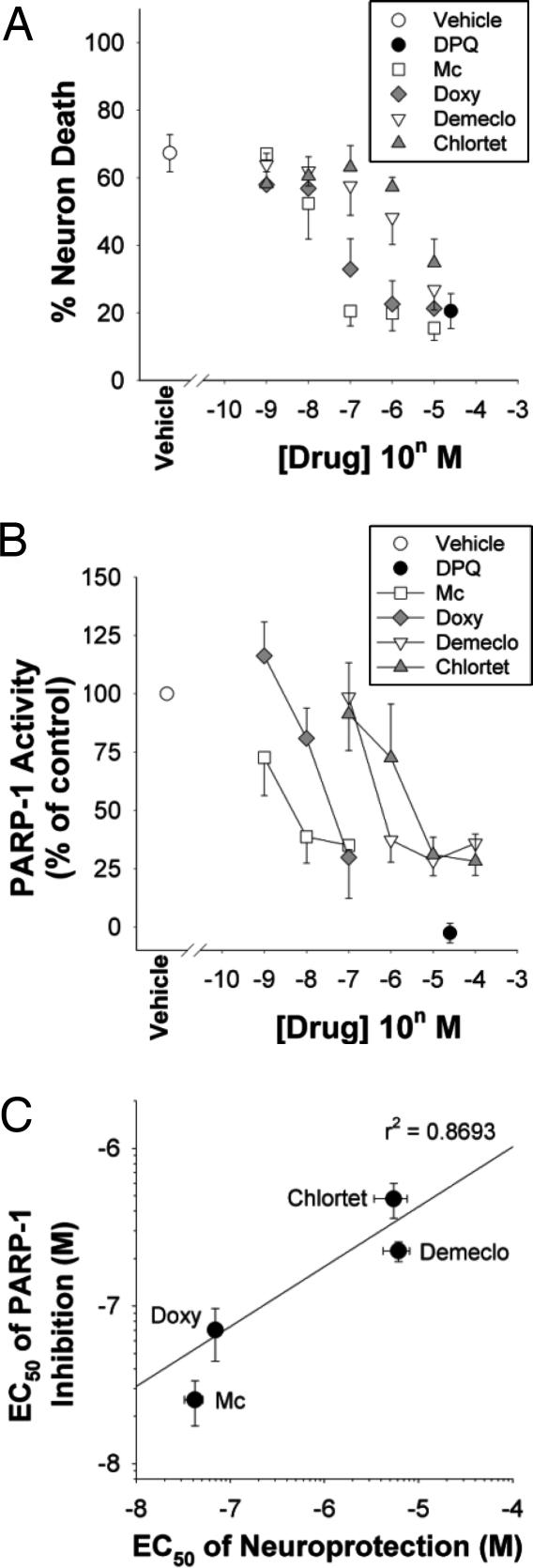
Doxycycline and other tetracycline derivatives have also been shown to have cytoprotective properties, although with somewhat lower potency than minocycline (1, 2, 33). Therefore, we examined four tetracycline derivatives to determine whether there might be a general relationship between their capacity as neuroprotective agents during genotoxic stress and their capacity to inhibit PARP-1 enzymatic activity. Tetracycline itself was neurotoxic at 5 μM and had no neuroprotective effects at concentrations lower than this (data not shown); however, each of the other four compounds examined prevented MNNG-induced neuronal death with efficacy equal to or approaching that of the PARP inhibitor DPQ (Fig. 4A). The rank order of potencies for these compounds was minocycline > doxycycline > demeclocycline > chlortetracycline. We then examined each of these compounds with respect to their potency as inhibitors of recombinant PARP-1 in a cell-free assay. The same rank order was observed (Fig. 4B), and a scatter-plot analysis showed a good correlation (r2 = 0.87) between these two measures (Fig. 4C).
Fig. 4.
Relative potency of tetracycline derivatives as neuroprotectants and PARP-1 inhibitors. (A) Neuron death evaluated 24 h after 30-min incubations with MNNG (75 μM) alone or in combination with the designated concentrations minocycline (Mino), doxycycline (Doxy), demeclocycline (Demeclo), chlortetracyceline (Chlortet), or the established PARP inhibitor DPQ. (B) Activity of isolated, recombinant PARP-1 in the presence of the same agents used in A. (C) Scatter plot showing the relative potencies of the tetracycline derivatives as neuroprotectants and PARP-1 inhibitors. Data are means ± SEM.
Discussion
These results identify a potent mechanism of minocycline neuroprotection. Using primary cultures, we first established that minocycline can protect neurons against PARP-1-mediated toxicity at submicromolar concentrations. Second, we verified the inhibitory effects of minocycline on biochemical markers of PARP-1 activation in the neuron cultures, and found that minocycline was again a highly potent inhibitor. Third, we identified a direct inhibitory effect of minocycline on PARP-1 at submicromolar concentrations in a cell-free assay. Comparison with other tetracycline derivatives suggested a general correlation between the potency of these agents as PARP-1 inhibitors and as neuroprotective agents in the setting of genotoxic stress.
The kinetic studies of PARP-1 inhibition suggest a competitive interaction between minocycline and NAD+. However, a maximal effect of minocycline was obtained at 100 nM, with higher concentrations having no further inhibitor effect. This could result from secondary interactions between minocycline (or minocycline by-products) and the histones or DNA that are also present in the reaction mixture, but our studies do not provide a clear explanation for this finding. It is notable, however, that NAD+ and minocycline share a carboxamide and aromatic ring structure (Fig. 5). A common structural feature of competitive PARP inhibitors is a carboxamide group attached to an aromatic ring or the carbamoyl group built in a polyaromatic heterocyclic skeleton (34). This structure is also present in each of the tetracycline derivatives with demonstrated PARP-1 inhibitory activity.
Fig. 5.
Structures of NAD+ and competitive PARP-1 inhibitors. An aromatic ring-linked carboxamide group (circled) or carbamoyl group built in a polyaromatic heterocyclic skeleton is shared by the natural PARP-1 substrate NAD+, the competitive PARP-1 inhibitors nicotinamide, DPQ, and PJ34, and the tetracycline derivatives. Nicotinic acid is not a PARP-1 inhibitor and differs from nicotinamide by the lack of the amide group.
Minocycline has been reported to have several other effects that could contribute to improved cell survival, but at far higher concentrations than shown here to inhibit PARP-1. These effects include up-regulation of mitochondrial bcl-2 expression at 2 μM minocycline (35); reduced mitochondrial calcium uptake, calcium-induced mitochondrial swelling, calcium-induced cytochrome-C release, and mitochondrial permeability transition at 10–100 μM (9, 36, 37); direct scavenging of reactive oxygen species at 3–100 μM (10); and inhibition of mitogen activated protein kinases at 10–100 μM minocycline (38, 39). By comparison, we found minocycline to block PARP-1-mediated neuron death at 10–100 nM in the present studies. Thus, under conditions causing PARP-1-mediated cell death, the primary mechanism of minocycline neuroprotection is likely to be PARP-1 inhibition. However, these results do not exclude the possibility that other effects of minocycline may also contribute to cell survival, particularly under conditions where cell death is not mediated primarily by PARP-1 activation.
In addition to direct effects on neuron survival, minocycline has also been shown to inhibit microglial proliferation, morphological changes, and secretory activity induced by excitotoxicity or brain ischemia (1, 4, 40, 41). These effects have been reported with minocycline concentrations of 20 nM in cell culture (40, 41). Our present studies may also be relevant to these antiinflammatory effects of minocycline, because microglial activation requires PARP-1 as a coactivator of NF-κB (14–17), and is blocked by PARP inhibitors and by PARP-1 gene deletion (18, 19, 42). Because minocycline and the other tetracycline derivatives are potent inhibitors of PARP-1, this suggests a mechanism by which these agents may exert both neuroprotective and antiinflammatory effects.
Materials and Methods
Cell culture reagents were purchased from CellGro and all other reagents were purchased from Sigma unless otherwise noted.
Neuron Cultures.
Cortical neurons were prepared from embryonic day-16 Swiss–Webster mice as described (21), under a protocol approved by the San Francisco Veterans Affaris Medical Center animal studies committee. In brief, mouse cortices were isolated, freed of meninges, dissociated with papain/DNase/trituration, and plated onto poly(d lysine)-coated glass coverslips or 24-well cell culture plates (BD Biosciences). After 1 day in culture, 10 μM cytosine arabinoside was added for 24 h to prevent glial proliferation. This medium was replaced with glial-conditioned media consisting of Eagle's MEM supplemented with 10% FBS and 2 mM glutamine that had been incubated with an cortical mouse astrocytes for 48–72 h. Culture medium was exchanged twice per week, and cultures were used at 7–10 days in vitro, at which time they contained <5% nonneuronal cells.
Drug Incubations.
Experiments were initiated by replacing the culture medium with artificial cerebrospinal fluid (aCSF) containing 3.1 mM KCl, 134 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgSO4, 0.25 mM KH2PO4, 15.7 mM NaHCO3, and 2 mM glucose (pH 7.25) equilibrated with 5% CO2 at 37°C. Osmolarity was adjusted to 290–300 mOsm monitored with a vapor pressure osmometer. Drugs were added from concentrated stocks prepared in aCSF immediately before use and adjusted to pH 7.2 when necessary. Exposures to MNNG or SIN-1 were performed at 37°C in a 5% CO2 atmosphere. DPQ (Calbiochem), PJ34 (Inotek), and tetracycline derivatives were added 10 min before the addition of MNNG or SIN-1. Incubations were terminated by washing and complete medium exchange with fresh aCSF containing DPQ, JP34, or minocycline at the same concentrations that were present during the MNNG or SIN-1 incubations.
Neuronal Death.
Neuronal death was assessed 20–24 h after the MNNG or SIN-1 incubations by the propidium iodide method (26) in which fluorescent propidium iodide enters cells with disrupted plasma membranes and binds to DNA. Nonfluorescent (live) neurons and fluorescent (dead) neurons were counted in four random fields per well, totaling at least 300 neurons per well. In some studies, neuron death was also evaluated by the lactate dehydrogenase method (43), in which the lactate dehydrogenase release corresponding to 100% neuron death was established by using prolonged MNNG incubations. The two methods gave essentially identical results.
NAD+ Measurements.
NAD+ was assayed by using an enzymatic recycling assay (44, 45) and normalized to total protein as measured by the bicinchonic acid method and BSA standards.
Immunocytochemistry.
Neurons on coverslips were fixed in trichloroacetic acid and ethanol as described (21). The fixed cells were incubated with rabbit antibody to poly(ADP-ribose) (Trevigen) at 1:2,000 dilution, followed by incubation with fluorescent anti-rabbit IgG (Molecular Probes). Nuclei were counterstained with propidium iodide, and photomicrographs were prepared with a confocal microscope.
PARP-1 Enzymatic Activity Assay.
Activity of recombinant PARP-1 was measured as described (46, 47). In brief, the reaction mixture contained 50 mM Tris·HCl (pH 8.0), 10 mM MgCl2, 10 mM DTT, 10% glycerol, 25 μg/ml calf thymus DNA, 25 μg/ml histones, 210 μM NAD+, 25 pCi 14C-NAD+ (248 mCi/mmol, Amersham Pharmacia), 4 units of purified recombinant PARP-1 (Trevigen), and tetracycline derivatives or PARP inhibitors at the designated concentrations. For Lineweaver–Burke analysis, the NAD+ concentrations were varied between 10–200 μM. After 75-min incubations, the reactions were terminated with trichloroacetic acid and the precipitated, protein-bound poly(ADP-ribose) was quantified by measurement of retained 14C.
PANT Assay for DNA Damage.
The PANT method was used to detect neurons with single strand DNA breaks, as described (48). Paraformaldehyde-fixed cultures were incubated with a reaction mixture containing 10 mM 2-mercaptoethanol, 20 μg/ml BSA, 20 μM of each dGTP, dCTP and dTTP; 1 μM of dATP, 19 μM of biotynylated dATP and 40 units/ml of DNA polymerase I. Biotinylated dATP incorporated into damaged DNA was detected by incubation with streptavidin-horseradish peroxidase and diaminobenzidine substrate. To evaluate for nonspecific labeling, some cultures were incubated with the PANT reaction mixture omitting DNA polymerase I.
Western Blots.
Western blots were prepared as described (19), using 80–100 μg of protein per lane. The membranes were probed with monoclonal anti-PARP-1 (clone 10H; Trevigen), and antibody binding was visualized with peroxidase-conjugated anti mouse IgG (Vector Laboratories) and the ECL WB Detection kit (Amersham Pharmacia). Total PAR immunoreactivity was measured in each lane from the major 116-kDa PARP-1 band to the top of the membrane, and quantified with the NIH image j program. Values for each lane were normalized to protein loading as measured by β-actin immunoreactivity, and expressed as a percent of PAR staining in the control condition prepared in each experiment. Immunoblots prepared in the absence of primary or secondary antibodies showed no signal.
Statistics.
Data are means ± SEM. Results were assessed by analysis of variance followed by the Student–Newman–Keuls post hoc test. The mean and SEM EC50 values were calculated by interpolation of the concentration–response curve resulting from each experiment using sigmaplot (Systat), with each full curve considered an “n” of 1.
Acknowledgments
We thank Drs. Stephen Massa and Gary Cecchini for helpful discussions and advice. This work was supported by the Department of Veterans Affairs (C.C.A. and R.A.S.), American Heart Association Grant 0525098Y (to T.M.K.), and National Institutes of Health Grant NS14543 (to R.A.S.).
Abbreviations
- PARP
poly(ADP-ribose) polymerase
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- DPQ
3, 4-dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone
- PJ34
N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-N,N-dimethylacetamide hydrochloride
- PANT
DNA-polymerase I-mediated biotin-dATP nick translation
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Yrjanheikki J., Keinanen R., Pellikka M., Hokfelt T., Koistinaho J. Proc. Natl. Acad. Sci. USA. 1998;95:15769–15774. doi: 10.1073/pnas.95.26.15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tikka T., Usenius T., Tenhunen M., Keinanen R., Koistinaho J. J. Neurochem. 2001;78:1409–1414. doi: 10.1046/j.1471-4159.2001.00543.x. [DOI] [PubMed] [Google Scholar]
- 3.Morimoto N., Shimazawa M., Yamashima T., Nagai H., Hara H. Brain Res. 2005;1044:8–15. doi: 10.1016/j.brainres.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 4.Yrjanheikki J., Tikka T., Keinanen R., Goldsteins G., Chan P. H., Koistinaho J. Proc. Natl. Acad. Sci. USA. 1999;96:13496–13500. doi: 10.1073/pnas.96.23.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox C., Dingman A., Derugin N., Wendland M. F., Manabat C., Ji S., Ferriero D. M., Vexler Z. S. J. Cereb. Blood Flow Metab. 2005;25:1138–1149. doi: 10.1038/sj.jcbfm.9600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu D. C., Jackson-Lewis V., Vila M., Tieu K., Teismann P., Vadseth C., Choi D. K., Ischiropoulos H., Przedborski S. J. Neurosci. 2002;22:1763–1771. doi: 10.1523/JNEUROSCI.22-05-01763.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y., Ma Z., Lin S., Dodel R. C., Gao F., Bales K. R., Triarhou L. C., Chernet E., Perry K. W., Nelson D. L., et al. Proc. Natl. Acad. Sci. USA. 2001;98:14669–14674. doi: 10.1073/pnas.251341998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M., Ona V. O., Li M., Ferrante R. J., Fink K. B., Zhu S., Bian J., Guo L., Farrell L. A., Hersch S. M., et al. Nat. Med. 2000;6:797–801. doi: 10.1038/77528. [DOI] [PubMed] [Google Scholar]
- 9.Zhu S., Stavrovskaya I. G., Drozda M., Kim B. Y., Ona V., Li M., Sarang S., Liu A. S., Hartley D. M., Wu du C., et al. Nature. 2002;417:74–78. doi: 10.1038/417074a. [DOI] [PubMed] [Google Scholar]
- 10.Kraus R. L., Pasieczny R., Lariosa-Willingham K., Turner M. S., Jiang A., Trauger J. W. J. Neurochem. 2005;94:819–827. doi: 10.1111/j.1471-4159.2005.03219.x. [DOI] [PubMed] [Google Scholar]
- 11.Virag L., Szabo C. Pharmacol. Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 12.Oei S. L., Keil C., Ziegler M. Biochem. Cell Biol. 2005;83:263–269. doi: 10.1139/o05-039. [DOI] [PubMed] [Google Scholar]
- 13.D’Amours D., Desnoyers S., D’Silva I., Poirier G. G. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 14.Ha H. C., Hester L. D., Snyder S. H. Proc. Natl. Acad. Sci. USA. 2002;99:3270–3275. doi: 10.1073/pnas.052712399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassa P. O., Hottiger M. O. Biol. Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 16.Kraus W. L., Lis J. T. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 17.Oliver F. J., Menissier-de Murcia J., Nacci C., Decker P., Andriantsitohaina R., Muller S., de la Rubia G., Stoclet J. C., de Murcia G. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullrich O., Diestel A., Eyupoglu I. Y., Nitsch R. Nat. Cell Biol. 2001;3:1035–1042. doi: 10.1038/ncb1201-1035. [DOI] [PubMed] [Google Scholar]
- 19.Kauppinen T. M., Swanson R. A. J. Immunol. 2005;174:2288–2296. doi: 10.4049/jimmunol.174.4.2288. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., Dawson V. L., Dawson T. M., Snyder S. H. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- 21.Alano C. C., Ying W., Swanson R. A. J. Biol. Chem. 2004;279:18895–18902. doi: 10.1074/jbc.M313329200. [DOI] [PubMed] [Google Scholar]
- 22.Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 23.Eliasson M. J., Sampei K., Mandir A. S., Hurn P. D., Traystman R. J., Bao J., Pieper A., Wang Z. Q., Dawson T. M., Snyder S. H., Dawson V. L. Nat. Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 24.Endres M., Wang Z. Q., Namura S., Waeber C., Moskowitz M. A. J. Cereb. Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Mandir A. S., Poitras M. F., Berliner A. R., Herring W. J., Guastella D. B., Feldman A., Poirier G. G., Wang Z. Q., Dawson T. M., Dawson V. L. J. Neurosci. 2000;20:8005–8011. doi: 10.1523/JNEUROSCI.20-21-08005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying W., Chen Y., Alano C. C., Swanson R. A. J. Cereb. Blood Flow Metab. 2002;22:774–779. doi: 10.1097/00004647-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Ha H. C., Snyder S. H. Proc. Natl. Acad. Sci. USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suto M. J., Turner W. R., Arundel-Suto C. M., Werbel L. M., Sebolt-Leopold J. S. Anticancer Drug Des. 1991;6:107–117. [PubMed] [Google Scholar]
- 29.Jagtap P., Soriano F. G., Virag L., Liaudet L., Mabley J., Szabo E., Hasko G., Marton A., Lorigados C. B., Gallyas F., Jr. Crit. Care Med. 2002;30:1071–1082. doi: 10.1097/00003246-200205000-00019. [DOI] [PubMed] [Google Scholar]
- 30.Hogg N., Darley-Usmar V. M., Wilson M. T., Moncada S. Biochem. J. 1992;281:419–424. doi: 10.1042/bj2810419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Augusto O., Gatti R. M., Radi R. Arch. Biochem. Biophys. 1994;310:118–125. doi: 10.1006/abbi.1994.1147. [DOI] [PubMed] [Google Scholar]
- 32.Pieper A. A., Blackshaw S., Clements E. E., Brat D. J., Krug D. K., White A. J., Pinto-Garcia P., Favit A., Conover J. R., Snyder S. H., Verma A. Proc. Natl. Acad. Sci. USA. 2000;97:1845–1850. doi: 10.1073/pnas.97.4.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang S. X., Lertvorachon J., Hou S. T., Konishi Y., Webster J., Mealing G., Brunette E., Tauskela J., Preston E. J. Biol. Chem. 2005;280:33811–33818. doi: 10.1074/jbc.M503113200. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J., Li J. H. In: Cell Death: the Role of PARP. Szabo C., editor. Boca Raton, FL: CRC; 2002. pp. 279–230. [Google Scholar]
- 35.Wang J., Wei Q., Wang C. Y., Hill W. D., Hess D. C., Dong Z. J. Biol. Chem. 2004;279:19948–19954. doi: 10.1074/jbc.M313629200. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Gomez F. J., Galindo M. F., Gomez-Lazaro M., Gonzalez-Garcia C., Cena V., Aguirre N., Jordan J. Neuroscience. 2005;133:959–967. doi: 10.1016/j.neuroscience.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Zhu S., Drozda M., Zhang W., Stavrovskaya I. G., Cattaneo E., Ferrante R. J., Kristal B. S., Friedlander R. M. Proc. Natl. Acad. Sci. USA. 2003;100:10483–10487. doi: 10.1073/pnas.1832501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikodemova M., Duncan I. D., Watters J. J. J. Neurochem. 2006;96:314–323. doi: 10.1111/j.1471-4159.2005.03520.x. [DOI] [PubMed] [Google Scholar]
- 39.Pi R., Li W., Lee N. T., Chan H. H., Pu Y., Chan L. N., Sucher N. J., Chang D. C., Li M., Han Y. J. Neurochem. 2004;91:1219–1230. doi: 10.1111/j.1471-4159.2004.02796.x. [DOI] [PubMed] [Google Scholar]
- 40.Tikka T. M., Koistinaho J. E. J. Immunol. 2001;166:7527–7533. doi: 10.4049/jimmunol.166.12.7527. [DOI] [PubMed] [Google Scholar]
- 41.Tikka T., Fiebich B. L., Goldsteins G., Keinanen R., Koistinaho J. J. Neurosci. 2001;21:2580–2588. doi: 10.1523/JNEUROSCI.21-08-02580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiarugi A., Moskowitz M. A. J. Neurochem. 2003;85:306–317. doi: 10.1046/j.1471-4159.2003.01684.x. [DOI] [PubMed] [Google Scholar]
- 43.Koh J. Y., Choi D. W. J. Neurosci. Methods. 1987;20:83–90. doi: 10.1016/0165-0270(87)90041-0. [DOI] [PubMed] [Google Scholar]
- 44.Szabo C., Zingarelli B., O’Connor M., Salzman A. L. Proc. Natl. Acad. Sci. USA. 1996;93:1753–1758. doi: 10.1073/pnas.93.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ying W., Sevigny M. B., Chen Y., Swanson R. A. Proc. Natl. Acad. Sci. USA. 2001;98:12227–12232. doi: 10.1073/pnas.211202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferro A. M., Olivera B. M. J. Biol. Chem. 1982;257:7808–7813. [PubMed] [Google Scholar]
- 47.Sevigny M. B., Silva J. M., Lan W. C., Alano C. C., Swanson R. A. Brain Res. Mol. Brain Res. 2003;117:213–220. doi: 10.1016/s0169-328x(03)00325-5. [DOI] [PubMed] [Google Scholar]
- 48.Chen J., Jin K., Chen M., Pei W., Kawaguchi K., Greenberg D. A., Simon R. P. J. Neurochem. 1997;69:232–245. doi: 10.1046/j.1471-4159.1997.69010232.x. [DOI] [PubMed] [Google Scholar]