Abstract
Alkylation and oxidation of cysteine residues significantly decrease the catalytic activity and stimulate the degradation of ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO). We analyzed the role of vicinal cysteine residues in redox regulation of RuBisCO from Synechocystis sp. strain PCC 6803. Cys172 and Cys192, which are adjacent to the catalytic site, and Cys247, which cross-links two large subunits, were replaced by alanine. Whereas all mutant cells (C172A, C192A, C172A-C192A, and C247A) and the wild type grew photoautotrophically at similar rates, the maximal photosynthesis rates of C172A mutants decreased 10 to 20% as a result of 40 to 60% declines in RuBisCO turnover number. Replacement of Cys172, but not replacement of Cys192, prominently decreased the effect of cysteine alkylation or oxidation on RuBisCO. Oxidants that react with vicinal thiols had a less inhibitory effect on the activity of either the C172A or C192A enzyme variants, suggesting that a disulfide bond was formed upon oxidation. Thiol oxidation induced RuBisCO dissociation into subunits. This effect was either reduced in the C172A and C192A mutant enzymes or eliminated by carboxypentitol bisphosphate (CPBP) binding to the activated enzyme form. The CPBP effect presumably resulted from a conformational change in the carbamylated CPBP-bound enzyme, as implied from an alteration in the electrophoretic mobility. Stress conditions, provoked by nitrate deprivation, decreased the RuBisCO contents and activities in the wild type and in the C192A and C247A mutants but not in the C172A and C172A-C192A mutants. These results suggest that although Cys172 does not participate in catalysis, it plays a role in redox regulation of RuBisCO activity and degradation.
The universal CO2-fixing enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (EC 4.1.1.39) catalyzes the primary reactions of photosynthesis and photorespiration by carboxylation and oxygenation of ribulose-1,5-bisphosphate (RuBP), respectively. The RuBisCO from cyanobacteria, algae (excluding some dinoflagellates), and higher plants is a hexadecamer consisting of eight large and eight small subunits organized as four protomers, each of which has two active sites at the interface between each large subunit pair (3, 4, 29). In the RuBisCO of many species, an intradimeric disulfide bond is formed under oxidizing conditions between the Cys247 residues of a large subunit pair (30). A sequence comparison of more than 500 RuBisCO genes revealed high levels of similarity, particularly among large subunits, and residues that form the catalytic site are entirely conserved (21). As a result of the enzymatic inefficiency manifested by a low catalytic turnover number (1.5 to 12 carboxylations s−1 catalytic site−1 [5, 24]), as well as a low affinity for substrate CO2 and competition between the CO2 and O2 substrates, it is accepted that the activity of RuBisCO limits both photosynthesis and photorespiration under various ecophysiological conditions (40).
Due to the pivotal role of RuBisCO in photosynthesis, the activity, synthesis, and degradation of this enzyme are regulated by several mechanisms (4, 17, 32, 40); one of the less familiar of these mechanisms is redox potential regulation. Early work (26, 39) demonstrated that thiol alkylating agents, such as p-mercury benzoate and iodoacetate, as well as HgCl2, inhibited the catalytic activity of the enzyme and increased the rate of enzyme proteolysis (36). A correlation was found between binding of the alkylating agent N-bromoacetylethanolamine phosphate to Cys172 on the large subunit of the enzyme and the inhibition of enzyme activity (34). Based on this finding, it has been suggested that Cys172 is involved in the catalytic site. Similarly, oxidizing conditions, both in vitro and in vivo, inhibited RuBisCO activity and stimulated its degradation, which in several instances was prevented by thiol reducing agents (8, 14, 27). This suggests that the redox state of one or several cysteine residues on the enzyme regulates its activity and is involved in its degradation. Because of its high cellular concentration (10), RuBisCO not only is a catalyst but also is a storage protein for fixed nitrogen, sulfur, and carbon skeletons. These compounds may be reutilized under nutritional deficiency, aging, and stress conditions (12, 15, 17). Garcia-Ferris and Moreno (14) extended this hypothesis and suggested that free radicals produced under stress conditions and during nutritional limitation and maturation oxidize sulfhydryl groups on the enzyme and thus trigger enzyme degradation. Moreno and Spreitzer (28) showed later that replacement of Cys172 by Ser affected hyperosmotic stress-induced turnover of RuBisCO from Chlamydomonas reinhardtii.
RuBisCO of the cyanobacterium Synechocystis sp. strain PCC 6803 contains 11 and 3 cysteine residues in the large and small subunits, respectively (2). Of these 14 residues, only Cys172 is entirely conserved in all hexadecameric enzymes (21). X-ray diffraction analysis of RuBisCO crystals from the cyanobacterium Synechococcus sp. strain PCC 6301 highlighted two pairs of cysteine residues in close proximity that may form an intramolecular disulfide bond. One pair, Cys172-Cys192 on the large subunit, is close to Lys175, which participates in the catalytic site, and to the activator Lys201 (29). However, no direct evidence for the existence of a disulfide bond between these residues has been obtained thus far. Besides this pair of cysteine residues, each pair of large subunits may cross-link via a Cys247-Cys247 disulfide bond (30).
In this study we used a mutant of Synechocystis sp. strain PCC 6803 (2), which enabled point mutagenesis of RuBisCO, in order to assess the roles of Cys172, Cys192, and Cys247 in regulation of RuBisCO activity and degradation.
MATERIALS AND METHODS
Growth conditions for cyanobacteria.
The Synechocystis sp. strain PCC 6803 wild type and mutants were grown on BG-11 medium (31) or on plates (with an additional 1.5% Bacto Agar [Difco]) under continuous white light (10 to 20 μmol m−2 s−1) at 32°C. A Synechocystis host, Syn6803Δrbc (2), in which the entire rbc operon had been replaced by the Rhodospirillum rubrum rbc gene, was grown under identical conditions except that the atmosphere was enriched with 5% (vol/vol) CO2 in air. The growth rate was monitored by determining the chlorophyll concentration and cell number. Cells were starved for nitrogen determinations by growing them on nitrate-deficient BG-11 medium for at least 7 days. In order to minimize nitrate carryover, the cultures were inoculated twice into nitrate-deficient medium. The nitrate concentration was monitored routinely by using Nitrate-Test sticks (Merck, Darmstadt, Germany) and never exceeded 10 mg liter−1, compared to 1,500 mg liter−1 in BG-11 medium. Photosynthesis rates were determined by measuring O2 exchange rates with a Clark-type O2 electrode (Hansatech, Norfolk, United Kingdom).
Point mutagenesis and genetic analysis.
All plasmid construction and propagation procedures were performed by using pBluescript (Stratagene, La Jolla, Calif.), whose XbaI restriction site within the polylinker was omitted, and Escherichia coli strain DH5α, as described previously (2). Cysteine residues 172, 192, and 247 were replaced by alanine by PCR by using the Megaprimer method (33), modified from the SOE method (gene splicing by overlap extension [16]). Each mutation was obtained by a two-step PCR with Pwo DNA polymerase (Roche) and three primers, one of which had the desired mutation and the other two of which spanned the XbaI and NsiI restriction sites that were used for insertion of the mutated DNA fragment back into the rbcL gene. The template DNA was pSynR4.0 (2). The following primers were used (changed codons are underlined, and replaced nucleotides are indicated by boldface type): for C172A, 5′-GGGTTTGATGGTAGCACCAAGCAGAG-3′; for C192A, 5′-GCTGTTTACGAAGCTCT CCGGGGTGG-3′; and for C247A, 5′-CATCATTTCTTCGGCGGTGCCAGCGGTG-3′. In addition, the upstream primer (252 bp from rbcL) was 5′-ACGCGTCGACGAATTCTATTAGAAAGTCCAA-3′, and downstream primer (1,241 bp from the ATG initiation codon), designed for the cDNA strand in the reverse orientation, was 5′-CAATTTACCAACCGCGGTACCGG-3′. The PCR product was cleaved with XbaI and NsiI, and the resulting 484-bp fragment was cloned into the corresponding sites in pSynR4.0, which was used to transform the Syn6803Δrbc cyanobacterial recipient by a previously described procedure (2). Air-grown colonies were obtained within 14 days. The RuBisCO DNA sequence was determined directly with transformant cyanobacterial colonies to verify the mutation and to eliminate the possibility of PCR-borne substitutions. The cells were permeabilized in 60 μl of saturated NaI for 10 min at 37°C, washed with 2 ml of H2O, and centrifuged, and the final pellet was suspended in TE buffer (10 mM Tris-Cl, 1 mM EDTA; pH 8.0). Aliquots (3 μl) were used for PCR-based sequencing. These analyses revealed full segregation of the mutations in all chromosome copies present in Synechocystis (1, 22).
RuBisCO isolation.
Cells from the mid-logarithmic phase were harvested by centrifugation, washed once, resuspended in 50 mM HEPES (pH 7.0)-5 mM dithiothreitol (DTT) containing protease inhibitor cocktail (P 2714 for general use; Sigma) at a final concentration of 5 to 10 μg of chlorophyll ml−1, and disrupted with a French press at 24,000 lb/in2. RuBisCO was partially purified by two steps of ammonium sulfate fractionation, which dissociated the carboxysomes (25) and removed compounds that could putatively associate with the enzyme. It remained soluble in a solution of 20% saturated ammonium sulfate and was precipitated by 60% saturated ammonium sulfate. The enzyme preparation was stored at −20°C. Prior to the assay the enzyme was dialyzed (molecular weight cutoff, 100,000) overnight against 5 mM HEPES (pH 7.0)-1 mM β-mercaptoethanol-1 mM phenylmethylsulfonyl fluoride to remove low-molecular-weight compounds. Since the yield of purified enzyme decreased substantially when gel filtration was used and since the kinetic properties of the partially purified enzyme could be determined accurately with 14C-labeled carboxypentitol bisphosphate (CPBP), all experiments were performed with the ammonium sulfate preparation.
RuBisCO assay.
Enzyme (concentration, 1 to 1.25 μM, as estimated by CPBP binding [see below]) was activated for 30 min at 30°C with 140 μM CO2 and 10 mM MgCl2 in 50 mM HEPES buffer (pH 7.0 to 8.0). Activated enzyme (20 to 50 pmol) was added to stoppered vials containing the reaction mixture (50 mM HEPES [pH 8.0], 20 mM MgCl2, 5 mM DTT) and 7.5 Wilbur Anderson units of carbonic anhydrase at 30°C. Various concentrations of RuBP and 14CO2 (6,000 to 12,000 Bq nmol−1) were added as required for each assay (Table 1 and see below). Each reaction was terminated after 2 min by adding 0.5 volume of 6 N acetic acid. Acid-stable material was collected and counted with a scintillation counter. Nonspecific counts (in the absence of RuBP or enzyme) were subtracted. Each experiment was performed in triplicate.
TABLE 1.
Kinetic constants of RuBisCO mutantsa
| Enzyme | Km(RuBP) (μM) | Km(CO2) (μM) | Vmax (min−1) |
|---|---|---|---|
| Wild type, reduced | 105 | 202 | 760 |
| Wild type, oxidized | 94 | 162 | 79 |
| C172A, reduced | 94 | 225 | 317 |
| C192A, reduced | 167 | 204 | 780 |
| C172A-C192A, reduced | 183 | 233 | 417 |
The enzyme was activated by incubation in the presence of 140 μM CO2 and 10 mM MgCl2 and either reduced for 20 min with 5 mM DTT or oxidized for 60 min with 100 μM iodosobenzoate at 30°C. Aliquots (20 μl) of activated enzyme were added to 380-μl reaction mixtures containing either various concentrations of RuBP and 0.66 mM 14CO2 (4 Bq nmol−1) or various 14CO2 concentrations and 0.5 mM RuBP in 50 mM HEPES buffer (pH 8.0). The buffer contained 7.5 Wilbur Anderson units of carbonic anhydrase, 5 mM DTT, and 20 mM MgCl2. The reaction was terminated after 2 min with 200 μl of 6 N acetic acid, and the acid-stable material was counted. The active site concentration of the reduced enzyme was determined by [14C]CPBP binding (see Materials and Methods for details). The data were analyzed by the method of Hanes-Woolf (35).
Determination of catalytic site concentration.
The molar concentration of catalytic sites of RuBisCO was determined by incubating the enzyme with 15 μM [14C]CPBP (66.6 × 107 Bq μmol−1) for 10 min at 30°C. [14C]CPBP was synthesized as previously described (24). [14C]CPBP-bound RuBisCO was separated from the free ligand by gel filtration with a 10-cm-long Sephadex G-100 column.
Redox and thermal manipulations.
RuBisCO, incubated in 50 mM HEPES buffer (pH 7.0 to 8.0) in the presence of 140 μM CO2 and 10 mM MgCl2, was exposed to the following concentrations of redox agents: 0.5 to 2.0 mM 5,5′-dithio-bis(2-nitrobenzoate) (DTNB); 20 mM cystamine (CSSC) or cysteamine (CSH); 25 μM CuSO4; 100 μM iodosobenzoate; and 1 mM H2O2. The reversibility of the oxidation effect was tested by addition of a reductant (usually 5 mM reduced DTT) to a preoxidized enzyme. RuBisCO in 50 mM HEPES buffer (pH 8.0) was either reduced with 5 mM DTT or oxidized with 25 μM CuSO4 for 20 min and then treated with 100 μM EDTA to sequester the copper ions. The various samples were incubated for 15 min at various temperatures, after which the enzyme was activated for 20 min with 140 μM CO2 and 10 mM MgCl2 at 30°C. To assay activity, the oxidized enzyme was reduced with 5 mM DTT.
Gel electrophoresis and immunochemical detection of RuBisCO.
RuBisCO-enriched fractions were analyzed by electrophoresis on either a 7% polyacrylamide nondenaturing gel at 3°C or a 12% polyacrylamide denaturing gel by using standard conditions (23) and by Western blotting. The RuBisCO holoenzyme and large subunit were identified by using polyclonal antibodies raised against Synechococcus RuBisCO (kindly provided by R. Tabita) and colorimetric visualization following secondary decoration with anti-rabbit immunoglobulin G linked to alkaline phosphatase (2).
RESULTS
To analyze the role of cysteine residues that are putatively involved in disulfide bond formation and particularly cysteine residues that are adjacent to the active site of RuBisCO, we generated four distinct mutations in the large subunit of the Synechocystis sp. strain PCC 6803 enzyme: C172A, C192A, C172A-C192A, and C247A. Transformed cells of the four RuBisCO mutants grew photoautotrophically at a rate equal to that of the wild type with continuous illumination or with cycles consisting of 9 h of light and 15 h of darkness at the ambient CO2 concentration (data not shown). To assess whether the replacement of cysteine residues affected the overall photosynthesis, the oxygen exchange rate was determined as a function of light intensity and inorganic carbon concentration. The photosynthesis rate at a saturating light intensity and a saturating inorganic carbon concentration was 10 to 20% less for the C172A and C172A-C192A mutants than for the wild type and the other mutants. No significant difference was found between the mutants and the wild type in terms of the photosynthetic affinity for inorganic carbon, for the light-limited rate of photosynthesis, or for the adaptation kinetics of predarkened cells to light (data not shown).
Redox effects on enzyme kinetic properties and stability.
The modified enzymes were isolated, and the kinetics of RuBP carboxylation as a function of substrate (CO2 and RuBP) concentration were determined. Replacement of Cys172 in two mutants (C172A and C172A-C192A) reduced the Vmax of the carboxylation reaction 45 to 60%, whereas the C192A and C247A mutations had no effect on this parameter. Although the C172A mutation did not affect the Km(RuBP), the C192A modification in both the C192A and C172A-C192A mutants increased this parameter from 105 to 167 and 183 μM, respectively. Still, these alterations in catalytic properties may not be significant because the physiological RuBP concentration usually saturates RuBisCO (40). In contrast to these changes, the Km(CO2) value was not affected by either of these mutations (Table 1).
One of the roles attributed to disulfide bonds is stabilization of the molecule framework. The proximity of C172 to C192 (29) suggests that the two molecules may form an intramolecular disulfide bond. Furthermore, a disulfide bridge between Cys247 residues in the large subunit homodimer has been shown to be present after enzyme oxidation (30). Therefore, we examined the effect of replacement of Cys and whether the redox state affects the thermal stability of the enzyme by analyzing carboxylase activity after incubation at temperatures ranging from 5 to 100°C. The wild type and the C172A and C247A enzyme variants were stable for 15 min at 50°C, whereas a higher temperature was destructive. The activity was one-half of the maximal value after incubation at 56°C, and only 10% of the activity persisted after incubation at 65°C. The C192A and C172A-C192A mutant enzymes were slightly more labile. Full activity was maintained if the temperature treatment did not exceed 35°C. The C192A mutant lost 50% of its activity at 45°C, and the C172A-C192A mutant lost 50% of its activity at 50°C (Fig. 1A). Oxidation of the wild-type and C247A enzymes with copper ions decreased their thermal stabilities (Fig. 1B). Copper ions facilitate intramolecular disulfide bond formation in the absence of free thiols (20), as was evident from dimerization of the large subunits through Cys247 (data not shown). Therefore, these results indicate that neither the C247-C247 disulfide bond nor the putative C172-C192 disulfide bond contribute to thermal stability of RuBisCO.
FIG. 1.
Thermal stability of RuBisCO from the wild type and mutants. (A) Enzymes under reducing conditions. (B) Comparison of reduced and oxidized enzymes. Reduction was achieved with 5 mM DTT, and oxidation was achieved with 25 μM CuSO4 for 20 min, followed by addition of 100 μM EDTA to sequester the copper ions. The redox-treated enzymes were incubated at different temperatures for 15 min. The enzymes were then activated by 140 μM CO2 and 10 mM MgCl2 at 30°C. Oxidized enzymes were rereduced with 5 mM DTT. The carboxylation reaction was performed in the presence of 0.5 mM RuBP and 0.66 mM 14CO2. Other details are described in Table 1. The activity of the enzyme treated at 30°C was defined as 100%. Symbols: ▪, wild-type enzyme, reduced; □, wild-type enzyme, oxidized; ⧫, C172A enzyme, reduced; ▾, C192A enzyme, reduced; •, C172A-C192A enzyme, reduced; ▴, C247A enzyme, reduced; ▵, C247A enzyme, oxidized.
Alkylation, oxidation, and reduction affect enzyme activity.
The activity of the hexadecameric enzyme RuBisCO is inhibited by alkylation (26, 34, 39), but whether this effect is due to active site perturbation remains unknown. To examine the involvement of Cys172 and Cys192, which are located in the vicinity of the catalytic site, in this inhibitory effect, we examined the effect of the alkylating agent iodoacetate. Iodoacetate at a concentration of 10 mM inhibited 65% of the activity of the wild-type and C192A enzymes but had little effect on enzyme variants lacking Cys172 (5% inhibition for the C172A enzyme and 15% inhibition for the C172A-C192A enzyme). Doubling the concentration of the alkylating agent had only a small additive inhibitory effect (Fig. 2). Since thiol oxidation was shown to inhibit RuBisCO activity (14), we examined the effects of the following three types of oxidizing agents on the enzyme mutants: (i) oxidants that may form intermolecular mixed disulfide bonds, including CSSC, DTNB, and oxidized glutathione (20); (ii) oxidants that react preferentially with vicinal thiols and produce intramolecular disulfide bonds, including iodosobenzoate and oxidized DTT (7, 19); and (iii) peroxide that produces a disulfide bond, which may be oxidized further to sulfenate, sulfinate, or sulfonate derivatives of cysteine (19). As the extent of inhibition was found to depend on the length of incubation and the type and concentration of the oxidant used (data not shown), conditions for oxidation were chosen so that the maximal inhibition could still be reversed. Oxidation of wild-type RuBisCO decreased the number of catalytic sites (as estimated by [14C]CPBP binding [data not shown]) and consequently the Vmax value, yet there was not a significant alteration in the affinity for CO2 and RuBP (Table 1 and Fig. 3). Oxidation of the wild-type enzyme with 20 mM CSSC for 20 min inhibited 70% of the enzyme activity. Prolonged incubation with this oxidant enhanced the inhibitory effect only slightly. Similar results were obtained with the C192A enzyme mutant. In contrast, the enzymes mutated at position 172 (C172A and C172A-C192A) were less affected by CSSC (only 40% inhibition). Most of the CSSC inhibitory effect could be restored by the reductants CSH and DTT (Fig. 4A). A low concentration of DTNB (0.5 mM) produced an effect similar to that of CSSC, whereas 2 mM DTNB eliminated the activity of all enzyme variants. Oxidized glutathione had no effect on either enzyme variant (data not shown).
FIG. 2.
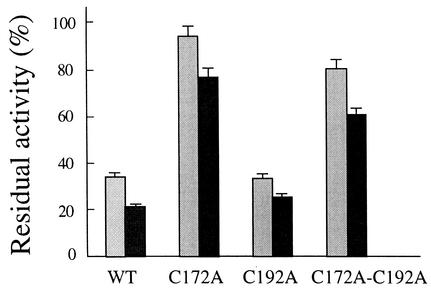
Effect of iodoacetate on RuBisCO activity. Preactivated RuBisCO isolated from a Synechocystis sp. strain PCC 6803 variant was incubated in the presence of 10 and 20 mM iodoacetate (light and dark gray bars, respectively). RuBisCO was activated and assayed as described in the legend to Fig. 1. WT, wild type.
FIG. 3.
Effect of iodosobenzoate on the kinetics of RuBisCO as a function of RuBP (A) and CO2 (B) concentrations. Wild-type RuBisCO was activated in the presence of 140 μM CO2 and 10 mM MgCl2. Activated enzyme was incubated in the presence of 100 μM iodosobenzoate (□) or 5 mM DTT (▪) for 60 min. The carboxylation assay was performed as described in Table 1.
FIG. 4.
Effects of reducing and oxidizing agents on RuBisCO activity. (A) Reduction with 20 mM CSH and oxidation with 20 mM CSSC for 20 min at 30°C. An aliquot of the oxidized enzyme was rereduced with 20 mM CSH. (B) Reduction with 5 mM DTT and oxidation with 100 μM iodosobenzoate for 60 min at 30°C. An aliquot of the oxidized enzyme was rereduced with 5 mM DTT. (C) Reduction with 5 mM DTT and oxidation with 1 mM H2O2 for 10 min at 30°C. An aliquot of the oxidized enzyme was rereduced with 5 mM DTT and subjected to an excess amount of catalase. RuBisCO activity was assayed as described in the legend to Fig. 1. The activity of each variant under reducing conditions was defined as 100%. WT, wild type.
Iodosobenzoate, which preferentially oxidizes dithiols (20, 38), inhibited 90 and 99.5% of the wild-type and C247A enzyme activities, respectively. The other enzyme mutants were inhibited to lesser extents by iodosobenzoate; the C192A enzyme activity was 60% inhibited, whereas the C172A enzyme activity was inhibited only 30%. The enzyme activity that was inhibited least (only 10% inhibition) was that of the C172A-C192A mutant. These inhibitory effects were partially restored upon reduction with DTT (Fig. 4B).
Wild-type RuBisCO is sensitive to H2O2 (8). Indeed, 10 min of incubation in the presence of 1 mM peroxide inhibited 97% of the enzyme activity. This inhibitory effect was fully restored by DTT reduction and peroxide decomposition with catalase (Fig. 4C). Prolonged incubation with H2O2 had no further inhibitory effect, but the reversibility of enzyme activity was hampered (data not shown). Oxidation of the C172A mutant enzyme with peroxide inhibited 40% of the activity, but unlike the wild-type enzyme, the inhibitory effect was irreversible. The C192A mutant enzyme was, however, only marginally inhibited by H2O2 oxidation (Fig. 4C).
RuBisCO degrades upon in vitro oxidation.
To examine whether the irreversible inhibition of activity upon oxidation (Fig. 4) was due to enzyme degradation, RuBisCO was analyzed by nondenaturing polyacrylamide gel electrophoresis. The wild-type and mutant enzymes were compared after reduction with 5 mM DTT or oxidation with 1 mM DTNB in the presence of protease inhibitors (see Materials and Methods). As shown in Fig. 5 to 7, the wild-type enzyme was degraded upon oxidation, but it was not degraded after alkylation with 10 mM iodoacetate (data not shown). However, this degradation could be prevented if the enzyme was first carbamylated and CPBP bound, which correlated with increased electrophoretic mobility (Fig. 5 to 7). Neither carbamylation nor CPBP binding separately could prevent the oxidation-induced decomposition of the enzyme or alter its electrophoretic mobility (Fig. 5). Interestingly, the degradation was largely eliminated in the C172A enzyme variant and to a lesser extent in the C192A enzyme variant (Fig. 6). Immunochemical analysis of RuBisCO and its degradation products revealed that following oxidation, the holoenzyme was decomposed and an immunoreactive band, which comigrated with phycobilins, appeared (Fig. 7A). This band was cut out, boiled in sample buffer, and loaded on a sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis gel beside the bands for the reduced holoenzyme and reduced and oxidized RuBisCO extracts (Fig. 7B). In all four lanes, a single immunoreactive band at the size of the RuBisCO large subunit was observed. Densitometric analysis of this band in the lanes containing the total reduced or oxidized RuBisCO extract (Fig. 7B, right two lanes) revealed that the amounts were comparable, indicating that the RuBisCO large subunit did not undergo observable fragmentation. These results suggest that oxidation of Cys172 or Cys192 triggers dissociation of RuBisCO into subunits.
FIG. 5.
Effects of activation, CPBP binding, and redox state on RuBisCO degradation and electrophoretic mobility. Following reduction with 5 mM DTT or oxidation with 1 mM DTNB, RuBisCO was divided between activated (20 mM NaHCO3 and 10 mM MgCl2 for 20 min) and nonactivated samples, and the samples were further divided between those treated with 10 μM CPBP and those used as a control. Protein (30 mg per lane) was loaded on a 7% polyacrylamide nondenaturing gel, and electrophoresis at a constant current of 15 mA was performed by using Tris-glycine buffer (pH 8.8). The gel was stained with Coomassie blue. The location of RuBisCO (indicated by arrows) was determined immunochemically. The dense bands at the bottom of the gel are bands for phycobiliproteins.
FIG. 7.
Immunochemical analysis of RuBisCO holoenzyme and oxidation-induced degradation product. (A) Western blot of a native 7% polyacrylamide gel that separated reduced and oxidized enzymes with and without CPBP. (B) Western blot of a sodium dodecyl sulfate-12% polyacrylamide gel comparing enzyme large subunits (LSU) from total reduced and oxidized extracts and from reduced and oxidized RuBisCO bands that appeared in the native gel. The boxes and arrows indicate bands that were reelectrophoresed after denaturation.
FIG. 6.
Effect of oxidation on RuBisCO degradation. Activated RuBisCO from the wild type (WT) and C172A (upper left gels) or from the wild type and C192A (upper right gels) was either oxidized with DTNB or reduced with DTT as described in the legend to Fig. 5. The CPBP binding and running conditions were as described in the legend to Fig. 5. Equal amounts of oxidized or reduced enzyme extract from each treatment were loaded. The graph at the bottom shows the calculated ratios of oxidized RuBisCO bands to reduced RuBisCO bands as determined densitometrically.
Deficiency in available nitrogen affects RuBisCO content and activity.
To examine whether Cys172 or Cys192 is involved in stress-induced degradation of the cyanobacterial RuBisCO, we analyzed the wild type and the four mutants (C172A, C192A, C172A-C192A, and C247A) after 7 days of growth in nitrate-deficient BG-11 medium. Despite the severe nitrate deficiency, the cultures grew at approximately 30% of the rate of a culture grown in complete medium, probably by reutilizing internal nitrogen resources. After 7 days, the cellular chlorophyll contents had hardly changed, but the total cellular protein contents had decreased up to 60% in the various strains and, as indicated by absorbance at 630 nm, the concentrations of the phycobilins had drastically decreased (data not shown). The cellular RuBisCO contents and activities in the wild type and in the C247A and C192A mutants declined to 15 to 20% of the levels obtained under nitrate-sufficient conditions. However, only 40% reductions in the RuBisCO content and activity occurred in the C172A-C192A mutant, and almost no changes in these parameters were observed in the C172A mutant (Fig. 8).
FIG. 8.
Effect of nitrate deficiency in the growth medium on RuBisCO activity (solid and open bars) and content (gray and cross-hatched bars). Synechocystis sp. strain PCC 6803 variants were grown in nitrate-deficient BG-11 medium (open and cross-hatched bars) or complete BG-11 medium (solid and gray bars) for 7 days. RuBisCO in crude extract was activated by 140 μM CO2-10 mM MgCl2-5 mM DTT for 20 min at 30°C and then assayed in the presence of 0.5 mM RuBP and 0.66 mM 14CO2. The concentration of RuBisCO catalytic sites was determined by [14C]CPBP binding. Other experimental conditions were identical to those described in the legend to Fig. 1. WT, wild type.
DISCUSSION
Of all the cysteine residues on the large subunit of hexadecameric RuBisCO enzymes, Cys172 is the most conserved, whereas Cys192 and Cys247 are found in many enzymes (21). Since Cys172 is spatially adjacent to the catalytic and activation sites (29), reduction and oxidation of its thiol group could affect enzyme function. The decrease in catalysis upon alkylation of Cys172 (Fig. 2), as shown previously (34), seemed to support this notion. Using both mutational and chemical approaches, we addressed the question of whether this effect was due to elimination of the thiol moiety or to hindrance of the catalytic site by the alkylating agent. We produced mutants with the C172A, C192A, C172A-C192A, and C247A substitutions and analyzed the effects on cell growth, photosynthetic capacity, and RuBisCO function. Since the four mutants grew much like the wild type, it was evident that replacement of either cysteine residue did not affect synthesis and assembly of a functional enzyme crucial for photoautotrophic growth. A similar observation was reported for a C172S substitution in RuBisCO of C. reinhardtii (28). The prominent activity of RuBisCO in the C172A and C192A cyanobacterial mutants (Table 1), the ability of these mutants to grow photoautotrophically, and the lack of interaction between these residues and RuBP in RuBisCO crystals (29) indicate quite explicitly that neither Cys172 nor Cys192 is involved in catalysis, as was previously suggested (34). However, the decreased inhibitory effect of RuBP carboxylation upon alkylation with iodoacetate or monothiol oxidation of C172A and C172A-C192A enzyme variants but not of C192A or C247A enzyme variants (Fig. 2 and 4) suggests that inhibition of RuBisCO carboxylation is due primarily to modification of Cys172. This effect coincides with a reduced number of functional active sites, as determined by [14C]CPBP binding, and consequently with a decreased Vmax of carboxylation (Fig. 3). This result is in accordance with the observation of Schloss et al. (34), who found a linear correlation between alkylation of Cys172 and enzyme inhibition, but differs from the finding of Moreno and Spreitzer (28), who did not observe a difference in the response to oxidation between C172S and wild-type RuBisCO variants. This discrepancy could result from the different experimental conditions, which were harsh in the case of Chlamydomonas RuBisCO (i.e., incubation with 2 mM DTNB for 2 h). Such conditions used with the Synechocystis enzyme caused enzyme dissociation (Fig. 5 to 7), and therefore, oxidation was carried out so that a maximal, yet reversible effect was obtained. The partial inhibition of RuBisCO C172A and C172A-C192A mutants by oxidants (Fig. 4) suggests that other thiol moieties on the enzyme may be affected. It is also not clear whether Cys192 is inaccessible for the alkylating and oxidizing agents or whether binding of these agents to Cys192 has no effect on the carboxylase reaction.
The inhibitory effect of iodosobenzoate (Fig. 3 and 4B), which preferentially oxidizes vicinal thiols (20, 38), and the reversible inhibitory effect of H2O2 on the wild-type enzyme (Fig. 4C) raise the possibility that formation of an intramolecular disulfide bond inhibits enzyme activity. The three-dimensional structure of RuBisCO from Synechococcus sp. strain PCC 6301 highlights two pairs of cysteine residues that may form such a bond (29). One bond involves Cys247, which covalently cross-links two large subunits of the enzyme (30), and the other may be generated between Cys172 and Cys192. Since oxidation of Cys247 in the C172A-C192A double mutant by iodosobenzoate had no effect on RuBisCO activity and since replacement of Cys247 by Ala did not prevent the inhibitory effect of iodosobenzoate (Fig. 4B), Cys247 might not be involved in redox regulation of RuBisCO activity. Conversely, the smaller effects of iodosobenzoate and H2O2 oxidation on the C172A and C192A enzyme variants and especially on the C172A-C192A enzyme variant (Fig. 4B and C) may suggest that there is a putative disulfide bond between Cys172 and Cys192 that, when formed (under oxidation conditions), inhibits enzyme activity. In such a case, replacement of either of the two Cys residues is expected to prevent the effect caused by iodosobenzoate. Indeed, iodosobenzoate had a small inhibitory effect on the double mutant, C172A-C192A, but it had greater yet unequal effects on C172A and C192A (Fig. 4B). The unequal effects could result from a difference in the accessibility of iodosobenzoate to Cys172 and Cys192 and some ability to oxidize monothiols despite the preference for dithiols. Another possibility is that under oxidizing conditions Cys172 may be able to form a disulfide bond with a nearby residue other than Cys192. Based on the virtual three-dimensional structure of the Synechocystis enzyme constructed by using the known X-ray structure of Synechococcus RuBisCO (29), the other residue could be Cys399. However, this possibility requires further proof.
Oxidation of RuBisCO by DTNB, CSSH, iodosobenzoate, and hydrogen peroxide was only partially reversible. The following reasons could account for this phenomenon: the redox potential of the solution was not sufficiently negative; oxidation by peroxide could form nonreducible oxygen derivatives, such as sulfonate (19); and thiol oxidation could lead to enzyme denaturation or stimulated degradation. Unlike Garcia-Ferris and Moreno (14), who showed that oxidation increases the susceptibility of RuBisCO to proteolysis by exogenous trypsin, we were able to demonstrate using a nondenaturing gel that cysteine oxidation resulted in enzyme disintegration into subunits even in the presence of protease inhibitors (Fig. 5 to 7). Therefore, the proteolytic degradation observed in the higher plant RuBisCO (14, 27) could have followed dissociation. This dissociation was prevented, however, by the C192A substitution and to a larger extent by the C172A substitution, which could be explained by (i) independent oxidation of each of these residues and formation of two mixed disulfide bonds, each of which is capable of stimulating dissociation, with a stronger effect of Cys172 due perhaps to greater accessibility to the oxidant; and (ii) formation of an intramolecular disulfide bond as a result of an interaction between the proximal oxidized monothiols. In such a case the two interacting residues should be close to one another, as implicated by the effect of iodosobenzoate on the wild-type enzyme compared to the reduced effect on the Cys mutants (Fig. 4B). Notably, the unequal effects on dissociation observed upon oxidation of C172A and C192A mutant enzymes resemble the unequal inhibitory effects of iodosobenzoate on enzyme activity.
The profound inhibition of dissociation by CPBP binding to the carbamylated form of oxidized RuBisCO and the change in electrophoretic mobility may be explained by a conformational change of the enzyme. Crystallographic studies have shown that RuBP or CPBP binding to activated RuBisCO stimulates closure of the catalytic site via movement of loop 6 in the large subunit in conjunction with a short strand of the N terminus across the substrate. The disordered C tail (residue Trp462 to the C terminus) in the open state is packed against loop 6 in the closed state and is locked by an electrostatic interaction between Asp473 and a highly positive pocket on the enzyme surface (9). This conformational alteration presumably affects the hydrodynamic features of the enzyme, thereby increasing its electrophoretic mobility (Fig. 5 to 7). Thus, oxidatively induced dissociation leading to RuBisCO degradation is eliminated by closure of the catalytic site after CPBP binding to the activated form of the enzyme.
Since no differences have been found between the various Cys mutants and wild type in terms of light-limited rates of photosynthesis or the adaptation kinetics of predarkened cells to light, regulation of RuBisCO by light is not mediated by reduction of Cys172, Cys192, or Cys247. Nevertheless, it remains unclear whether the in vitro effects of redox agents on RuBisCO activity also occur in vivo. It has been shown that under extreme conditions, including severe cold (6, 18, 37), osmotic stress (11, 13), nutrient deficiency (12, 15), and accumulation of free radicals (27), RuBisCO is oxidized (the number of thiols decreases and the number of disulfide bonds increases), which leads to a decline in activity and degradation. The Cys mutants were instrumental in this respect for examination of RuBisCO stability and degradation under stress conditions. Severe nitrogen starvation results in a decrease in RuBisCO content and activity (15, 17), as was shown for wild-type cells, as well as the C247A and C192A mutants. However, RuBisCO mutated at Cys172 (C172A and C172A-C192A mutants) was hardly affected by the stress (Fig. 8), which was in accordance with the hypothesis that stress-induced Cys oxidation of RuBisCO stimulates enzyme degradation (14, 27). It is also possible that the lack of stress-induced enzyme degradation in mutant C172A resulted from the substantial decline (∼60%) in enzyme catalytic turnover. Since the C172A and C172A-C192A mutants grew in nitrate-deficient medium at rates comparable to the wild-type rate, RuBisCO was most likely not a limiting factor in growth that prevented stress-induced degradation. Furthermore, other photosynthetic components, such as phycobilisomes, were decomposed (data not shown), indicating that elimination of Cys172 from RuBisCO had not perturbed stress-induced degradation of other cell constituents.
Although none of the conserved Cys172, Cys192, and Cys247 residues in RuBisCO of Synechocystis sp. strain PCC 6803 is vital for catalysis, Cys172 appears to be the main target for redox modulation of enzyme activity and enzyme degradation.
Acknowledgments
This research was supported by United States-Israel Binational Science Foundation (BSF) grant 95-00472/1, by the New Energy and International Technology Development Organization (NEDO)/Research Institute of Innovative Technology for the Earth (RITE, Japan), and by Israeli Science Foundation grant 641/02.
REFERENCES
- 1.Amichay, D., M. Sheffer, and M. Gurevitz. 1992. Restoration of the wild-type locus in an RuBP carboxylase/oxygenase mutant of Synechocystis PCC 6803 via targeted gene recombination. Mol. Gen. Genet. 235:247-252. [DOI] [PubMed] [Google Scholar]
- 2.Amichay, D., R. Levitz, and M. Gurevitz. 1993. Construction of a Synechocystis PCC 6803 mutant suitable for the study of variant hexadecameric ribulose bisphosphate carboxylase/oxygenase enzymes. Plant Mol. Biol. 23:465-476. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, I. 1996. Large structures at high resolution: the 1.6 Å crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complex with 2-carboxyarabinitol bisphosphate. J. Mol. Biol. 259:160-174. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, T. J., and G. H. Lorimer. 1987. Rubisco: structure, mechanisms, and prospects for improvements, p. 131-218. In M. D. Hatch and N. K. Boardman (ed.), The biochemistry of plants, vol. 10. Academic Press, New York, N.Y.
- 5.Badger, M. R., and T. J. Andrews. 1987. Co-evolution of Rubisco and CO2 concentrating mechanisms. Prog. Photosynth. Res. 9:601-609. [Google Scholar]
- 6.Bruggemann, W. 1995. Long-term chilling of young tomato plants under low light. VI. Differential chilling sensitivity of ribulose-1,5-bisphosphate carboxylase/oxygenase is linked to the oxidation of cysteine residues. Plant Cell Physiol. 36:733-736. [Google Scholar]
- 7.Danon, A., and S. P. Mayfield. 1994. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science 266:1717-1719. [DOI] [PubMed] [Google Scholar]
- 8.Desimone, M., A. Henke, and E. Wagner. 1996. Oxidative stress induces partial degradation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase in isolated chloroplasts of barley. Plant Physiol. 111:789-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duff, A. P., T. J. Andrews, and P. M. G. Curmi. 2000. The transition between the open and closed states of Rubisco is triggered by the inter-phosphate distance of the bound bisphosphate. J. Mol. Biol. 298:903-916. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, R. J. 1979. The most abundant protein in the world. Trends Biochem. Sci. 4:241-244. [Google Scholar]
- 11.Ferreira, R. B., and D. D. Davies. 1989. Conversion of ribulose-1,5-bisphosphate carboxylase to an acidic and catalytically inactive form by extracts of osmotically stressed Lemna minor fronds. Planta 179:448-455. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira, R. B., and A. R. N. Teixeira. 1992. Sulfur starvation in Lemna leads to degradation of ribulose bisphosphate carboxylase without plant death. J. Biol. Chem. 267:7253-7254. [PubMed] [Google Scholar]
- 13.Ferreira, R. B., and N. M. Shaw. 1989. Effect of osmotic stress on protein turnover in Lemna minor fronds. Planta 179:456-465. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Ferris, C., and J. Moreno. 1993. Redox regulation of enzymatic activity and proteolytic susceptibility of ribulose-1,5-bisphosphate carboxylase/oxygenase from Euglena gracilis. Photosynth. Res. 35:55-66. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Ferris, C., and J. Moreno. 1994. Oxidative modification and breakdown of ribulose-1,5-bisphosphate carboxylase/oxygenase induced in Euglena gracilis by nitrogen starvation. Planta 193:208-215. [Google Scholar]
- 16.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 17.Huffaker, R. C. 1982. Biochemistry and physiology of leaf proteins, p. 370-400. In D. Boulter and B. Parthier (ed.), Encyclopedia of plant physiology, new series, vol. 14A. Springer-Verlag, Berlin, Germany.
- 18.Huner, N. P. A., and F. D. H. MacDowall. 1978. Evidence for an in vivo conformational change in ribulose bisphosphate carboxylase-oxygenase from Puma rye during cold adaptation. Can. J. Biochem. 56:1154-1161. [DOI] [PubMed] [Google Scholar]
- 19.Jocelyn, P. 1972. Biochemistry of the SH groups. Academic Press, London, United Kingdom.
- 20.Jocelyn, P. 1987. Spectrometric assay of thiols. Methods Enzymol. 143:44-67. [DOI] [PubMed] [Google Scholar]
- 21.Kellogg, E. A., and N. D. Juliano. 1997. The structure and function of Rubisco and their implications for systematic studies. Am. J. Bot. 84:413-428. [PubMed] [Google Scholar]
- 22.Labarre, J., F. Chauvat, and P. Thuriaux. 1989. Insertional mutagenesis by random cloning of antibiotic resistance genes into the genome of the cyanobacterium Synechocystis PCC 6803. J. Bacteriol. 171:3449-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Marcus, Y., and M. Gurevitz. 2000. Activation of cyanobacterial RuBP-carboxylase/oxygenase is facilitated by inorganic phosphate via two distinct mechanisms. Eur. J. Biochem. 267:5995-6003. [DOI] [PubMed] [Google Scholar]
- 25.Marcus, Y., J. A. Berry, and J. Pierce. 1992. Photosynthesis and photorespiration in a mutant of the cyanobacterium Synechocystis PCC 6803 lacking carboxysomes. Planta 187:511-516. [DOI] [PubMed] [Google Scholar]
- 26.Mayaudon, J., A. A. Benson, and M. Calvin. 1957. Ribulose-1,5-diphosphate from and CO2 fixation by Tetraginia expansa leaves extract. Biochim. Biophys. Acta 23:342-351. [DOI] [PubMed] [Google Scholar]
- 27.Mehta, R. A., T. W. Fawcett, D. Porath, and A. K. Mattoo. 1992. Oxidative stress causes rapid membrane translocation and in vivo degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Biol. Chem. 267:2810-2816. [PubMed] [Google Scholar]
- 28.Moreno, J., and R. J. Spreitzer. 1999. C172S substitution in the chloroplast-encoded large subunit affects stability and stress-induced turnover of ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Biol. Chem. 274:26789-26793. [DOI] [PubMed] [Google Scholar]
- 29.Newman, J., and S. Gutteridge. 1993. The X-ray structure of Synechococcus ribulose-bisphosphate carboxylase/oxygenase-activated quaternary complex at 2.2-Å resolution. J. Biol. Chem. 268:25876-25886. [PubMed] [Google Scholar]
- 30.Ranty, B., G. Lorimer, and S. Gutteridge. 1991. An intra-dimeric crosslink of large subunits of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase is formed by oxidation of cysteine 247. J. Biochem. 200:353-358. [DOI] [PubMed] [Google Scholar]
- 31.Rippka, R., J. Druelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Genetic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Microbiol. 111:1-61. [Google Scholar]
- 32.Salvucci, M. E., and W. L. Ogren. 1996. The mechanism of Rubisco activase: insights from studies of the properties and structure of the enzyme. Photosynth. Res. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. Bio/Techniques 8:404-407. [PubMed] [Google Scholar]
- 34.Schloss, J. V., C. D. Stringer, and F. C. Hartman. 1978. Identification of essential lysyl and cysteinyl residues in spinach ribulose bisphosphate carboxylase/oxygenase modified by the affinity label N-bromoacetylethanolamine phosphate. J. Biol. Chem. 253:5707-5711. [PubMed] [Google Scholar]
- 35.Segel, I. H. 1975. Enzyme kinetics. John Wiley, New York, N.Y.
- 36.Takabe, T., and T. Akazawa. 1975. The role of sulphydryl groups in the ribulose-1,5-bisphosphate carboxylase and oxygenase reactions. Arch. Biochem. Biophys. 169:686-694. [DOI] [PubMed] [Google Scholar]
- 37.Tenaud, M., and J. P. Jacquot. 1987. In vitro thiol dependent redox regulation of purified ribulose-1,5-bisphosphate carboxylase. J. Plant Physiol. 130:315-326. [Google Scholar]
- 38.Valle, E. M., N. Carrillo, and R. H. Vallejos. 1982. Functional sulfhydryl groups of ferredoxin-NADP+ oxidoreductase. Biochim. Biophys. Acta 681:412-418. [Google Scholar]
- 39.Weissbach, A., B. L. Horecker, and J. Hurwitz. 1956. The enzymatic formation of phosphoglyceric acid from ribulose diphosphate and carbon dioxide. J. Biol. Chem. 218:795-810. [PubMed] [Google Scholar]
- 40.Woodrow, I. E., and J. B. Berry. 1988. Enzymatic regulation of photosynthetic CO2 fixation in C3 plants. Annu. Rev. Plant Physiol. 39:533-559. [Google Scholar]