Abstract
Starch is the ultimate storage molecule formed in the photosynthetic fixation of carbon dioxide by chloroplasts. Starch accumulates during the day and is degraded at night to intermediates that are exported to heterotrophic organs. The mechanism by which diurnal cycles control the transitory biosynthesis and degradation of chloroplast starch has long remained a mystery. We now report evidence that a dual-specificity protein phosphatase, DSP4, binds to starch granules during the day and dissociates at night. Disruption of the DSP4 gene resulted in a dramatic increase in the level of starch in mutant Arabidopsis plants. Moreover, although composition was apparently unchanged, the morphology of the starch granule was significantly altered compared to the wild type counterpart. Two regulatory factors linked to light (i.e., pH and redox status) changed both the activity and the starch-binding capacity of DSP4. The results further revealed that DSP4 represents a major fraction of granule-bound phosphatase activity during the day but not at night. Our study suggests that DSP4 acts as a bridge between light-induced redox changes and protein phosphorylation in the regulation of starch accumulation.
Keywords: redox regulation, starch metabolism, starch-binding phosphatase
As the ultimate storage form of photosynthetically fixed carbon, starch plays a central role in plants. During the day, chloroplasts convert carbon dioxide into soluble sugars that, with the collective action of starch synthases and branching enzymes, are converted to a mixture of linear and branched glucans (amylose and amylopectin) that determine the properties of the starch (1). At night, the starch granules are degraded by a combination of enzymes (2). The degradation products are transported out of the chloroplast and further exported to heterotrophic tissues in the form of sucrose (in most plants), thereby providing a carbon source for growth and energy. In this way, starch built up during the day and degraded at night meets the energy needs of the plant. The extent of starch synthesis and degradation in the chloroplast is tightly regulated by the light–dark cycle and by a number of environmental factors. However, little is known about the molecular mechanisms underlying the regulation of starch accumulation.
Protein phosphorylation and dephosphorylation represents a ubiquitous mechanism for regulating cellular processes (3), including the enzymes of glycogen breakdown and synthesis. Furthermore, the protein kinases and phosphatases constituting the signaling pathways regulating these latter processes are now textbook material. By contrast, information on the regulation of starch metabolism by protein phosphorylation is scant.
In an earlier effort to identify protein phosphatases in Arabidopsis that could dephosphorylate phosphotyrosine, we isolated both tyrosine-specific (4) and dual-specificity protein phosphatases (DSPs) (ref. 5 and L.N.S. and S.L., unpublished data) that may participate in a number of physiological and developmental processes (3). Several DSP members resemble laforin, the dual-specificity phosphatase that regulates glycogen accumulation in animal cells and that causes Lafora disease, a condition leading to excessive glycogen accumulation (6). The laforin-like Arabidopsis DSP members have been named DSP4, -5, and -6 (http://nature.berkeley.edu/luanlab/PTPs/List.htm). DSP4 was also known as AtPTPKIS1 as a result of its interaction with a protein kinase (7).
Based on work in animal cells, we explored the possibility that laforin-like phophatases might regulate starch metabolism in plants. We isolated a dsp4 Arabidopsis mutant that displayed a dramatic increase in the accumulation of leaf starch and observed that its gene product, DSP4, was the predominant phosphatase activity bound to chloroplast starch granules. The evidence suggests that DSP4 functions in control of starch accumulation and, itself, is regulated by light via redox and pH as documented for Calvin cycle enzymes (8). Protein phosphorylation is reported to regulate carbon fixation at the enzyme level (9) and via a thioredoxin-dependent protein kinase (10). The present results add a unique dimension in linking the regulation of starch metabolism to protein phosphorylation and redox via a single phosphatase, DSP4.
Results and Discussion
The DSP4 Protein Is Located in the Chloroplast.
The Arabidopsis genome encodes at least 25 protein tyrosine phosphatases, most belonging to the DSP subfamily. Unlike other DSP members, the laforin-like DSP4 has a putative transit peptide for chloroplast targeting according to the prediction program targetp (Fig. 1A). Therefore, we tested whether the DSP4 protein is targeted to the chloroplast by employing GFP fusion expressed in transgenic plants. The precise subcellular localization of the DSP4::GFP fusion protein was resolved with laser scanning confocal microscopy (Fig. 1C vs. Fig. 1 B and D–F). As seen in Fig. 1G, the emission spectrum recorded from the chloroplast was characteristic of GFP (peak at 543 nm) when measured from any of the green areas of the image. These studies confirmed that DSP4 is located in the chloroplast.
Fig. 1.
Chloroplast localization of DSP4-GFP. (A) Structural domains of DSP4 protein: green, predicted transit peptide; red, catalytic domain; blue – starch binding domain. (B–F) Chloroplast localization of the DSP4 protein visualized by GFP reporter protein. (B) GFP control in leaves. (C and D) Scans in the red and green channels for chlorophyll fluorescence (C) and DSP4-GFP (D). (E and F) Overlaid images showing DSP4-GFP localization in the chloroplasts at two different magnifications. Green is the true GFP fluorescence after linear unmixing (See Materials and Methods), red is chloroplast autofluorescence. (Scale bars, 5 μm in B and C and 20 μm in E and F.) (G) Emission spectrum across the green image area after linear unmixing. The major GFP emission peak is at 543 nm.
Disruption of DSP4 Enhances Starch Accumulation.
In view of the known role of laforin in regulating glycogen metabolism in animals, we used a reverse genetics approach to test the possibility that laforin-like DSP4, -5, and -6 are involved in starch metabolism in Arabidopsis. We identified T-DNA insertional mutants for each of the Arabidopsis laforin-like phosphatases and monitored the starch level in each. One of the mutants, dsp4, showed significant changes in the level of leaf starch and was, therefore, selected and characterized in detail.
Individual plants of the SALK_126784 T-DNA insertion line (Arabidopsis Biological Resource Center, University of Ohio, Columbus; ref. 11) were brought to homozygosity by selfing and identified by PCR analysis. The T-DNA in this line was inserted in the eighth intron (Fig. 2A), leading to disruption of DSP4 expression as confirmed by RT-PCR (Fig. 2B Left). A wild-type genomic fragment was transformed back into the mutant to restore the expression of DSP4 (Fig. 2B Right). The dsp4 plants were smaller, and their flowering was slightly delayed compared to the wild-type (Fig. 2C and data not shown). Similar phenotypic changes, i.e., stunted growth and delayed flowering, were described for the sex1 mutant and considered a consequence of altered starch metabolism (12).
Fig. 2.
Isolation and phenotypic analysis of dsp4 mutant. (A) DSP4 gene model with T-DNA insertion position (open triangle). (B) (Upper) DSP4 mRNA levels quantified by RT-PCR in the wild type, dsp4 mutant (35 cycles), and three complemented lines (dsp4/DSP4) (31 cycles). −, a negative control without primers. (Lower) The same cDNA amplified with Actin2 primers (25 cycles). (C) Phenotype of soil-grown wild type, dsp4/DSP4, dsp4 and sex1plants before flowering.
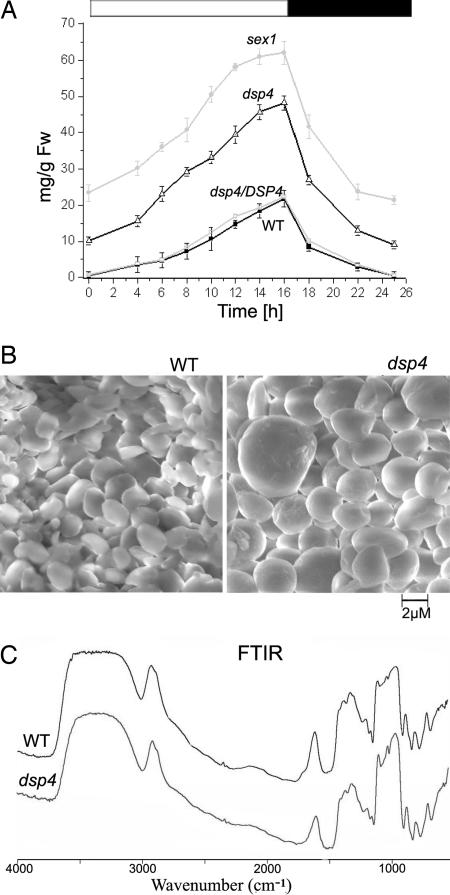
We found that, like sex1, the dsp4 mutant accumulated leaf starch to significantly higher levels than the wild-type and complemented lines. The sex1 Arabidopsis mutant is known to have a disrupted water glucan dikinase that is required for the phosphorylation and degradation of starch (13, 14). Accordingly, the sex1 mutant plants accumulated higher levels of starch than the wild type (Fig. 3A). Similarly, the dsp4 mutant accumulated up to 2.5-fold more starch than the wild type. The starch level in dsp4 followed diurnal pattern similar to the wild type: the highest level was found at the end of the light period and the lowest at the end of the dark period (Fig. 3A). The starch levels in several complemented lines were similar to wild type (data not shown). This result confirms that the changes in starch accumulation in the dsp4 mutant resulted from disruption of the DSP4 gene.
Fig. 3.
Altered starch level and granule morphology in the dsp4 mutant. (A) Starch content of wild type, dsp4, dsp4/DSP4, and sex1 throughout the diurnal cycle. Several independent complemented lines were analyzed, only one line is presented for clarity. The open bar atop the figure indicates the light period, the filled bar indicates time in darkness. Each data point represents three repeats using different batch of plants. (B) Scanning electron microscopy of the starch granules of the wild type and dsp4. (C) Fourier transform infrared absorbance pattern of the wild type and dsp4 starches. Relative absorbance is presented on the y axis.
The dsp4 Mutant Produces Starch Granules with Distinct Morphology.
The structural properties of starch granules are species-specific. A typical granule in Arabidopsis leaves is observed as a flattened disk under a scanning electron microscope (15). This morphology has been preserved in many Arabidopsis mutants showing variation in starch accumulation (i.e., sex1, sseiii), although some mutants may have altered granule diameter (15, 16). Interestingly, the dsp4 starch granules were both larger and more spherical in shape relative to wild type. We also frequently observed much larger granules, in certain cases with irregular protrusions on the surface (Fig. 3B).
A starch granule is comprised of crystalline rings laid down in concentric circles during synthesis that show a particular diffraction pattern when examined by polarized light microscopy. In our analysis, we observed that, unlike the wild type, dsp4 starch granules lacked a typical maltese cross pattern displayed in scattering polarized light (data not shown). Significant change in the shape and diffraction pattern of the mutant starch granules suggested that the rate of starch synthesis had exceeded the capacity for shaping the granule by the relevant enzymes.
We also examined the mutant starch granules for differences in composition by Fourier transform infrared spectroscopy (FTIR), a method known to elucidate molecular details of starch. Absorption in the 1,500–400 nm range provides information on the different intramolecular bonds and structures; absorption in the 4,000–1,500 wavelength range is due to various functional groups (such as -OH, C=O, and CH3). Our analyses revealed no difference in the profile of wild-type and dsp4 starch (Fig. 3C). Therefore, we concluded that the extent of starch branching and composition of the glucan polymers are similar in wild-type and mutant plants.
In Vitro Binding of DSP4 to Starch Is Regulated by pH and Redox.
The results above suggested that the DSP4 enzyme could bind to starch and, in this way, regulate other proteins associated with the synthesis or degradation of the granule. The presence in DSP4 protein of conserved amino acids essential for starch binding in laforin supports this idea (Fig. 4A). We performed an in vitro starch-binding assay and found that the DSP4 protein can, indeed, bind to starch isolated from plant sources (Fig. 4 B and C). The protein seemed to attach to starch directly as prior treatment of isolated granules with proteases had no effect on subsequent binding (data not shown). Starch binding activity of DSP4 was, however, affected by pH. Fig. 4B shows that optimal binding occurred at pH 7.5, whereas less was bound at pH 7.0 and 8.0. No binding was detected at pH 8.8 (or at pH 6.5, data not shown). As the pH of chloroplast stroma increases from the nocturnal value of about pH 7 to a pH of 8 during the day, these results suggest that DSP4 would bind more actively to starch during the day than at night.
Fig. 4.
DSP4-GST binding to starch in vitro. (A) Protein alignment of starch-binding domains of Homo sapiens laforin (NP_005661) and DSP4. Conserved residues involved in binding are highlighted. (B) Starch-binding of DSP4 and its dependence on pH. The total GST-DSP4 used in the binding assay was used as a loading control (Ctr). (C) Starch binding of GST-DSP4 in 50 mM Tris, pH 7.5 (Ctr); of catalytically inactive DSP4 (C198A); and of GST-DSP4 in presence of 1.0 mM H2O2 or 5.0 mM GSSG. “So” denotes the soluble phase (supernatant), and “St” denotes the starch pellets. Similar results were obtained with His6x-DSP4 protein and His6x antibody (not shown).
Redox is another regulatory factor associated with the day/night cycle of the chloroplast. Light reduces and thereby regulates numerous enzymes of the stroma via the ferredoxin/thioredoxin system in reactions that are reversed at night. We tested whether the binding of DSP4 to starch is affected by redox status. As seen in Fig. 4C, the addition of an oxidant, hydrogen peroxide (H2O2) or oxidized glutathione (GSSG), abolished binding of DSP4 to the starch granule. Furthermore, when the catalytic cysteine residue (CYS198) was mutated to alanine, the mutant protein lost starch-binding capacity (Fig. 4C), suggesting that this cysteine may be involved either in binding itself or in regulating binding.
DSP4 Binds to Starch During the Day and Dissociates at Night in Vivo.
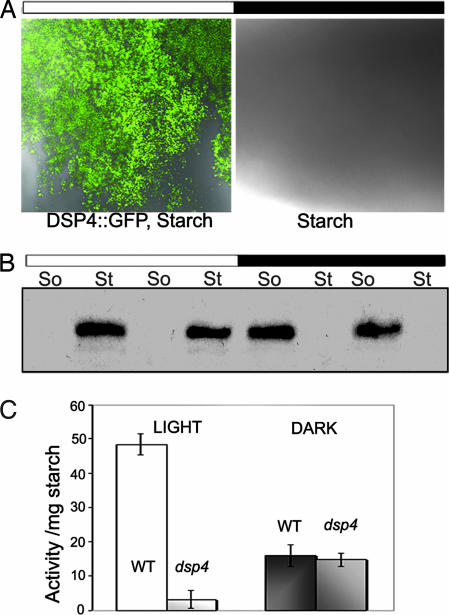
We addressed the question of whether DSP4 binds to starch in vivo using DSP4-GFP transgenic plants. Starch isolated from plants maintained under a normal (long day) diurnal cycle was examined for GFP fluorescence. DSP4-GFP was bound to granules isolated during the day but was not detected on the dark counterparts (Fig. 5A). To determine whether DSP4 protein was degraded or simply dissociated from the starch granule in the dark, we probed the level of DSP4 bound to isolated starch granules (St) vs. DSP4 in the soluble phase (So) by Western blot using a GFP antibody. The results clearly showed that DSP4 was present on the granule in the light and in the soluble phase in the dark, thus confirming the light-dependence of DSP4 binding (Fig. 5B). We found that bound DSP4-GFP dissociated within 30 min after start of the dark period (data not shown). This observation suggests that binding is modified by mechanisms closely linked to light and dark, e.g., change in stromal pH and redox status as suggested by the in vitro results (Fig. 4B).
Fig. 5.
DSP4-GFP binding to starch in vivo. The amount of DSP4-GFP on the starch granules was detected by confocal microscopy (A) or GFP antibody (B). (A) The binding of DSP4-GFP to starch isolated in the day or night. Starch granules were isolated and observed under confocal microscope. Images were obtained by two subsequent scans with blue excitation light for GFP detection and DIC for starch visualization. The figure is typical of three independent experiments. (B) The amount of starch-bound DSP4-GFP (St) or DSP4-GFP in the soluble phase (So) was analyzed by Western blots. Samples at two sequential time points in the light period (8 and 14 h) and dark period (2 and 4 h) were analyzed. At each time point, 5 mg of starch or 150–200 μg of total protein from the soluble phase were subjected to SDS/PAGE and Western blotting. (C) Starch-bound phosphatase activity in the wild type and dsp4 after 14-h illumination in the day (Light) and after 2-h in the darkness (Dark). The mean values and standard deviations of two experiments for each data point are shown.
DSP4 Is a Major Phosphatase Associated with the Starch Granule During the Day.
The above results suggest that DSP4 binds to starch and regulates its metabolism during photosynthesis. To test whether other phosphatases also associate with the starch granule, we assayed total phosphatase activity bound to starch isolated from wild type and dsp4 mutant plants during the diurnal cycle (Fig. 5C). The results indicate that disruption of the DSP4 gene in mutant plants almost completely abolished phosphatase activity bound to starch during the day. The phosphatase-bound activity at night was much lower than the daytime activity, and, furthermore, did not appear to be affected by disruption of DSP4. This result suggests that DSP4 is a major active phosphatase bound to Arabidopsis transitory starch in the day, whereas other phosphatase(s) associate with granules at night.
DSP4 Phosphatase Activity Is Redox-Regulated.
Using a recombinant DSP4, we tested whether its activity is regulated by pH and redox, factors found to affect its starch binding. Consistent with an earlier study on the tomato homologue (7), Arabidopsis DSP4 displayed high activity toward pNPP (Fig. 6A). As found for starch binding, mutation of the catalytic center cysteine (C198) abolished enzyme activity. When tested in different buffers, DSP4 displayed optimal activity at pH 7.0 with less, but significant activity at pH 7.5 and marginal activity at pH 8.0 (data not shown). Activity of the enzyme was markedly decreased on addition of H2O2 or GSSG (Fig. 6 B and C). This result is consistent with studies on other protein tyrosine phosphatases and DSPs in which it has been found that the enzyme is active only when the reaction center cysteine is in the reduced form. In the present case, redox affected not only enzymatic, but also the starch binding activity. It remains to be seen how redox and pH interact to influence these properties. It is noted that a shift in pH optimum is known to accompany the reductive activation of fructose bisphosphatase, a Calvin cycle enzyme, by thioredoxin (17).
Fig. 6.
Redox control of DSP4 activity in vitro. (A) Time course of enzyme activity (DSP4 vs.C198A mutant). (B and C) Inhibition of the DSP4 activity by GSSG (B) or H2O2 (C). The protein was fully reduced by 5 mM DTT. DTT was subsequently removed for further assays in the presence of the H2O2 or GSSG for 15 min. The plotted activity is % of the control (in the absence of oxidants). The mean values and standard deviations of three experiments for each data point are shown.
Concluding Comments.
Starch metabolism in the chloroplast is of primary importance to plants from the standpoint of carbon and energy flow. Accordingly, regulatory mechanisms that tightly regulate the biosynthesis and degradation of transitory starch would be expected to occur in photosynthetic cells. Although protein phosphorylation was discovered in the late 1950s in studies on glycogen breakdown, very little is known about the potential role of protein phosphorylation in the metabolism of starch. The present study shows that a protein phosphatase, DSP4, functions in the accumulation of starch and in remodeling the morphology of the granule. The evidence for diurnal control of the binding of DSP4 to starch granules provides a further link between protein phosphorylation and the diurnal cycle controlling starch accumulation. Additionally, based on the sensitivity of phosphatase activity to oxidants (GSSG and H2O2), DSP4 apparently serves as a single regulator to link redox and protein phosphorylation. The diurnal accumulation of starch thus seems to incorporate three regulatory elements: protein phosphorylation, redox, and pH. Redox is known to regulate other enzymes functional in starch metabolism, e.g., thioredoxin controls the activity of ADP-glucose pyrophosphorylase (18, 19), the activity and binding of α-glucan, water dikinase (SEX1) to starch granules (20), and possibly the activity of β-amylase (21). However, to our knowledge, this is the first case in which redox has been linked to an enzyme involved with protein phosphorylation for the regulation of starch metabolism.
In addition to providing new information, our study raises a number of major questions to be pursued in the future. (i) Determination of the nature of the redox system linked to DSP4 and the potential regulatory properties of conserved cysteines residing outside the enzyme’s active site. (ii) Identification of the protein(s) interacting with DSP4, i.e., enzymes of starch synthesis and breakdown as well as unknown regulatory components. (iii) Elucidation of the sequence of regulatory events. (iv) Relation of the current chloroplast results to the amyloplast and chloroplast work of Tetlow et al. (22) in which several enzymes of starch synthesis were found to be phosphorylated via an unidentified mechanism(s). Regulatory parallels between chloroplasts and amyloplasts are of interest as the organelles resemble one another with respect to their thioredoxin systems (23). (v) Functional similarities between DSP4 and laforin, an animal phosphatase that interacts with the R5 subunit of PP1 and GSK3 (24, 25), the phosphatase and kinase that regulate glycogen synthase, and the overall similarities in phosphorylation-linked events in the synthesis of glycogen and starch. (vi) A final question concerns glycogen, i.e., whether redox is also associated with glycogen metabolism in animals.
While we were in the process of submitting this manuscript, an article was published online in which some of the above findings were described (26).
Materials and Methods
Plant Materials and RNA Analysis.
The dsp4 homozygous KO line was isolated from T4 seeds of the SALK_126784 T-DNA insertion line (11) by PCR screen of genomic DNA from individual plants using the T-DNA left border and DSP4 gene specific primers. Plants were grown on 1/2 strength MS (Murashugie and Skoog) with 1× Gamborg vitamins, or in the soil in a growth room at 20–22°C with 75% relative humidity and a 16-h photoperiod, unless otherwise stated. The quantum irradiance was 170 μmol·m−2·s−1. For all experiments, wild-type and mutant plants were harvested at a similar developmental stage before flowering. The starch excess 1 (sex1–1) allele is the CS3093 ethylmethane sulfonate generated homozygous mutant line of the STARCH EXCESS 1 protein (At1g10760) in Col-0 background, and its starch content is the least affected of the sex mutants studied.
For complementation of dsp4 mutant line, the genomic DNA fragment containing 5′ promoter region and 3′ flanking sequence of the gene was cloned into pENTR (TOPO; Invitrogen) vector and subsequently transferred to pMDC99 binary vector (27) according to manufacturer’s protocol (Invitrogen). The binary plasmids were introduced into Agrobacterium tumefaciens strain containing the pGV3101. The transformed Agrobacterium strain was used to transform dsp4 plants by flower-dip method (28).
The procedure for RNA isolation and RT-PCR was described (29). Primers for RT-PCR were the same as those used for the GFP construct (see below) Actin primers were used as a control (30).
Subcellular Localization of DSP4.
Full-length DSP4 coding region was amplified with primers: DSP4cds_F CACCATGAATTGTCT-TCAGAATCTTC and DSP4cds_R AACTTCTGCCTCAGAACAAGTCTC (lacking the stop codon) and subcloned into pENTR-TOPO entry vector. GFP was fused to the C terminus of DSP4 coding region in pMDC83 and the construct was used to transform Arabidopsis plants (28, 31).
A Zeiss LSM 510 confocal microscope fitted with Meta module was used to examine the GFP fluorescence at excitation wavelengths of 488 and 543 nm simultaneously. Images were collected and recorded by using the Zeiss lsm 510 software (version 3.2). The Meta module and the linear unmixing capability of the software allow for accurate discrimination between the GFP chromophore originated fluorescence and chlorophyll green channel autofluorescence. The Meta module can detect the full spectra of multiple fluorophores simultaneously by using user-generated library of reference spectra. These spectra included the distinct spectral signature from tissue background autofluorescence and GFP control from GFP-transformed control plants.
Starch Analysis and Starch-Binding Assays.
Mature leaves were harvested at specified time points of the diurnal cycle from wild-type and mutant plants. The starch content was measured according to the procedure of Wingler et al. (30), except that after treatment with amyloglucosidase, glucose 6-phosphate was quantified as in ref. 19. Starch grains were viewed with a Phillips (Eindhoven, The Netherlands) XL30 Field Emission Gun scanning electron microscope at an acceleration potential of 10 kV as described (29) but without gold coating. A Nicolet (avator-360) Fourier transform infrared spectrometer in the range of 4,000–500 cm−1 was used to record the infrared spectra for the wild-type and dsp4 starch samples.
Starch granules for microscopic analyses were isolated as in ref. 16. Briefly, 10–15 g of plant material was homogenized with a mortar and pestle, dissolved in starch isolation buffer containing 50 mM Tris (pH 7.4), 100 mM NaCl, 0.05% Tween 20, 1 mM EDTA and protease inhibitors (1 μM leupeptin, 5 μM pepstatin, 100 units of aprotinin). The tissue lysate was filtered through two layers of Miracloth and centrifuged for 5 min at 3,000 × g (4°C). The starch pellet was solubilized in the isolation buffer and centrifuged at 9,000 × g for 10 min through 95% Percol in the same buffer. The pellet was washed repeatedly to remove any green residue.
For in vitro starch binding, washed corn starch (20 mg/ml) was incubated with 5 μg DSP4-GST or GST (glutatione S-transferase) in 50 mM Tris·HCl buffer with various pH (see Fig. 4) for 30 min at room temperature, followed by centrifugation for 10 min at 5,000 × g, 4°C. The pellet was washed in the same buffer several times and resuspended in 50 μl of buffer. Samples were subjected to SDS/PAGE and Western blot analysis with GST antibody.
Protein Expression and Phosphatase Assay.
The cDNA sequence corresponding to the mature DSP4 protein (after the putative transit peptide cleavage site) was amplified with Pfu polymerase and subcloned into a pGEX-KGatt, a Gateway recombination system tailored pGEX-KG vector (GE Healthcare, Amersham Pharmacia) with thrombin cleavage site. The resulting clones were transformed into Escherichia coli BL21 strain and DSP4-GST recombinant protein was purified as described (29). To generate the catalytic cysteine mutant, two PCR primers (DS4_C198A_F ACATATGTGCACGCCACTGCTGGAATG and DS4_C198A_R CATTCCAGCAGTGGCGTGCACATATGT) were used to replace Cys-198 (TGC) to Ala (GCC) in the DSP4 protein. The site-directed mutagenesis was performed by using QuikChange kit (Stratagene) according to the manufacturer’s instructions. The mutation was confirmed by sequencing and the mutant protein was produced as described for the wild-type protein.
For phosphatase activity assay, DSP4 protein or its mutant (100–200 ng) was incubated with 2.5 or 5 mM para-nitrophenyl phosphate (pNPP) (Sigma) in 1 ml of buffer (50 mM Tris, pH 7.0, containing 1 mM DTT) at 30°C for the times specified. The reaction was terminated by addition of NaOH. Absorbance was measured at 410 nm.
Note Added in Proof.
After this article was submitted, a paper was published that confirmed and extended evidence that chloroplast β-amylase is regulated by redox via thioredoxin (32).
Acknowledgments
We thank The Arabidopsis Biological Resource Center (Ohio State University, Columbus) for mutant seeds; Dr. Mark Curtis (University of Zürich, Zürich) for the Gateway binary vectors; and Drs. Joshua Wong, Steve Ruzin, and Gordon Vrdoljak for helpful discussions and technical assistance. This research is supported by a grant from the National Science Foundation (to S.L.).
Abbreviation
- DSP
dual-specificity protein phosphatase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Tetlow I. J., Morell M. K., Emes M. J. J. Exp. Bot. 2004;55:2131–2145. doi: 10.1093/jxb/erh248. [DOI] [PubMed] [Google Scholar]
- 2.Smith A. M., Zeeman S. C., Smith S. M. Annu. Rev. Plant Biol. 2005;56:73–98. doi: 10.1146/annurev.arplant.56.032604.144257. [DOI] [PubMed] [Google Scholar]
- 3.Luan S. Annu. Rev. Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 4.Xu Q., Fu H. H., Gupta R., Luan S. Plant Cell. 1998;10:849–857. doi: 10.1105/tpc.10.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta R., Huang Y., Kieber J., Luan S. Plant J. 1998;16:581–589. doi: 10.1046/j.1365-313x.1998.00327.x. [DOI] [PubMed] [Google Scholar]
- 6.Minassian B. A. Pediatr. Neurol. 2001;25:21–29. doi: 10.1016/s0887-8994(00)00276-9. [DOI] [PubMed] [Google Scholar]
- 7.Fordham-Skelton A. P., Chilley P., Lumbreras V., Reignoux S., Fenton T. R., Dahm C. C., Pages M., Gatehouse J. A. Plant J. 2002;29:705–715. doi: 10.1046/j.1365-313x.2002.01250.x. [DOI] [PubMed] [Google Scholar]
- 8.Buchanan B. B., Balmer Y. Annu. Rev. Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 9.Xu W., Zhou Y., Chollet R. Plant J. 2003;34:441–452. doi: 10.1046/j.1365-313x.2003.01740.x. [DOI] [PubMed] [Google Scholar]
- 10.Saze H., Ueno Y., Hisabori T., Hayashi H., Izui K. Plant Cell Physiol. 2001;42:1295–1302. doi: 10.1093/pcp/pce182. [DOI] [PubMed] [Google Scholar]
- 11.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 12.Blennow A., Nielsen T. H., Baunsgaard L., Mikkelsen R., Engelsen S. B. Trends Plant Sci. 2002;7:445–450. doi: 10.1016/s1360-1385(02)02332-4. [DOI] [PubMed] [Google Scholar]
- 13.Yu T. S., Kofler H., Hausler R. E., Hille D., Flugge U. I., Zeeman S. C., Smith A. M., Kossmann J., Lloyd J., Ritte G., et al. Plant Cell. 2001;13:1907–1918. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritte G., Lloyd J. R., Eckermann N., Rottmann A., Kossmann J., Steup M. Proc. Natl. Acad. Sci. USA. 2002;99:7166–7171. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeeman S. C., Tiessen A., Pilling E., Kato K. L., Donald A. M., Smith A. M. Plant Physiol. 2002;129:516–529. doi: 10.1104/pp.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., Myers A. M., James M. G. Plant Physiol. 2005;138:663–674. doi: 10.1104/pp.105.060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schürmann P., Wolosiuk R. A. Biochim. Biophys. Acta. 1978;522:130–138. doi: 10.1016/0005-2744(78)90329-7. [DOI] [PubMed] [Google Scholar]
- 18.Ballicora M. A., Frueauf J. B., Fu Y., Schürmann P., Preiss J. J. Biol. Chem. 2000;275:1315–1320. doi: 10.1074/jbc.275.2.1315. [DOI] [PubMed] [Google Scholar]
- 19.Hendriks J. H., Kolbe A., Gibon Y., Stitt M., Geigenberger P. Plant Physiol. 2003;133:838–849. doi: 10.1104/pp.103.024513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikkelsen R., Mutenda K. E., Mant A., Schurmann P., Blennow A. Proc. Natl. Acad. Sci. USA. 2005;102:1785–1790. doi: 10.1073/pnas.0406674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balmer Y., Koller A., del Val G., Manieri W., Schürmann P., Buchanan B. B. Proc. Natl. Acad. Sci. USA. 2003;100:370–375. doi: 10.1073/pnas.232703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tetlow I. J., Wait R., Lu Z., Akkasaeng R., Bowsher C. G., Esposito S., Kosar- Hashemi B., Morell M. K., Emes M. J. Plant Cell. 2004;16:694–708. doi: 10.1105/tpc.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balmer Y., Vensel W. H., Cai N., Manieri W., Schürmann P., Hurkman W. J., Buchanan B. B. Proc. Natl. Acad. Sci. USA. 2006;103 doi: 10.1073/pnas.0511040103. 10.1073/pnas.0511040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Sanchez M. E., Criado-Garcia O., Heath K. E., Garcia-Fojeda B., Medrano-Fernandez I., Gomez-Garre P., Sanz P., Serratosa J. M., Rodriguez de Cordoba S. Hum. Mol. Genet. 2003;12:3161–3171. doi: 10.1093/hmg/ddg340. [DOI] [PubMed] [Google Scholar]
- 25.Lohi H., Ianzano L., Zhao X. C., Chan E. M., Turnbull J., Scherer S. W., Ackerley C. A., Minassian B. A. Hum. Mol. Genet. 2005;14:2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- 26.Niittylä T., Comparot-Mossa S., Lued W.-L., Messerlie G., Trevisanf M., Seymourh M. D. J., Gatehouseh J. A., Villadseni D., Smith S. M., Chend J., et al. J. Biol. Chem. 2006;281:11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 27.Curtis M. D., Grossniklaus U. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clough S. J., Bent A. F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 29.Gupta R., Ting J. T., Sokolov L. N., Johnson S. A., Luan S. Plant Cell. 2002;14:2495–2507. doi: 10.1105/tpc.005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingler A., Fritzius T., Wiemken A., Boller T., Aeschbacher R. A. Plant Physiol. 2000;124:105–114. doi: 10.1104/pp.124.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1995. [Google Scholar]
- 32.Spara F. Costa, A., Lo Schiavo F., Pupillo P., Trost P. Plant Physiol. 2006 doi: 10.1104/pp.106.079186. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]