Abstract
Twenty-six FliF monomers assemble into the MS ring, a central motor component of the bacterial flagellum that anchors the structure in the inner membrane. Approximately 100 amino acids at the C terminus of FliF are exposed to the cytoplasm and, through the interaction with the FliG switch protein, a component of the flagellar C ring, are essential for the assembly of the motor. In this study, we have dissected the entire cytoplasmic C terminus of the Caulobacter crescentus FliF protein by high-resolution mutational analysis and studied the mutant forms with regard to the assembly, checkpoint control, and function of the flagellum. Only nine amino acids at the very C terminus of FliF are essential for flagellar assembly. Deletion or substitution of about 10 amino acids preceding the very C terminus of FliF resulted in assembly-competent but nonfunctional flagella, making these the first fliF mutations described so far with a Fla+ but Mot− phenotype. Removal of about 20 amino acids further upstream resulted in functional flagella, but cells carrying these mutations were not able to spread efficiently on semisolid agar plates. At least 61 amino acids located between the functionally relevant C terminus and the second membrane-spanning domain of FliF were not required for flagellar assembly and performance. A strict correlation was found between the ability of FliF mutant versions to assemble into a flagellum, flagellar class III gene expression, and a block in cell division. Motile suppressors could be isolated for nonmotile mutants but not for mutants lacking a flagellum. Several of these suppressor mutations were localized to the 5′ region of the fliG gene. These results provide genetic support for a model in which only a short stretch of amino acids at the immediate C terminus of FliF is required for flagellar assembly through stable interaction with the FliG switch protein.
Bacteria swim by virtue of a complex organelle, the flagellum. Motility is conferred by rotating a long filament that acts as a propeller and is connected via a flexible hook and a rod to a membrane-embedded motor (37) that uses the proton motive force as an energy source (19, 24). At least 20 proteins are assembled into the highly complex flagellar structure, and another 30 are used for its construction (23). Correct assembly and function of this macromolecular structure require complex regulation on the levels of both gene expression and protein interaction (36).
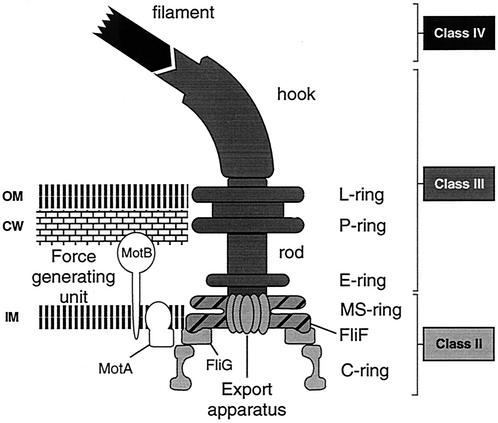
A key component of the flagellar structure is the MS ring (Fig. 1). It is the first substructure to be assembled and anchors the flagellum in the cytoplasmic membrane (17). Twenty-six FliF monomers assemble into this ring structure (9, 40, 43), forming a central pore where the integral membrane components of flagellar export are located (4, 16, 27). The MS ring also interacts with the axial extension, the rod, on the periplasmic side and with the C ring in the cytoplasm (Fig. 1) (6). The latter is required for torque generation and for the switching of flagellar rotation in response to signals from the environment (20, 42). The C ring is composed of three proteins, FliG, FliM, and FliN (6, 25, 41), and is mounted onto the membrane-integral MS ring through a direct interaction between FliG and FliF in a 1:1 stoichiometry (12, 18, 21, 25, 30). It has been shown that this interaction occurs between the C terminus of FliF and the N terminus of FliG (5, 15, 25), and deletion analysis of FliG has revealed that only the N-terminal 46 amino acids of FliG are required for interaction with FliF (15). However, for FliF, the specific requirements for FliG interaction have not been analyzed. The data presented here provided a first indication of the precise position of the FliG interaction site in the FliF C terminus.
FIG. 1.
Diagram of the C. crescentus flagellum. The flagellar structure is adapted from references 12 and 22. Flagellar components encoded by class II genes are light gray, components encoded by class III genes are dark gray, and the filament (class IV) is black. Class I is assigned to genes that are required for the activation of class II genes. The flagellar substructures are indicated on the right. Approximately 26 FliF protein subunits form the MS ring structure in the inner membrane (light gray with hatching). Rotation is conferred by proton flow through stator protein MotA of the force-generating unit in interaction with the rotor C ring protein FliG (22). Abbreviations: IM, inner membrane; CW, cell wall (peptidoglycan layer); OM, outer membrane.
Along with enteric bacteria, Caulobacter crescentus has been used as a model system by which to understand the assembly process of the flagellar structure and its regulation by the cell cycle. Both gain and loss of motility are integral parts of the C. crescentus developmental cycle, which is, in turn, intimately connected with cell proliferation (10). A flagellum is assembled in the predivisional cell at the pole opposite the stalk. As a consequence, cell division generates two distinct daughter cells, a motile, flagellated swarmer cell and a nonflagellated stalked cell. After a defined period of chemotactic activity, the swarmer cell differentiates into a stalked cell and restarts the division cycle. During this process, the flagellum is ejected into the medium. The FliF protein presumably plays a key role in both cell cycle-dependent assembly and ejection of the flagellum. FliF is expressed very early during the C. crescentus cell cycle and is specifically localized to the swarmer pole of the predivisional cell, where the flagellum is assembled (12). Since the MS ring forms in the absence of any other flagellar component (17), FliF is a good candidate by which to target flagellar assembly to the correct pole. Coincident with flagellar release, the MS ring protein is proteolytically removed, suggesting that timed removal of the MS ring anchor protein may be a crucial step in flagellar ejection (12).
As in other bacteria, expression of flagellar genes and assembly of their products into the nascent structure are tightly linked in C. crescentus. Most genes coding for flagellar components are expressed in a four-tiered regulatory cascade (classes I to IV; Fig. 1). The expression of a given class of flagellar genes requires the physical presence and correct assembly of all of the components encoded by the preceding gene class (45). Moreover, the first flagellar intermediate composed of proteins encoded by class II genes acts also as a checkpoint for cell division (13, 29, 32). Thus, mutations in fliF that prevent flagellar assembly block expression of class III and IV flagellar genes and inhibit normal cell division.
The C. crescentus FliF membrane topology predicted that most of the protein is exposed to the periplasm and that a short N-terminal part and about 100 amino acids at the C terminus are exposed to the cytoplasm (12). Removal of 26 amino acids from the C terminus not only prevented cell cycle-dependent turnover of FliF but also abolished flagellar assembly (12). This implied that the immediate C terminus of FliF accommodates both a cell cycle turnover signal and important assembly determinants. A fusion of the FliF protein lacking the C-terminal 26 amino acids (Δ5; Fig. 2) to the FliG switch protein restored flagellar assembly and motility, indicating that this portion of FliF is mainly required for the interaction with the FliG switch protein (12). The observation that the FliF protein is degraded normally in the absence of the FliG switch protein led to the conclusion that FliF-FliG interaction is not a prerequisite for FliF turnover (12). However, it is possible that the site of FliG interaction overlaps the sequence that targets FliF to the proteolytic machinery.
FIG. 2.
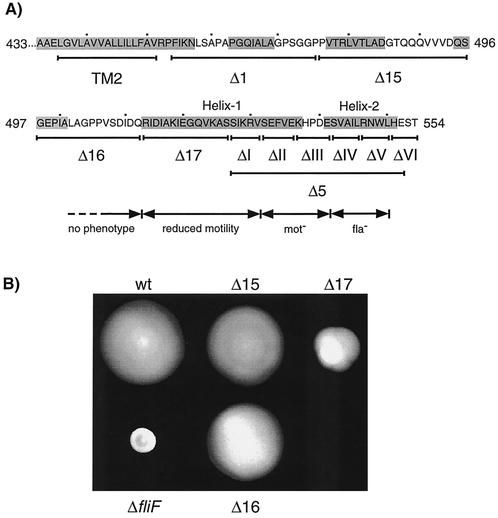
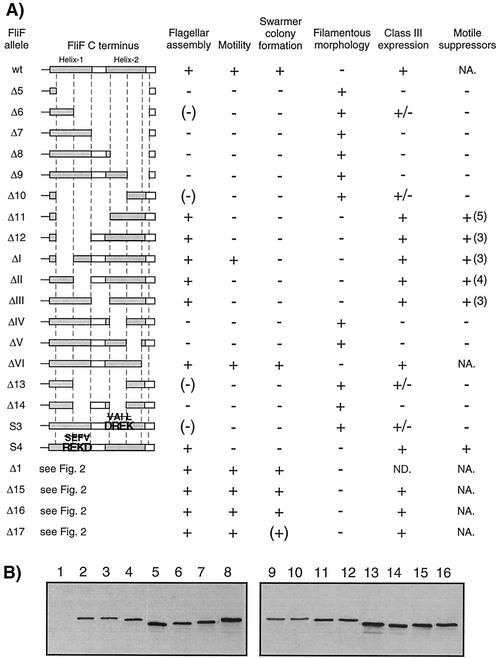
Positions of deletions in the FliF C terminus and resulting motility phenotypes. (A) Positions of in-frame deletions Δ1, Δ15, Δ16, Δ17, Δ5, ΔI, ΔII, ΔIII, ΔIV, ΔV, and ΔVI in the cytoplasmic C terminus of FliF are marked with lines below the amino acid sequence. Deletions Δ1 and Δ5 have been described before (12). The second transmembrane domain (TM2), as proposed in reference 12, is indicated. The shaded sequences are regions that were proposed to form α-helices by the secondary-structure prediction program PDHsec. The two α-helices at the immediate C terminus are referred to in the text as helices 1 and 2, and the area between them is referred to as the loop region. The numbers beside the amino acid sequence indicate positions in the FliF protein. It has been taken into account that the experimentally proven start codon of FliF is 18 codons upstream of the one predicted in the GenBank database entry (12). (B) Semisolid agar plate assay of the motility of fliF null mutant strain LS1218 (ΔfliF) complemented with a wild-type copy of fliF (wt) or with the fliF deletion allele Δ15, Δ16, or Δ17. The fliF copy used for complementation was integrated into the chromosome at the fliF locus. The swarming capacity of strain LS1218 complemented with fliF Δ1 and Δ5 has been shown in reference 12.
To understand the requirements for proteolysis and FliG interaction in more detail, we have dissected the entire cytoplasmic C terminus of FliF by mutational analysis. In this study, we analyzed the mutant versions with regard to FliG interaction and motor function. Surprisingly, only nine amino acids close to the FliF C terminus were essential for flagellar assembly. Moreover, deletions in the 29 amino acids upstream of this essential region allowed flagellar assembly but led to motility defects that were more pronounced the closer the mutations were to the region essential for assembly. Motile suppressors of these mutants mapped to both the fliF and fliG genes, suggesting that the motility phenotypes observed were at least partially due to defective FliF-FliG interaction.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Escherichia coli DH10B and S17-1 were used as host strains for molecular cloning experiments and as donor strains for conjugational transfer of plasmids into Caulobacter. E. coli strains were grown at 37°C in Luria-Bertani broth (34) supplemented with kanamycin (50 μg/ml) or tetracycline (12.5 μg/ml) when necessary. C. crescentus strains were grown at 30°C either in PYE complex medium (31) or in M2 minimal glucose medium (14) supplemented with kanamycin (5 μg/ml), tetracycline (2.5 μg/ml), or nalidixic acid (20 μg/ml) when necessary. For motility assays, cells were spotted onto soft agar plates containing 0.3% agar and left at room temperature for 1 week.
TABLE 1.
Strains used in this study
| Strain | Genotype or description | Reference, source, and/or method of construction |
|---|---|---|
| E. coli | ||
| DH10B | F−mrcA Δ(mrr hsdRMS mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara leu) 7697 araD139 galU galK nupG rpsL | GIBCO BRL |
| S17-1 | M294::RP4-2 (Tet::Mu) (Kan::Tn7) | 38 |
| C. crescentus | ||
| LS1218 | NA1000 ΔfliF | 12 |
| LS1528 | ΔfliF::fliF (wild-type control for all strains carrying fliF derivatives) | LS1218::pUJ70; 12 |
| LS1944 | ΔfliF/pfliFΔ1360-1425 (FliFΔ454-475 Δ1) | LS1218/pUJ99; 12 |
| NA1000 | Synchronizable holdfast mutant derivative of wild-type strain CB15 | 3 |
| UJ355 | ΔfliF::fliFΔ1579-1593 (FliFΔ527-531 ΔI) | LS1218::pBG1 |
| UJ356 | ΔfliF::fliFΔ1594-1608 (FliFΔ532-536 ΔII) | LS1218::pBG2 |
| UJ357 | ΔfliF::fliFΔ1609-1623 (FliFΔ537-541 ΔIII) | LS1218::pBG3 |
| UJ358 | ΔfliF::fliFΔ1624-1638 (FliFΔ542-546 ΔIV) | LS1218::pBG4 |
| UJ359 | ΔfliF::fliFΔ1639-1650 (FliFΔ547-550 ΔV) | LS1218::pBG5 |
| UJ360 | ΔfliF::fliFΔ1651-1662 (FliFΔ551-554 ΔVI) | LS1218::pBG6 |
| UJ434 | ΔfliF::fliFΔ1579-1656 (FliFΔ527-552 Δ5) | LS1218::pBG7 |
| UJ553 | ΔfliF::fliFΔ1594-1656 (FliFΔ532-552 Δ6) | LS1218::pBG8 |
| UJ554 | ΔfliF::fliFΔ1609-1656 (FliFΔ537-552 Δ7) | LS1218::pBG9 |
| UJ555 | ΔfliF::fliFΔ1624-1656 (FliFΔ542-552 Δ8) | LS1218::pBG10 |
| UJ556 | ΔfliF::fliFΔ1639-1656 (FliFΔ547-552 Δ9) | LS1218::pBG11 |
| UJ857 | ΔfliF::fliFΔ1579-1638 (FliFΔ527-546 Δ10) | LS1218::pSG1 |
| UJ858 | ΔfliF::fliFΔ1579-1623 (FliFΔ527-541 Δ11) | LS1218::pSG2 |
| UJ859 | ΔfliF::fliFΔ1579-1608 (FliFΔ527-536 Δ12) | LS1218::pSG3 |
| UJ881 | Mot− suppressor of UJ355 | This study |
| UJ882 | Mot− suppressor of UJ355 | This study |
| UJ883 | Mot− suppressor of UJ355 | This study |
| UJ887 | Mot− suppressor of UJ356 | This study |
| UJ970 | ΔfliF::fliFΔ1594-1638 (FliFΔ532-546 Δ13) | LS1218::pSG4 |
| UJ971 | ΔfliF::fliFΔ1594-1608, 1624-1638 (FliFΔ532-536, 542-546 Δ14) | LS1218::pSG5 |
| UJ974 | ΔfliF::fliF-S3 (FliFV543D A544R I545E L546K) | LS1218::pSG8 |
| UJ975 | ΔfliF::fliF-S4 (FliFS532R F534K V535D) | LS1218::pSG9 |
| UJ1077 | ΔfliF::fliFΔ1426-1488 (FliFΔ476-496 Δ15) | LS1218::pSG10 |
| UJ1078 | ΔfliF::fliFΔ1489-1536 (FliFΔ497-512 Δ16) | LS1218::pSG11 |
| UJ1079 | ΔfliF::fliFΔ1537-1578 (FliFΔ513-526 Δ17) | LS1218::pSG12 |
| UJ1236 | NA1000 Δ(fliF-fliG) | This study |
| UJ1471 | Mot− suppressor of UJ858 | This study |
| UJ1479 | Mot− suppressor of UJ975 | This study |
| UJ1480 | Mot− suppressor of UJ975 | This study |
| UJ1482 | Mot− suppressor of UJ975 | This study |
| UJ1512 | Δ(fliF-fliG)/p(fliF-fliG)UJ881 | UJ1236/pBG38 |
| UJ1513 | Δ(fliF-fliG)/p(fliF-fliG)UJ882 | UJ1236/pBG39 |
| UJ1514 | Δ(fliF-fliG)/p(fliF-fliG)UJ883 | UJ1236/pBG40 |
| UJ1518 | Δ(fliF-fliG)/p(fliF-fliG)UJ887 | UJ1236/pBG44 |
| UJ1522 | Δ(fliF-fliG)/p(fliF-fliG)LS1528 | UJ1236/pBG60 |
| UJ1523 | Δ(fliF-fliG)/p(fliF-fliG)UJ1471 | UJ1236/pBG48 |
| UJ1531 | Δ(fliF-fliG)/p(fliF-fliG)UJ1479 | UJ1236/pBG56 |
| UJ1532 | Δ(fliF-fliG)/p(fliF-fliG)UJ1480 | UJ1236/pBG57 |
| UJ1534 | Δ(fliF-fliG)/p(fliF-fliG)UJ1482 | UJ1236/pBG59 |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pBG1 | fliFΔ1579-1593 (ΔI) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG2 | fliFΔ1594-1608 (ΔII) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG3 | fliFΔ1609-1623 (ΔIII) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG4 | fliFΔ1624-1638 (ΔIV) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG5 | fliFΔ1639-1650 (ΔV) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG6 | fliFΔ1651-1662 (ΔVI) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG7 | fliFΔ1579-1656 (Δ5) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG8 | fliFΔ1594-1656 (Δ6) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG9 | fliFΔ1609-1656 (Δ7) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG10 | fliFΔ1624-1656 (Δ8) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG11 | fliFΔ1639-1656 (Δ9) in pBGS18T as BamHI-EcoRI fragment | This study |
| pBG22 | Sequence flanking Δ(fliF-fliG) in pNPTS128 as BamHI-EcoRI fragment | This study |
| pBG38 | (fliF-fliG)UJ881 in pMR10 as BamHI-EcoRV fragment | This study |
| pBG39 | (fliF-fliG)UJ882 in pMR10 as BamHI-EcoRV fragment | This study |
| pBG40 | (fliF-fliG)UJ883 in pMR10 as BamHI-EcoRV fragment | This study |
| pBG44 | (fliF-fliG)UJ887 in pMR10 as BamHI-EcoRV fragment | This study |
| pBG48 | (fliF-fliG)UJ1471 in pMR10 as BamHI-EcoRV fragment | This study |
| pBG56 | (fliF-fliG)UJ1479 in pMR10 as BamHI-EcoRV fragment | This study |
| pBG57 | (fliF-fliG)UJ1480 in pMR10 as BamHI-EcoRV fragment | This study |
| pBG59 | (fliF-fliG)UJ1482 in pMR10 as BamHI-EcoRV fragment | This study |
| pBG60 | (fliF-fliG)LS1528 in pMR10 as BamHI-EcoRV fragment | This study |
| pBGS18T | Kanr derivative of pUC18 with oriT (suicide vector in C. crescentus) | M. R. K. Alley |
| pCM4 | flgF promoter in pRKlac290 as HindIII/EcoRI fragment | 28 |
| pMR10 | Kanr low copy number, broad-host-range vector | C. Mohr and R. Roberts |
| pNPTS128 | Kanr derivative of pLITMUS28 with sacB and oriT | M. R. K. Alley |
| pRKlac290 | RK2-based lacZ transcriptional fusion vector (Tetr) | 7 |
| pSG1 | fliFΔ1579-1638 (Δ10) in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG2 | fliFΔ1579-1623 (Δ11) in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG3 | fliFΔ1579-1608 (Δ12) in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG4 | fliFΔ1594-1638 (Δ13) in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG5 | fliFΔ(1594-1608 1624-1638) (Δ14) in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG8 | fliF S3 in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG9 | fliF S4 in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG10 | fliFΔ1426-1488 (Δ15) in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG11 | fliFΔ1489-1536 (Δ16) in pBGS18T as BamHI-EcoRI fragment | This study |
| pSG12 | fliFΔ1537-1578 (Δ17) in pBGS18T as BamHI-EcoRI fragment | This study |
| pUJ70 | fliF in pBGS18T as BamHI-EcoRI fragment | 12 |
DNA manipulation techniques and construction of fliF mutant alleles.
Standard cloning and PCR protocols were used (2, 34). All of the PCR products used for cloning were amplified with high-fidelity Pfu polymerase (Stratagene). All fliF mutant alleles were generated by two-step PCR (GeneSOEing) (44) with pUJ70 as the template, primer #157 (5′-GCC GTC ACC AAC TAC GAG-3′) and the reverse primer (5′-GTC AGC GAC ATC GAC CAG-3′) as flanking primers, and the following mutagenesis primers: #158 (5′-CGA CGC CTT CAC CTG ACC CTC G-3′) and #159 (5′-GGT CAG GTG AAG GCG TCG TCC GAG TTT GTC GAG AAG-3′) for fliFΔ1579-1593 (ΔI), #172 (5′-CAC GCG TTT GAT CGA CGA GGC C-3′) and #173 (5′-CGT CGA TCA AAC GCG TGA AGC ATC CCG ACG AGT C-3′) for fliFΔ1594-1608 (ΔII), #160 (5′-GCG ACG GAC TCG ACA AAC TCG GAC AC-3′) and #161 (5′-GTT TGT CGA GTC CGT CGC GAT CC-3′) for fliFΔ1609-1623 (ΔIII), #174 (5′-CTC GTG CAG CCA GTT ACG CTC GTC GGG ATG CTT CTC-3′) and #175 (5′-CGT AAC TGG CTG CAC GAG-3′) for fliFΔ1624-1638 (ΔIV), #176 (5′-CCA TCA GGT GGA CTC GTG CAG GAT CGC GAC GGA CT-3′) and #177 (5′-CAC GAG TCC ACC TGA TGG CTA TGA-3′) for fliFΔ1639-1650 (ΔV), #178 (5′-CGA GCT TCA TAG CCA TCA CAG CCA GTT ACG CAG GAT-3′) and #179 (5′-TGA TGG CTA TGA AGC TCG-3′) for fliFΔ1651-1662 (ΔVI), #189 (5′-TCA TAG CCA TCA GGT CGA CGA GGC CTT CAC CT-3′) and #190 (5′-TCG ACC TGA TGG CTA TGA-3′) for fliFΔ1579-1656 (Δ5), #220 (5′-TCA TAG CCA TCA GGT CGA CAC GCG CTT GAT CGA C-3′) and #190 (see above) for fliFΔ1594-1656 (Δ6), #221 (5′-TCA TAG CCA TCA GGT CGA CTC GAC AAA CTC GGA CAC-3′) and #190 (see above) for fliFΔ1609-1656 (Δ7), #222 (5′-TCA TAG CCA TCA GGT CGA CTC GTC GGG ATG CTT C-3′) and #190 (see above) for fliFΔ1624-1656 (Δ8), #223 (5′-TCA TAG CCA TCA GGT CGA CAG GAT CGC GAC GGA CTC-3′) and #190 (see above) for fliFΔ1639-1656 (Δ9), #282 (5′-GGT CAG GTG AAG GCC TCG CGT AAC TGG CTG CAC GA-3′) and #281 (5′-CGA GGC CTT CAC CTG ACC-3′) for fliFΔ1579-1638 (Δ10), #283 (5′-GGT CAG GTG AAG GCC TCG TCC GTC GCG ATC CTG CG-3′) and #281 (see above) for fliFΔ1579-1623 (Δ11), #284 (5′-GGT CAG GTG AAG GCC TCG AAG CAT CCC GAC GAG TC-3′) and #281 (see above) for fliFΔ1579-1608 (Δ12), #310 (5′-CTC GTG CAG CCA GTT ACG CAC GCG CTT GAT CGA CG-3′) and #175 (see above) for fliFΔ1594-1638 (Δ13), #311 (5′-CGT CGA TCA AGC GCG TGA AGC ATC CCG ACG AGC G-3′) and #312 (5′-CAC GCG CTT GAT CGA CG-3′) for fliFΔ(1594-1608 1624-1638) (Δ14), #317 (5′-GTC CGA CCG GGA GAA GCG TAA CTG GCT GCA CGA G-3′) and #318 (5′-TAC GCT TCT CCC GGT CGG ACT CGT CGG GAT GCT TC-3′) for fliF S3, #320 (5′-CTC GTC CTT CTC CCG CAC GCG CTT GAT CGA CGA G-3′) and #347 (5′-CGC GTG CGG GAG AAG GAC GAG AAG CAT CCC-3′) for fliF S4, #353 (5′-GGT CCT TCG GGC GGT CCG GGT GAA CCG ATC GCG C-3′) and #374 (5′-CGG ACC GCC CGA AGG ACC-3′) for fliFΔ1426-1488 (Δ15), #355 (5′-GTG GTG GTC GAT CAG TCC CGT ATC GAC ATC GCC AAG-3′) and #375 (5′-GGA CTG ATC GAC CAC CAC-3′) for fliFΔ1489-1536 (Δ16), and #358 (5′-GTC AGC GAC ATC GAC CAG TCG ATC AAG CGC GTG TC-3′) and #376 (5′-CTG GTC GAT GTC GCT GAC-3′) for fliFΔ1537-1578 (Δ17). The BstEII-EcoRI fragment of the PCR products was used to replace the equivalent wild-type fragment of pUJ70 to generate the plasmids listed in Table 2. The integrity of cloned PCR fragments was confirmed by DNA sequencing by the dideoxy-chain termination method (35) with an ABI Prism 310 automatic sequence analyzer (Perkin-Elmer). The plasmids were transferred into C. crescentus strain LS1218 by conjugation and integrated into the chromosome by homologous recombination. The correct site of integration was confirmed by PCR.
Microscopy techniques.
Cell morphology and swimming was observed by light microscopy with a Nikon Labphot-2 or an Olympus AX70 microscope. Pictures were taken with a charge-coupled device camera (Hamamatsu C4742-95) connected to the Olympus microscope. Flagellar assembly and structure were investigated by electron microscopy. Exponentially growing cells in M2 minimal glucose medium were concentrated 10 times and fixed with negative stain as described before (1). Pictures were taken with a Philips 401 electron microscope.
Class III promoter activity.
The activity of a flagellar class III promoter was determined in the various fliF mutant backgrounds by introducing plasmid pCM4 into all mutant strains. This plasmid carries a transcriptional fusion of the class III flgF promoter with the reporter gene lacZ. Cells were grown exponentially in PYE complex medium to an optical density at 660 nm of 0.3 to 0.6, and β-galactosidase activity was determined as described previously (26). The experiment was repeated four times for each strain. In each series of experiments, flgF promoter activity was measured independently in the NA1000 wild-type background and the β-galactosidase activities of the mutant strains were indicated as relative to that of the wild type.
Construction of a chromosomal fliF-fliG deletion.
An in-frame deletion of the fliF and fliG genes was generated by a two-step PCR with the universal primer 5′-TGT AAA ACG ACG GCC AGT-3′ and primer #392 (5′-CCA GTC GAA AGT GAA GGC-3′) as flanking primers, mutagenesis primers #390 (5′-ATA GAG CAG CTC ACT GGG-3′) and #391 (5′-CCC AGT GAG CTG CTC TAT TGA GGG GCA TGC GAT GAC-3′), and genomic DNA of strain NA1000 as the template. The PCR fragment was digested with BamHI and NcoI and subcloned into the same sites of pNPTS128, resulting in plasmid pBG22. This plasmid was used to delete the chromosomal wild-type copies of fliF and fliG via a two-step recombination procedure (11). Loss of the fliF-fliG locus in the resulting strain, UJ1236, was confirmed by PCR and DNA sequence analysis. Motility was fully restored by plasmid pBG60, which contains a wild-type copy of the fliF-fliG locus.
Characterization of Mot− suppressors.
Motile suppressors of nonmotile strains were obtained by isolating cells from swarms protruding from compact colonies on soft agar plates. To determine if the suppressor mutation was in either fliF or fliG, the locus containing the endogenous promoter and both genes was amplified by PCR from genomic DNA of the suppressor strain by using the universal primer (see above) and primer #496 (5′-GCG AAC TTG CGG TGA GGG-3′). The PCR product was digested with BamHI-EcoRV and cloned into the same sites of pMR10, generating the plasmids listed in Table 3. Five independent ligation products were transferred into UJ1236 by conjugation and tested for motility by microscopy. If at least one of the resulting strains was motile, the suppressor mutation was considered to be in fliF or fliG. Point mutations in fliF or fliG were mapped by DNA sequence analysis.
TABLE 3.
Summary of Mot− suppressor study
| Mot+ suppressor strain | Original mot mutant | Complemented Δ(fliF-fliG) strain | Plasmid used for complementation | Mutation |
|---|---|---|---|---|
| UJ881 | UJ355 (ΔI) | UJ1512 | pBG38 | FliF S526W |
| UJ882 | UJ355 (ΔI) | UJ1513 | pBG39 | FliF S526L |
| UJ883 | UJ355 (ΔI) | UJ1514 | pBG40 | FliF S526L |
| UJ887 | UJ356 (ΔII) | UJ1518 | pBG44 | FliG M78V |
| UJ1471 | UJ858 (Δ11) | UJ1523 | pBG48 | FliG V68M |
| UJ1479 | UJ975 (S4) | UJ1531 | pBG56 | FliG M71K |
| UJ1480 | UJ975 (S4) | UJ1532 | pBG57 | FliF D535V |
| UJ1482 | UJ975 (S4) | UJ1534 | pBG59 | FliF D535V |
RESULTS
Only a short core region at the immediate C terminus of FliF is required for flagellar assembly and function.
Previous work had shown that deletion of 26 amino acids at the C-terminal end of FliF (Δ5; Fig. 2A) abolished flagellar assembly, while a deletion of 22 amino acids immediately following the second transmembrane domain of FliF (Δ1; Fig. 2A) had no effect on flagellar assembly or function (12). These phenotypes had been observed for plasmid-borne copies of the fliF mutant alleles. To make sure that the Fla− phenotype was not the result of an increased copy number of the fliF Δ5 allele, we introduced this allele into the chromosomal fliF locus of ΔfliF strain LS1218. In agreement with the phenotype described earlier, the resulting strain, UJ434, was unable to swim (data not shown), formed tight colonies on soft agar plates (data not shown), had a filamentous morphology (see Fig. 5), was unable to assemble a flagellum (data not shown), and had no class III promoter activity (see Fig. 7). To determine if other parts of the C-terminal cytoplasmic domain of FliF are required for flagellar assembly or function, three additional deletion alleles (Δ15 to Δ17) were constructed and introduced into strain LS1218 (Fig. 2A). Light and electron microscopy analysis revealed that the resulting mutant strains, UJ1077 to UJ1079, had normal cell morphology (data not shown), were able to swim (data not shown), assembled polar flagella (data not shown), and had wild-type class III promoter activity (see Fig. 7). As expected, all three mutant strains formed swarmer colonies on soft agar plates. However, while strains UJ1077 (Δ15) and UJ1078 (Δ16) formed wild-type-size colonies, strain UJ1079 (Δ17) formed a swarmer colony of intermediate size (Fig. 2B). This suggested that the entire cytoplasmic region upstream of Δ5 is dispensable for flagellar assembly while mutations close to this core region involved in flagellar assembly affect motor performance.
FIG. 5.
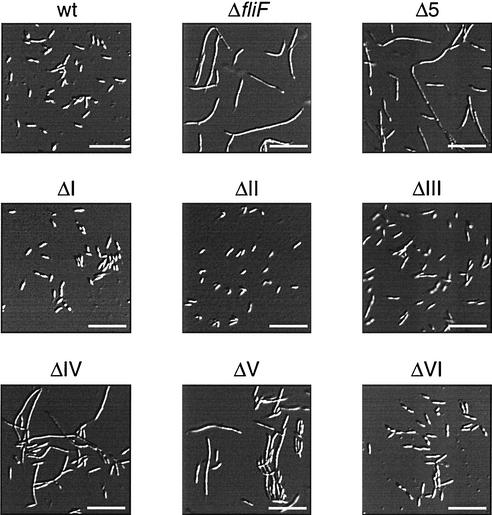
Light microscopy analysis of fliF null mutant strain LS1218 (ΔfliF) complemented with either a wild-type copy of fliF (wt) or a copy of fliF deletion allele ΔI, ΔII, ΔIII, ΔIV, ΔV, or ΔVI. Bars, 10 μm.
FIG. 7.
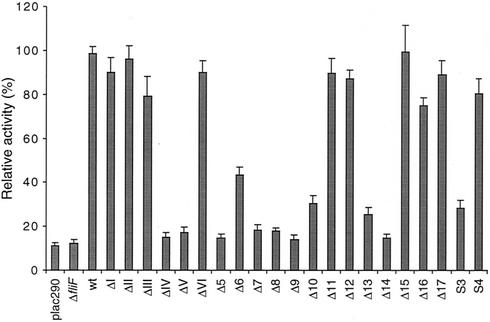
Relative activity of the flagellar class III flgF promoter in C. crescentus fliF null mutant strain LS1218 (ΔfliF) complemented with various fliF mutant alleles. Plasmid pCM4, which contains a transcriptional fusion of the flgF promoter with the reporter gene lacZ was introduced into all mutant strains. Promoter activity was determined by measuring β-galactosidase activity. The fliF alleles are indicated below each bar. Background β-galactosidase activity was determined by using the vector control plac290 in strain NA1000. The height of each bar represents the relative promoter activity of the corresponding mutant strain compared to the activity of the C. crescentus NA1000 wild-type (wt) strain. The error bars represent the standard errors.
High-resolution mutational analysis of the immediate C terminus of FliF.
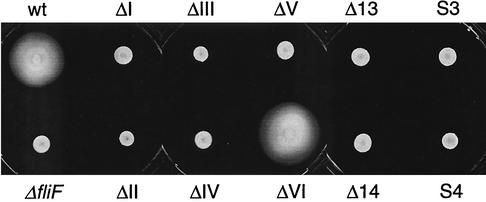
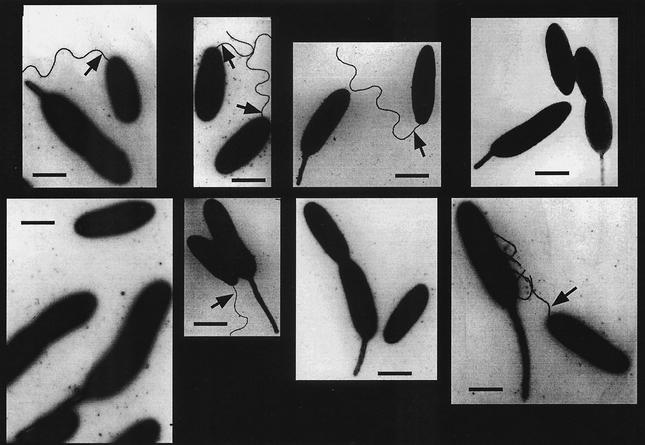
No structural information is available for the C terminus of C. crescentus FliF or any of its homologues. Secondary-structure prediction analysis (PDHsec at http://dodo.cpmc.columbia.edu/predictprotein) (33) proposed the expected α-helix in the second membrane-spanning domain, as well as long and short α-helices (helices 1 and 2, respectively) in the last 42 amino acids of the FliF C terminus that are separated by a short loop with a central proline residue (Fig. 2A). To define the requirements of helices 1 and 2 for flagellar function and assembly in more detail, fliF mutant alleles were constructed with short successive deletions covering the complete Δ5 region (ΔI to ΔVI; Fig. 2 and 3). Light microscopy analysis revealed that only mutants UJ355 (ΔI) and UJ360 (ΔVI) were able to swim, while mutants UJ356 to UJ359 (ΔII to ΔV) were completely paralyzed (Fig. 3A). The swimming behavior of mutant strain UJ360 (ΔVI) was indistinguishable from that of the wild type. In contrast, mutant strain UJ355 (ΔI) showed reduced swimming activity, with fewer swimming cells and reduced swimming speed. In good agreement with this observation, UJ355 (ΔI) formed compact colonies on soft agar plates, similar to completely nonmotile mutant strains UJ356 to UJ359 (ΔII to ΔV) (Fig. 4). Mutant strain UJ360 (ΔVI), on the other hand, showed no swimming deficiency and formed a swarmer colony of wild-type size (Fig. 4). Apart from their different motility behaviors, the deletion mutants had striking differences in cell morphology: While strains UJ355 (ΔI), UJ356 (ΔII), UJ357 (ΔIII) and UJ360 (ΔVI) had normal cell length, strains UJ358 (ΔIV) and UJ359 (ΔV) were very filamentous, indicating a cell division defect. This is similar to the cell division phenotype of fliF null mutant strain LS1218 and of strain UJ434, which lacks the entire Δ5 region (Fig. 5). Analysis of the mutant strains by electron microscopy revealed that strains UJ355 (ΔI), UJ356 (ΔII), UJ357 (ΔIII), and UJ360 (ΔVI) were able to assemble a flagellar structure (Fig. 6). All of these strains had flagella that were localized properly to the swarmer cell pole and were ejected normally during the swarmer-to-stalked cell transition. In contrast, no flagella were found in mutant strains UJ358 (ΔIV) and UJ359 (ΔV) (Fig. 6). In all of the mutant strains, flagellar assembly correlated with class III promoter expression. Mutants UJ355 (ΔI), UJ356 (ΔII), UJ357 (ΔIII), and UJ360 (ΔVI) had normal class III promoter activity, whereas only background promoter activity was measured for UJ358 (ΔIV) and UJ359 (ΔV) (Fig. 7). In summary, these data suggested that only a short region in the FliF C-terminal domain covered by deletions ΔIV and ΔV and corresponding to putative α-helix 2 is essential for flagellar assembly. The stretch of amino acids covered by deletions ΔI to ΔIII, while not being required for assembly, seems to be important for flagellar rotation. Deletion of the last four amino acids at the C terminus (ΔVI) had no effect on either assembly or function. Finally, a strong correlation was observed among failure of flagellar assembly, class III flagellar gene expression, and filamentous cell morphology.
FIG. 3.
Summary of all mutations in the cytoplasmic C terminus of FliF and of the corresponding phenotypic analysis described in this study. (A) A schematic diagram of the last 30 amino acids of the C terminus is shown on the left. The shaded areas are the two regions that were proposed to form α-helices by the secondary-structure prediction program PDHsec. The area between the two predicted α-helices is referred to as the loop region in the text. The corresponding mutant names are on the left. Flagellar assembly: flagella detected by electron microscopy (see Fig. 6). A plus indicates that most of the swarmer cells were flagellated, a minus indicates that no swarmer cells were flagellated, and a minus in parentheses indicates that very few swarmer cells were flagellated. Motility: cell motility determined by light microscopy. A plus indicates that swimming cells were observed, and a minus indicates that no swimming cells were present. Swarmer colony formation: motility assayed on semisolid agar plates. A plus indicates formation of a large swarmer colony of wild-type size, a minus indicates a small, compact colony, and a plus in parentheses indicates a swarmer colony of intermediate size (see Fig. 2 and 4). Filamentous morphology: cell morphology determined by light microscopy (see Fig. 5). A plus indicates that the cells were filamentous, and a minus indicates that the cells had wild-type morphology. Class III expression: activity of the flgF flagellar class III promoter (see Fig. 7). A plus indicates wild-type promoter activity, a minus indicates background promoter activity, and a plus-and-minus sign indicates intermediate promoter activity. Motile suppressors: isolation of motile suppressors from nonmotile mutants. A plus indicates that motile suppressors could be isolated, and a minus indicates that no motile suppressors were found. The numbers in parentheses are the numbers of independent suppressor strains isolated. Abbreviations: ND, not determined; NA, not applicable. (B) Immunoblot analysis of strains expressing FliF mutant forms using an anti-FliF antibody. Extracts (normalized for cell numbers) of the following strains were used: lane 1, LS1218 (ΔfliF); lane 2, NA1000 (wild type); lane 3, LS1528 (wild-type fliF); lane 4, UJ556 (fliF Δ9); lane 5, UJ857 (fliF Δ10); lane 6, UJ970 (fliF Δ13); lane 7, UJ971 (fliF Δ14); lane 8, UJ974 (fliF S3); lane 9, LS1218 (ΔfliF); lane 10, NA1000 (wild type); lane 11, UJ358 (fliF ΔIV); lane 12, UJ359 (fliF ΔV); lane 13, UJ553 (fliF Δ6); lane 14, UJ434 (fliF Δ5); lane 15, UJ554 (fliF Δ7); lane 16, UJ555 (fliF Δ8).
FIG. 4.
Semisolid agar plate assay used to determine the swimming capacity of fliF null mutant strain LS1218 (ΔfliF) complemented with either a wild-type copy of fliF (wt) or a copy of fliF deletion allele ΔI, ΔII, ΔIII, ΔIV, ΔV, or ΔVI. Motile strains form a large swarmer colony, and nonmotile strains form a small, compact colony.
FIG. 6.
Electron microscopy analysis of fliF null mutant strain LS1218 (ΔfliF) complemented with either a wild-type copy of fliF (wt) or a copy of fliF deletion allele ΔI, ΔII, ΔIII, ΔIV, ΔV, or ΔVI. Arrows indicate flagellar structures. The contrast of the flagella was manually enhanced. Bars, 1 μm.
To confirm that putative α-helix 2 is the only region of the FliF C terminus required for flagellar assembly, deletion Δ5 was gradually shortened toward either end (Δ6 to Δ12; Fig. 3A) and deletions around the turn region between helices 1 and 2 (Δ13 and Δ14) were constructed. The phenotypic analysis of these mutant strains is schematically summarized in Fig. 3A. Strains UJ553 to UJ556 (Δ6 to Δ9), in which fliF lacked the second half of the helix 2 coding region, were unable to swim, were deficient in flagellar assembly, and showed a filamentous cell morphology (Fig. 3A). While most UJ553 (Δ6) cells had no flagellum, a few flagellated cells were reproducibly detected by electron microscopy (data not shown). In good agreement with this, an intermediate class III promoter activity was measured for this strain (Fig. 7). It is not clear if this is due to frequent spontaneous fla+ mutations appearing in this genetic background or if the FliF Δ6 protein is able to partially interact with other motor components and assemble into nonfunctional flagellar structures. No flagella were observed for mutant strains UJ554 to UJ556 (Δ7 to Δ9), and class III promoter activity in these strains was comparable to background levels (Fig. 7).
Deletions Δ10, Δ11, and Δ12 were shortened from the FliF C terminus toward the beginning of the Δ5 region (Fig. 3A). As expected, strain UJ857 (Δ10), which lacked half of the helix 1 region, was unable to swim, had a filamentous cell morphology, and was deficient in flagellar assembly (Fig. 3A). As observed for UJ553 (Δ6), most of the cells carrying this allele had no flagellum but few flagellated cells were reproducibly detected and a low level of class III promoter activity was measured (Fig. 7). Strains UJ858 (Δ11) and UJ859 (Δ12) had normal morphology (Fig. 3A), assembled flagella (data not shown), and showed normal class III promoter activity (Fig. 7) but were nonmotile (data not shown). Strains UJ970 (Δ13) and UJ971 (Δ14) (Fig. 3A) also had filamentous, nonmotile cells that were deficient in flagellar assembly (Fig. 3A and 4). Similar to UJ553 (Δ6) and UJ857 (Δ10), strain UJ970 (Δ13) showed an intermediate phenotype with a small subfraction of flagellated cells (data not shown) and a low level of class III promoter activity (Fig. 7).
To further define the requirement of helices 1 and 2 for flagellar assembly and motility, several hydrophobic amino acids in either region were replaced with charged residues (S3 and S4; Fig. 3A). Alteration S3 in the proposed helix 2 coding region resulted in cells (UJ974) with a Fla− phenotype and filamentous morphology (Fig. 3A). A small subfraction of cells carrying allele S3 were flagellated (Fig. 3A), and UJ974 cultures showed a low level of class III promoter activity (Fig. 7). Changes at the end of helix 1 did not disturb flagellar assembly or cell morphology but produced a paralyzed flagellum in strain UJ975 (S4) (Fig. 3A). Thus, amino acid substitutions in the helix 1 and 2 regions confirmed the results obtained by deletion analysis and supported our earlier conclusions that only the region covered by helix 2 is strictly required for flagellar assembly and that changes in helix 1 cause failure of flagellar function. To exclude the possibility that reduced protein stability rather than functional impairment produced the observed motility defects of the fliF mutants, cellular levels of all FliF derivatives, which were unable to support productive flagellar assembly, were assayed by immunoblot analysis. The levels of all mutant proteins were either similar to or slightly higher than that of the wild type, arguing against the possibility that a limited MS ring protein concentration caused the observed phenotype (Fig. 3B).
Motile suppressors of nonmotile fliF mutants.
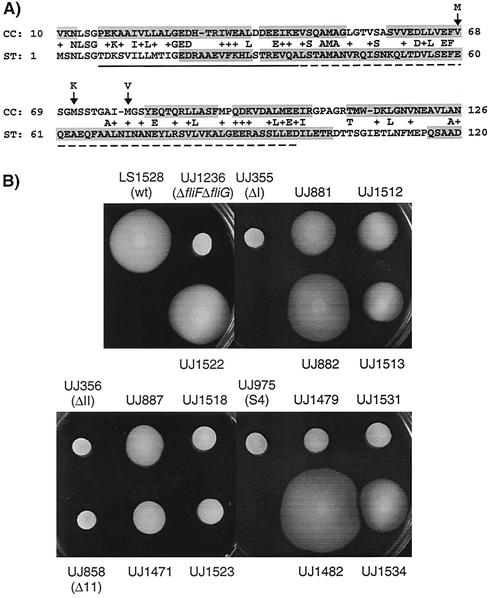
Most of the mutations at the end of fliF that were analyzed in this study led to strains that either were not able to assemble a flagellum or produced a paralyzed structure. To define the molecular basis of these defects, we attempted to isolate motile suppressors of all nonmotile mutant strains. Nonmotile mutants were inoculated on semisolid agar plates, and spontaneous motile suppressors were isolated from emerging flairs or from the edge of the swarmer colony. No motile suppressors could be isolated for any of the nonflagellated mutants with changes in the helix 2 region (Fig. 3A). In contrast, motile suppressors were readily obtained for strains carrying mutations in the helix 1 or turn region of fliF (Fig. 3A). All motile suppressor strains were able to swim, as determined by light microscopy. However, only the three suppressors of UJ355 (ΔI) (UJ881, UJ882, and UJ883), one suppressor of UJ356 (ΔII) (UJ887), one suppressor of UJ858 (Δ11) (UJ1471), and two suppressors of UJ975 (S4) (UJ1480 and UJ1482) were able to form swarmer colonies on semisolid agar plates that were larger than their nonmotile ancestors (Fig. 8). Of these, only strains UJ882, UJ883, UJ1480, and UJ1482 formed colonies comparable in size to wild-type strains (Fig. 8).
FIG. 8.
(A) Alignment of the N-terminal FliG sequences from C. crescentus (CC) and S. enterica serovar Typhimurium (ST) by using BLAST. The numbers on the sides are amino acid positions according to the sequences in the GenBank database. The solid line below the sequence indicates the amino acids essential for FliF binding in S. enterica serovar Typhimurium, and the broken line indicates amino acids that are required for proper motility (deletion mutants have reduced motility) (15). Arrows point to the amino acids that were altered in the motile suppressor strains of C. crescentus, and the letters above indicate the substituted amino acids. The shaded sequences are regions that were proposed to form α-helices by the secondary-structure prediction program PDHsec. (B) Swarming on semisolid agar of the motile suppressors with mutations in fliF or fliG, the original fliF mutants, and the Δ(fliF-fliG) tester strain complemented with the fliF-fliG loci of the suppressor strains. The strains shown are LS1528 (wild type [wt]), UJ1236 (ΔfliF-fliG), UJ1522 (UJ1236 with the fliF-fliG locus from wild-type strain LS1528), UJ355 (ΔI), UJ881 (motile suppressor of UJ355), UJ1512 (UJ1236 with the fliF-fliG locus from UJ881), UJ882 (motile suppressor of UJ355), UJ1513 (UJ1236 with the fliF-fliG locus from UJ882), UJ356 (ΔII), UJ887 (motile suppressor of UJ356), UJ1518 (UJ1236 with the fliF-fliG locus from UJ887), UJ858 (Δ11), UJ1471 (motile suppressor of UJ858), UJ1523 (UJ1236 with the fliF-fliG locus from UJ1471), UJ975 (S4), UJ1479 (motile suppressor of UJ975), UJ1531 (UJ1236 with the fliF-fliG locus from UJ1479), UJ1482 (motile suppressor of UJ975), and UJ1534 (UJ1236 with the fliF-fliG locus from UJ1482). Strains UJ883 and UJ1480 are not shown here because they contain the same mutations as strains UJ882 and UJ1482, respectively.
It has been proposed that the FliF C terminus directly interacts with the N-terminal part of the FliG protein (5, 12, 15, 25, 30). It thus appeared likely that the nonmotile phenotype observed in some of the fliF mutants is due to a disturbed interaction between FliF and FliG and that suppressor mutations restoring motility would be localized in either of the two genes. To test this possibility, we generated an in-frame deletion of fliF and fliG in strain UJ1236 and used this strain for complementation with the fliF-fliG wild-type locus or with the same locus of the motile suppressor strains. Light microscopy analysis revealed that 8 of the 22 constructs used for complementation restored motility. These included the fliF-fliG loci of all motile suppressor strains that were able to form swarmer colonies on soft agar plates (strains UJ881, UJ882, UJ883, UJ887, UJ1471, UJ1480, and UJ1482) and of an additional strain originating from UJ975 (UJ1479). This result implied that the original suppressor mutations in these strains were located in either fliF or fliG. Five of these eight complemented strains were able to form swarmer colonies on semisolid agar plates (Fig. 8). However, all of the complemented strains formed swarmer colonies that were smaller than those of the corresponding original suppressor strains. In contrast, complementation of Δ(fliF-fliG) strain UJ1236 with a wild-type copy of fliF and fliG (UJ1522) gave rise to a swarmer colony of the same size as the wild type (Fig. 8).
To map the suppressor mutations, the sequence of the fliF-fliG locus was determined for all motile suppressor strains. A mutation in either fliF or fliG was found in each of the suppressor strains, which were successful donors in the complementation experiment described above. All motile suppressors of UJ355 (ΔI) contained a point mutation in codon 526 of the fliF gene, immediately upstream of the ΔI deletion. In one strain, the mutation led to a Ser-to-Trp exchange (UJ881), and in two strains (UJ882 and UJ883), the mutation caused the Ser residue to be replaced with a Leu residue. In both motile suppressors of UJ975 that were able to form swarmer colonies of wild-type size (UJ1480 and UJ1482), aspartate codon 535 of fliF S4, which had been introduced by site-directed mutagenesis (Fig. 3A), had reverted to the original valine codon. Three point mutations were mapped in the fliG gene, and all were located within a region of 10 codons. A Met-to-Val substitution in codon 78 of suppressor strain UJ887, a Val-to-Met substitution in codon 68 of UJ1471, and a Met-to-Lys substitution in codon 71 of strain UJ1479 were found to be responsible for the suppression. No mutations in fliF or fliG were found in the fragments from motile suppressors, which did not restore motility to the Δ(fliF-fliG) strain upon complementation. The results obtained for the suppressor strains are summarized in Table 3.
DISCUSSION
One of the key components involved in both flagellar assembly and ejection in C. crescentus is the FliF protein, which forms the MS ring in the cytoplasmic membrane. It is the platform on which the entire flagellar structure is built and directs flagellar assembly to the pole opposite the stalk. At the same time, the MS ring anchors the flagellum in the inner membrane and is proteolytically removed at flagellar ejection. It has previously been shown that the FliF C terminus harbors functional determinants for both flagellar assembly and MS ring turnover (12). Here we have analyzed this large cytoplasmic domain with respect to flagellar assembly and function. A stretch of only nine amino acids close to the immediate C terminus of FliF was found to be essential for flagellar assembly. Any deletion or amino acid substitution that altered this short region disrupted flagellar assembly and induced a cell division block. This was not due to increased degradation of the FliF mutant proteins, since none of the strains expressing the fliF mutant alleles had reduced FliF concentrations. Since the length of deletions ΔIV and ΔV limited the resolution of the mutational analysis of this region, it is possible that the region essential for assembly is even shorter than nine amino acids. It was shown earlier that covalent fusion of C. crescentus FliF lacking 26 amino acids at the C terminus to FliG restored flagellar assembly and function (12). These results and studies with E. coli and Salmonella enterica serovar Typhimurium (5, 15, 25) strongly suggested that the C terminus of FliF is required for FliG binding. Our data indicate that a core region of only nine or fewer amino acids at the immediate C terminus of FliF represents the site of FliF interaction with FliG.
Mutations upstream of the core region for flagellar assembly interfered with flagellar function. The severity of the phenotype correlated with the distance of the mutations from the core binding region. Removal or substitution of up to 10 amino acids immediately upstream of the core region resulted in a paralyzed flagellum, while deletions in the next 19 amino acids further upstream produced functional flagella but cells that had lost the ability to efficiently swarm on semisolid agar, indicating either a chemotaxis defect or, more likely, impaired flagellar rotation in viscous medium. Large deletions even further upstream covering more than the first half of the FliF C-terminal domain exposed to the cytoplasm had no effect at all on flagellar assembly or performance under the conditions tested.
Analysis of the FliF C-terminal sequence with the secondary-structure prediction program PDHsec proposed two α-helices separated by a short loop in the 42 amino acids of the immediate C terminus of FliF (Fig. 2). It is interesting that all of the mutations that caused reduced motility or a paralyzed flagellum were located in helix 1 or in the turn region, while all fla mutations were within putative helix 2. Even though the secondary-structure prediction is hypothetical and no structural data for this domain of FliF are available, this overlap indicated that secondary-structure elements at the very C terminus of FliF might be critical for FliF-FliG interaction and for motor function. While the helix 2 region most likely constitutes the direct interface with FliG, the role of the helix 1 region is less clear, but it could be important for correct folding and spatial positioning of the downstream FliG binding site or it could directly contribute to FliG interaction.
The three suppressor mutations that were found in fliG were located within 10 codons (Fig. 8). Mutations in the homologous region of the S. enterica serovar Typhimurium FliG protein caused only weak motility defects (15). Similar to helix 1 in FliF, this region is adjacent to the essential amino acids required for FliF interaction and flagellar assembly (15) and could thus also constitute a domain that (i) is critical for correct folding and positioning of the core FliF binding element at the N terminus of FliG or (ii) interacts directly with FliF.
Evidence for a direct interaction of the helix 1 region of FliF with FliG comes from motile suppressors that were isolated for the mutant strain carrying fliF allele S4, which has three hydrophobic amino acids replaced with charged residues at the end of helix 1 (Fig. 3A). In two independently isolated suppressors, reversion of Asp535 to Val in FliF S4 fully restored motility. Thus, of the few amino acids altered in S4, only Asp535 at the end of the helix 1 region seems to be responsible for the motility defect. In another motile suppressor of the strain carrying fliF allele S4, Met71 of FliG was replaced with Lys. Since this residue has the opposite charge of the critical Asp residue at the end of helix 1 of FliF S4, it is possible that suppression is the result of electrostatic interactions and that Val535 of FliF directly interacts with Met71 of FliG via hydrophobic interactions in the wild-type situation.
Most of the 22 motile suppressor strains isolated had neither a mutation in fliF nor a mutation in fliG. Identifying these suppressor mutations will give additional information about the role of FliF in flagellar structure and function. The respective mutations could be in genes coding for components of the flagellar structure or the motor that interact, either directly or indirectly, with FliF. Alternatively, the mutations could be of a regulatory nature affecting a sensor that is able to detect perturbations in the final flagellar structure and repress flagellar rotation. Candidates for putative regulators of flagellar rotation in response to flagellar integrity are proteins PleD and FliL. Both of these proteins have been suggested to be involved in the turning on and off of flagellar rotation (13, 39). For instance, the atypical response regulator PleD (8) is known to block flagellar rotation in the absence of the swarmer pole-specific sensor kinase PleC (39). Since the PleC sensor colocalizes with the flagellar structure, it is conceivable that PleC and PleD constitute a control element that keeps the motor off until the flagellum is entirely built and ready to rotate. Further studies exploring the nature of the as yet unmapped suppressor mutations conferring motility on fliF mutant strains will help to shed light on the existence of such a control mechanism.
Acknowledgments
We thank E. Amstutz for editing assistance, A. Schauerte for experimental support, and S.-I. Aizawa for helpful discussions and critical reading of the manuscript.
This work was supported by Swiss National Science Foundation fellowship 31-59050.99 to U.J.
REFERENCES
- 1.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379-391. [DOI] [PubMed] [Google Scholar]
- 2.Dieffenbach, C. W., and G. S. Dveksler. 1995. PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan, F., K. Ohnishi, N. R. Francis, and R. M. Macnab. 1997. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26:1035-1046. [DOI] [PubMed] [Google Scholar]
- 5.Francis, N. R., V. M. Irikura, S. Yamaguchi, D. J. DeRosier, and R. M. Macnab. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. USA 89:6304-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 7.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecht, G. B., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homma, M., S. Aizawa, G. E. Dean, and R. M. Macnab. 1987. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 84:7483-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenal, U. 2000. Signal transduction mechanisms in Caulobacter crescentus development and cell cycle control. FEMS Microbiol. Rev. 24:177-191. [DOI] [PubMed] [Google Scholar]
- 11.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenal, U., and L. Shapiro. 1996. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 15:2393-2406. [PMC free article] [PubMed] [Google Scholar]
- 13.Jenal, U., J. White, and L. Shapiro. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J. Mol. Biol. 243:227-244. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, R. C., and B. Ely. 1977. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics 86:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kihara, M., G. U. Miller, and R. M. Macnab. 2000. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 182:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kihara, M., T. Minamino, S. Yamaguchi, and R. M. Macnab. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 183:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubori, T., N. Shimamoto, S. Yamaguchi, K. Namba, and S.-I. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433-446. [DOI] [PubMed] [Google Scholar]
- 18.Kubori, T., S. Yamaguchi, and S.-I. Aizawa. 1997. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J. Bacteriol. 179:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen, S. H., J. Adler, J. J. Gargus, and R. W. Hogg. 1974. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc. Natl. Acad. Sci. USA 71:1239-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd, S. A., H. Tang, X. Wang, S. Billings, and D. F. Blair. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lux, R., N. Kar, and S. Khan. 2000. Overproduced Salmonella typhimurium flagellar motor switch complexes. J. Mol. Biol. 298:577-583. [DOI] [PubMed] [Google Scholar]
- 22.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macnab, R. M. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131-158. [DOI] [PubMed] [Google Scholar]
- 24.Manson, M. D., P. Tedesco, H. C. Berg, F. M. Harold, and C. Van der Drift. 1977. A proton motive force drives bacterial flagella. Proc. Natl. Acad. Sci. USA 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marykwas, D. L., S. A. Schmidt, and H. C. Berg. 1996. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J. Mol. Biol. 256:564-576. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohr, C. D., U. Jenal, and L. Shapiro. 1996. Flagellar assembly in Caulobacter crescentus: a basal body P-ring null mutation affects stability of the L-ring protein. J. Bacteriol. 178:675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muir, R. E., and J. W. Gober. 2001. Regulation of late flagellar gene transcription and cell division by flagellum assembly in Caulobacter crescentus. Mol. Microbiol. 41:117-130. [DOI] [PubMed] [Google Scholar]
- 30.Oosawa, K., T. Ueno, and S.-I. Aizawa. 1994. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 176:3683-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramakrishnan, G., J. L. Zhao, and A. Newton. 1994. Multiple structural proteins are required for both transcriptional activation and negative autoregulation of Caulobacter crescentus flagellar genes. J. Bacteriol. 176:7587-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapiro, L. 1995. The bacterial flagellum: from genetic network to complex architecture. Cell 80:525-527. [DOI] [PubMed] [Google Scholar]
- 37.Silverman, M., and M. Simon. 1974. Flagellar rotation and the mechanism of bacterial motility. Nature 249:73-74. [DOI] [PubMed] [Google Scholar]
- 38.Simon, R., U. Prieffer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 39.Sommer, J. M., and A. Newton. 1989. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J. Bacteriol. 171:392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sosinsky, G. E., N. R. Francis, D. J. DeRosier, J. S. Wall, M. N. Simon, and J. Hainfeld. 1992. Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc. Natl. Acad. Sci. USA 89:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang, H., T. F. Braun, and D. F. Blair. 1996. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261:209-221. [DOI] [PubMed] [Google Scholar]
- 42.Toker, A. S., and R. M. Macnab. 1997. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 273:623-634. [DOI] [PubMed] [Google Scholar]
- 43.Ueno, T., K. Oosawa, and S. Aizawa. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227:672-677. [DOI] [PubMed] [Google Scholar]
- 44.Vallejo, A. N., R. J. Pogulis, and L. R. Pease. 1994. In vitro synthesis of novel genes: mutagenesis and recombination by PCR. PCR Methods Appl. 4:S123-S130. [DOI] [PubMed] [Google Scholar]
- 45.Wu, J., and A. Newton. 1997. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol. Microbiol. 24:233-239. [DOI] [PubMed] [Google Scholar]