Abstract
Rhizobium etli CFN42 contains six plasmids. Only one of them, p42a, is self-conjugative at high frequency. This plasmid is strictly required for mobilization of the symbiotic plasmid (pSym). To study the transfer mechanism of p42a, a self-transmissible cosmid clone containing its transfer region was isolated. Its sequence showed that most of the tra genes are highly similar to genes of Agrobacterium tumefaciens pTiC58 and other related plasmids. Four putative regulatory genes were identified; three of these (traI, traR, and cinR) belong to the LuxR-LuxI family. Mutagenesis of these genes confirmed their requirement for p42a transfer. We found that the conjugative transfer of p42a is dependent on quorum sensing, and consequently pSym transfer also was found to be similarly regulated, establishing a complex link between environmental conditions and pSym transfer. Although R. etli has been shown to produce different N-acyl-homoserine lactones, only one of them, a 3-oxo-C8-homoserine lactone encoded by the traI gene described here, was involved in transfer. Mutagenesis of the fourth regulatory gene, traM, had no effect on transfer. Analysis of transcriptional fusions of the regulatory genes to a reporter gene suggests a complex regulation scheme for p42a conjugative transfer. Conjugal transfer gene expression was found to be directly upregulated by TraR and the 3-oxo-C8-homoserine lactone synthesized by TraI. The traI gene was autoregulated by these elements and positively regulated by CinR, while cinR expression required traI. Finally, we did not detect expression of traM, indicating that in p42a TraM may be expressed so weakly that it cannot inhibit conjugal transfer, leading to the unrepressed transfer of p42a.
Rhizobium species are gram-negative soil bacteria that are able to fix nitrogen and to establish a symbiotic relationship with leguminous plants. Besides the chromosome, their genome is usually constituted by large plasmids which carry genetic material relevant for very diverse functions, such as utilization of plant metabolites, aromatic compounds, and diverse sugars (14), in addition to nodulation and nitrogen fixation, which is the best-studied role of rhizobial plasmids.
Rhizobium etli CFN42 induces the formation of nitrogen fixing nodules in roots of Phaseolus vulgaris (bean) plants. In this strain, the genomic material is distributed between one chromosome and six plasmids (p42a to p42f). One of these plasmids, p42d, has been identified as the symbiotic plasmid (pSym), because it carries most of the genes required for nodulation and nitrogen fixation (43). Some functional characteristics have previously been ascribed to these plasmids (4). The conjugative abilities of the six plasmids have been evaluated by using an Agrobacterium tumefaciens strain lacking the Ti plasmid as a recipient. It was found that p42a is self-transmissible at a high frequency (10−2). Transfer of pSym was also detected but was found to be fully dependent on the presence of p42a. Thus, p42a functions as a helper for the transfer of the pSym, which is a mobilizable but not self-transmissible plasmid. The mechanism for pSym transfer requires its cointegration with p42a, and this cointegration is apparently accomplished through two mechanisms: one dependent on and another independent of recA. Transfer of the other plasmids of R. etli CFN42 has not been detected (5).
Distribution of symbiotic plasmids is a fundamental issue in the Rhizobium soil population. Some transfer genes have been found in rhizobial plasmids such as pNGR234a, the symbiotic plasmid of Rhizobium sp. strain NGR234. These genes were identified through homology to the conjugal transfer genes of Agrobacterium Ti plasmids (13); however, clear data regarding the transfer frequency of this plasmid have not been published. The horizontal transfer of genetic material between bacteria of the genus Rhizobium is a frequent event and has been demonstrated previously under laboratory conditions. Indeed, self-transmissible symbiotic plasmids have been observed in Rhizobium leguminosarum (28). Indirect evidence also suggests pSym transfer in soil (16), and recently, several studies have demonstrated the presence of similar plasmids in different genomic backgrounds (31). An identical organization of plasmid-borne symbiotic genes was found in isolates belonging to four distinct 16S ribosomal DNA species (22). These studies suggest the occurrence of horizontal transfer during the diversification of natural populations of rhizobia. From an applied perspective, it has been possible to increase the effectiveness of the Rhizobium-legume symbiosis with an R. leguminosarum strain through selective transfer of symbiotic plasmids (10).
In Agrobacterium, another genus of the Rhizobiaceae, the tumor-inducing plasmid (pTi) can be transferred between bacterial populations, which remain in the soil after infection of the plant tissues. Opines produced during the infection by the plant are the signal that turns on the expression of the genes required for their utilization. Additionally, they also induce the expression of the transcriptional regulator of the transfer process, traR, which activates traI expression. This regulatory effect is mediated by the AccR repressor (2, 20). The TraR and TraI proteins are LuxR-LuxI-type regulators that activate the expression of target genes in a quorum-sensing-dependent manner. The TraI protein is an acylated homoserine lactone (acyl-HSL) synthase that synthesizes 3-oxo-C8-HSL, and TraR is the autoinducer-responsive transcriptional regulator (40, 25). Both control the expression of at least five promoters of genes involved in plasmid transfer, and consequently, pTi transfer is quorum sensing dependent. A third regulatory protein, TraM, antagonizes TraR activity (26, 35).
The tumor-inducing plasmid pTiC58 of A. tumefaciens seems to be phylogenetically close to p42a. Previous studies from our laboratory have shown that these plasmids are incompatible (15). In addition, Bittinger et al. have identified a region in p42a that contains genes with similarity to vir genes of pTi (3).
Recent data suggest that the cinI locus, belonging to the LuxI family, is involved in conjugative transfer of pRL1JI in R. leguminosarum. Surprisingly, insertion mutations in cinI in both the donor and the recipient are required to decrease conjugation frequency (34). BisR and TriR, two LuxR-type transcriptional regulators, seem to participate in conjugal transfer gene expression, through an elaborate regulatory cascade (53). Also, two quorum-sensing systems have been identified in Sinorhizobium meliloti (36). Based on sequence homologies, those authors propose that a traR-traM locus is involved in conjugation. Nevertheless, data regarding the transfer mechanism for Rhizobium plasmids are still scarce.
In this paper we present the structural and functional characterization of the mobilization region of p42a of R. etli. Although the arrangement of the mobilization region is highly similar to that of pTi, there are important differences among the regulatory mechanisms. Knowledge regarding the regulation of p42a transfer will be an important factor to better understand the distribution of symbiotic information, due to its fundamental role in the conjugative transfer of this plasmid.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are described in Table 1. R. etli strains were grown at 30°C on PY medium (39). Escherichia coli and A. tumefaciens strains were grown on Luria-Bertani medium (38) at 37 and 30°C, respectively. When required, antibiotics were added at the following concentrations: nalidixic acid, 20 μg/ml; spectinomycin, 75 μg/ml; kanamycin, 15 μg/ml; neomycin, 60 μg/ml; rifampin, 100 μg/ml; streptomycin, 50 μg/ml; tetracycline, 2 μg/ml for Rhizobium and 10 μg/ml for E. coli; gentamicin, 15 μg/ml; ampicillin, 200 μg/ml; and carbenicillin, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| R. etli | ||
| CFN42 | Wild type, contains 6 plasmids (p42a to p42f) | 44 |
| CFN2001 | CFN42 derivative (p42a− p42d−) | 32 |
| CFNX182 | CFN42 derivative (p42a−) | 4 |
| CFNX187 | CFNX182 complemented with p42a::Tn5mob | 4 |
| CFNX668 | CFN42 derivative, traM::pSUPΩSp | This work |
| CFNX669 | CFN42 derivative, traI::pSUPΩSp | This work |
| CFNX670 | CFN42 derivative, traR::loxPSp | This work |
| CFNX671 | CFN42 derivative, cinR::loxPSp | This work |
| CFNX672 | CFN42 derivative containing pCT7 | This work |
| CFNX673 | CFN42 derivative containing pCT8 | This work |
| CFNX674 | CFN42 derivative containing pCT9 | This work |
| CFNX675 | CFN42 derivative containing pCT10 | This work |
| CFNX676 | CFNX669 derivative containing pCT7 | This work |
| CFNX677 | CFNX669 derivative containing pCT8 | This work |
| CFNX678 | CFNX669 derivative containing pCT9 | This work |
| CFNX679 | CFNX669 derivative containing pCT10 | This work |
| CFNX680 | CFNX670 derivative containing pCT7 | This work |
| CFNX681 | CFNX670 derivative containing pCT8 | This work |
| CFNX682 | CFNX670 derivative containing pCT9 | This work |
| CFNX683 | CFNX670 derivative containing pCT10 | This work |
| CFNX684 | CFNX671 derivative containing pCT7 | This work |
| CFNX685 | CFNX671 derivative containing pCT8 | This work |
| CFNX686 | CFNX671 derivative containing pCT9 | This work |
| CFNX687 | CFNX671 derivative containing pCT10 | This work |
| CFNX688 | CFN42 containing p42a::Tn5mob and p42d::Tn5GDYN1 | This work |
| CFNX689 | CFNX669 derivative containing p42d::Tn5mob | This work |
| CFNX690 | CFNX670 derivative containing p42d::Tn5mob | This work |
| CFNX691 | CFNX671 derivative containing p42d::Tn5mob | This work |
| CFNX692 | CFN42 derivative containing pC-13 and p42d::Tn5GDYN1 | This work |
| Agrobacterium | ||
| GM19023 | C-58 cured of its native plasmids | 46 |
| NT1 | C-58 harboring plasmid pTiC58Trac::Tn517-52 | 1 |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 47 |
| HB101 | supE44 hsd20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 47 |
| S17-1 | C600::RP-4-2 (Tc::Mu) (Km::Tn7) | 49 |
| S17-1/pSUP5011 | Source of Tn5mob | 49 |
| S17-1/pDR21 | Source of Tn5GDYN1 | 12 |
| Plasmids | ||
| pLAX | pBluescript derivative carrying an 8.5-kb ApaI-XbaI fragment containing from the middle of trbC to the end of trbI of pTiC58 (Apr) | S. K. Farrand, unpublished data |
| pUCM | pCU18 derivative carrying the traM gene of pTiC58 (Apr) | 26 |
| pSEP-1 | pUC18 derivative carrying the traI gene of pTiC58 (Apr) | S. K. Farrand, unpublished data |
| pBADTR3 | pBAD22 derivative carrying a NcoI-HindIII fragment containing the traR gene of pTiC58 (Apr) | S. K. Farrand, unpublished data |
| pTraC1-DG | pET-14b derivative carrying a 2.6-kb NdeI-EcoRI fragment containing the traC, traD, and traG genes of pTiC58 (Apr) | 11 |
| ptraAF | pET-14b derivative carrying a 4.7-kb NdeI-BamHI fragment containing the traA, traF, and traB of pTiC58 (Apr) | 11 |
| pSK+ | Sequencing vector (Apr) | Stratagene, La Jolla, Calif. |
| pBBMCS53 | ΔplacZpBBRIMCS-5 derivative carrying the promoterless uidA gene from pWM5 (Gmr) | 18 |
| pJQ200SK+ | Suicide cloning vector (Gmr) | 42 |
| pJMS2 | pMS102 derivative, loxPSp interposon (Spr) | 37 |
| pSUP202ΩSp | pSUP202 derivative containing an ΩSp cassette in HindIII site | L. Girard, unpublished data |
| pBBRIMCS5 | Broad-host-range cloning vector (Gmr) | 30 |
| pZLR4 | PBBR1MCS5 derivative (Gmr) containing traG::lacZ reporter fusion and the traR gene from pTiC58 | 7 |
| pRK7813 | Broad-host-range cosmid vector, oriV-oriT from RK2, lacZα (Tcr) | 29 |
| pC-13 | pLAFR1 derivative containing the complete mobilization region of p42a | This work |
| pCT1 | pSUP202Ωsp derivative carrying a 254-bp traM EcoRI fragment obtained by PCR with oligonucleotides UtraM-E and LtraR-E | This work/PICK> |
| pCT2 | pSUP202ΩSp derivative carrying a 359-bp traI EcoRI fragment obtained by PCR with oligonucleotides UtraI-E and LtraI-E | This work |
| pCT3 | pSK+ derivative carrying a 0.9-kb XhoI-EcoRI fragment (generated by PCR) containing a fragment of the wild-type traR gene | This work |
| pCT4 | pSK+ derivative carrying a 1.1-kb HindIII-XhoI fragment containing the complete cinR gene and a fragment of traR | This work |
| pCT5 | pSK+ derivative carrying 1.1-kb HindIII-XhoI fragment containing cinR::loxPSp in the SalI site and a fragment of wild-type traR | This work |
| pCT6 | pSK+ derivative carring a 0.9-bp XhoI-EcoRI (generated by PCR) fragment containing traR::loxPSp in the SalI site | This work |
| pCT53 | pJQ200SK+ carrying a 2-kb SpeI fragment containing cinR::loxPSp and wild-type traR, constructed by ligation of the vector with SpeI-XhoI fragments from pCT5 and pCT3 | This work |
| pCT64 | pJQ200SK+ carrying a 2-kb SpeI fragment containing traR::loxPSp and wild-type cinR, constructed by ligation of the vector with SpeI-XhoI fragments from pCT6 and pCT4 | This work |
| pCT7 | pBBMCS53 traI::uidA | This work |
| pCT8 | pBBMCS53 traR::uidA | This work |
| pCT9 | pBBMCS53 traM::uidA | This work |
| pCT10 | pBBMCS53 cinR::uidA | This work |
| pCT11 | pBBRIMCS5 derivative carrying 143-bp HindIII-EcoRI fragment containing the oriT | This work |
| pDCKE 7ΔB | pRK415K derivative carrying a 250-bp BamHI-EcoRI fragment containing the active oriT of pTiC58 | 8 |
| pCT12 | pBBRIMCS5 derivative carrying the complete traI gene | This work |
| pCT13 | pRK7813 derivative carrying the complete traR gene | This work |
| pCT14 | pRK7813 derivative carrying the complete cinR gene | This work |
| pCF424 | pBBRIMCS5 derivative containing traM of pNGR234a | He and Fuqua, submitted for publication |
When conditioned medium was required, 3-ml cultures of wild-type R. etli were grown to stationary phase. The cultures were centrifuged, and the supernatants were filtered through a 0.22-μm-pore-size Millipore filter and added to cultures of other strains.
Bacterial matings.
Conjugation between E. coli and R. etli was done biparentally, using E. coli S17-1 as the donor. Transconjugants were selected with the appropriate antibiotics. Conjugation experiments were performed on PY plates at 30°C, using overnight cultures grown to stationary phase. Donors and recipients were mixed in a 1:2 ratio and incubated overnight. The mixtures were collected and suspended in 1 ml of 10 mM MgSO4-0.01% (vol/vol) Tween 40. Serial dilutions were plated on suitable selective media. The transfer frequency was expressed as the number of transconjugants per output donor.
Transfer experiments to determine dependence on cellular density were performed with donor cultures grown to low density (104 cells/ml) and recipient cultures grown to high density (106cells/ml). Cultures were mixed, and samples of 100 μl of the mixtures were plated on PY and incubated at 30°C. Every 4 h, two samples were collected and suspended in 1 ml of 10 mM MgSO4-0.01% (vol/vol) Tween 40. Serial dilutions were plated on appropriate media to select either transconjugants containing p42a, transconjugants containing pSym, or the donors.
Hybridization experiments.
p42a was purified by the method of Hirsch et al. (23), digested with EcoRI, and used as a probe in colony hybridization experiments (21). A total of 2,200 colonies from a cosmid library of the complete genome of R. etli CFN42 (24) were analyzed. Forty-two colonies which showed homology to the probe were selected for further analysis.
Southern hybridizations.
Cosmid DNA was digested with EcoRI as specified by the manufacturer (Amersham Ltd.). Digested DNA was separated by electrophoresis in 1% agarose gels and transferred to nylon (Hybond N+) as described by Southern (50). Seven bands purified from several vectors containing the transfer genes of pTiC58 (Table 1) were labeled with [α-32P]dCTP by random priming with Amersham's Rediprime system and used as probes. Hybridizations were done as previously described (17).
N-Acyl-HSL detection.
Autoinducers were detected through thin-layer chromatography (TLC) analysis with the reporter plasmid pZLR4 (48). This plasmid contains the traR gene and the traG::lacZ reporter fusion from pTiC58, independently cloned into the broad-host-range vector pBBR1MCS5 (7). Extracts of R. etli derivatives were prepared from 5-ml cultures grown in YM medium (51).
PCR amplification and cloning.
The oligonucleotides used in this study are listed in Table 2. All of the primers were purchased from Unidad de Síntesis Química IBT-UNAM. PCR amplification was carried out with recombinant Taq DNA polymerase as specified by the manufacturer in a Mastercycler 5330 (Eppendorf). PCR conditions consisted of 30 cycles of 92°C for 1 min, 56°C for 1 min, and 72°C for 1 min. PCR products were purified with a Geneclean III kit (Bio101). Vectors were purified by standard protocols (47). T4 polynucleotide ligase was used as indicated by the manufacturer (Amersham Ltd.).
TABLE 2.
Oligonucleotides used in this work
| Primera | Sequenceb |
|---|---|
| −traI-E | TAGAATTCCTTGCGGTTCGGGTTT |
| +traI-E | TCGAATTCACGGCTGTCTCCTT |
| −traR-E | TGGAATTCTCGGTTCGGGGGATTTA |
| +traR-S | CCGTCGACCATCGCCGTTTCAGGTG |
| −traM-E | CTGAATTCGTGCATGGCACCGATTA |
| +traM-S | ATGTCGACGAGGAGCCGACGGTGTT |
| −cinR-E | CTGAATTCTATCGACCGTCGCAACT |
| +cinR-S | GCGTCGACAAAGCCCTCCCGAATGA |
| UtraM-E | TTGAATTCGTCCAGGATTTCCGAGA |
| LtraM-E | TTGAATTCGCAGACGAAACAAACGG |
| UtraI-E | TTGAATTCGCTGCTCCCCGAGGGC |
| LtraI-E | TCGAATTCAGGATGCCAGCGACGG |
E corresponds to EcoRI sites, and S corresponds to SalI sites.
Nucleotides corresponding to the restriction sites are underlined.
Transcriptional fusions.
A PCR product containing the complete traI gene was obtained with oligonucleotides +traI-E and −traI-E (Table 2) and cloned in pSK+ (Table 1). A 991-bp EcoRI-ClaI fragment from this clone was subcloned into plasmid pBBMCS53 (Table 1) to construct a traI::uidA reporter fusion containing the first 15 amino acids (aa) of repA, the intergenic region between repA and traI, and the first 195 aa of traI.
A traR::uidA fusion was generated by cloning a PCR product obtained by using the oligonucleotides +traR-E and −traR-S (Table 2). This product, containing the complete traR gene, was cloned in pSK+ (Table 1). A 514-bp EcoRI-HindIII fragment from this clone, containing 59 bp of noncoding region and the first 150 aa of traR, was subcloned into plasmid pBBMCS53 (Table 1).
A traM::uidA fusion was constructed by cloning a 747-bp EcoRI-SalI fragment, obtained from a PCR product generated by using the oligonucleotides −traM-E and +traM-S, into plasmid pBBMCS53 (Table 1). This fragment contains 580 bp of noncoding region and the first 49 aa of traM.
A cinR::uidA fusion was constructed by cloning a PCR product obtained by using oligonucleotides +cinR-S and −cinR-E (Table 2). This product, containing the complete cinR gene, was cloned in pSK+ (Table 1). A 469-bp SmaI-ClaI fragment from this clone, containing the last 6 aa of trbI, the noncoding region between trbI and cinR, and the first 150 aa of traR, was subcloned into plasmid pBBMCS53 (Table 1).
Measurement of β-glucuronidase activity.
Cultures of R. etli derivatives harboring transcriptional fusions were grown to stationary phase. Quantitative uidA activity was measured in 1-ml culture samples with p-nitrophenyl glucuronide as a substrate (18).
Construction of mutant derivatives.
traM and traI mutants were constructed by interruption with pSUPΩSp (Table 1) introduced by single recombination. traM was mutagenized by using plasmid pCT1 (pSUP202ΩSp with a 254-bp EcoRI fragment of traM [Table 1]). Recombination creates two incomplete copies of the gene. One of them lacks 43 bp of the carboxy-terminal end, ending at nucleotide position 296, while the other lacks 44 bp of the amino-terminal end, starting at nucleotide position 44. traI was mutagenized by using plasmid pCT2 (pSUP202ΩSp with a 359-bp EcoRI fragment of traI [Table 1]). One of the interrupted copies lacks 78 bp of the carboxy-terminal end, ending at nucleotide position 543, while the other lacks 258 bp of the amino-terminal end, starting at nucleotide position 258 (Table 1).
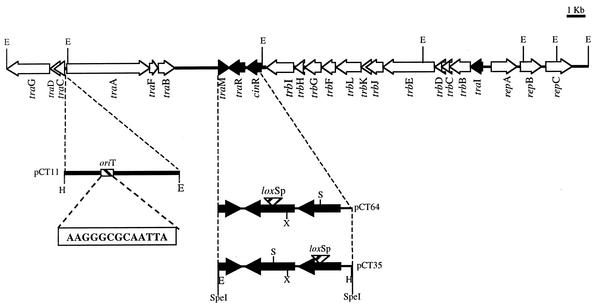
The traR and cinR mutants were generated by insertion of a lox::PSp cassette by using plasmids pCT64 and pCT53, respectively, to generate double recombinants (Table 1). cinR was interrupted at the SalI site at nucleotide position 414 (Fig. 1). traR was interrupted at the SalI site at nucleotide position 248 (Fig. 1).
FIG. 1.
Physical map of pC-13, containing the transfer region of p42a. Arrows indicate the transcriptional directions of the identified genes. The regulator genes traI, traR, cinR, and traM are shown by black arrows. pCT11 contains the 143-bp mobilizable region, carrying the oriT of p42a. The 13 bp of the nic site sequence are shown. pCT53 and pCT64 contain the regions used to generate mutations in the traR and cinR genes. H, HindIII; E, EcoRI; X, XhoI; S, SalI.
Nucleotide sequence accession number.
The sequence of the complete tra region of p42a of R. etli, cloned in pC-13, was deposited in the GenBank database under accession number AF528525.
RESULTS
Identification of a self-transmissible cosmid clone from p42a.
Plasmid p42a has been shown to be self-transmissible (5). Its high transfer frequency remains constant, even if it is mobilized from a different background such as A. tumefaciens GMI9023 (Table 3), thus indicating that no additional transfer regulatory elements are located in a replicon other than p42a. To identify the transfer region of p42a, a collection of clones from this plasmid was isolated from a cosmid library covering the whole R. etli CFN42 genome (24) through the selection of clones showing a positive signal in Southern blot colony hybridization with the complete p42a as a probe. Forty-two clones were selected, digested with EcoRI, and hybridized against transfer genes of pTiC58 from A. tumefaciens (Table 1) under low-stringency conditions. Most of the probes (traAFB, traCDG, trbC-I, and traI) showed positive signals with a 27.1-kb cosmid clone. This cosmid was called pC-13. We did not find hybridization signals for traM and traR.
TABLE 3.
Transfer frequencies of R. etli plasmids p42a, pSym, and pC-13 from different donorsa
| Transfer | Donor | Relevant genotype | Recipient | Transfer frequencyb |
|---|---|---|---|---|
| p42a | CFNX187 | p42a::Tn5mob | GMI9023 | 1.0 × 10−2 |
| GMI9023/p42a | p42a::Tn5mob | GMI9023 | 1.0 × 10−2 | |
| CFNX187/pCF424 | p42a::Tn5mob, traM (pNGR234a) | GMI9023 | 6.3 × 10−4 | |
| CFNX668 | traM::pSUPΩSp | CFN2001(p42a− pSym−) | 1.0 × 10−2 | |
| CFNX669 | traI::pSUPΩSp | CFN2001(p42a− pSym−) | NDc | |
| CFNX670 | traR::loxPSp | CFN2001(p42a− pSym−) | ND | |
| CFNX671 | cinR::loxPSp | CFN2001(p42a− pSym−) | ND | |
| CFNX669/pCT12 | traI::pSUPΩsp, pBBR1MCS5/traI | CFN2001(p42a− pSym−) | 8.7 × 10−3 | |
| CFNX670/pCT13 | traR::loxPSp, pRK7813-traR | CFN2001(p42a− pSym−) | 7.3 × 10−3 | |
| CFNX671/pCT14 | cinR::loxPSp, pRK7813-cinR | CFN2001(p42a− pSym−) | 8.5 × 10−3 | |
| p42a tra region, cloned in pC-13 | CFN42/pC-13 | Cosmid with all p42a tra genes | GMI9023 or DH5α | 1.2 × 10−2 |
| GMI9023/pC-13 | Cosmid with all p42a tra genes | GMI9023 or DH5α | 1.0 × 10−2 | |
| Cloned oriT from p42a | CFN42/pCT11 | pBBRIMCS5::oriT of p42a | DH5α | 1.0 × 10−2 |
| CFN42/pBBRIMCS5 | pBBRIMCS5 (vector) | DH5α | ND | |
| CFNX182/pCT11 | p42a−/pBBRIMCS5::oriT of p42a | DH5α | ND | |
| Cloned oriT from pTiC58 | CFN2001/p42a+ pDCKE.7ΔB | p42a::GDYN1, oriT of pTiC58 | HB101 | 2.5 × 10−4 |
| CFN2001/pDCKE.7ΔB | pDCKE.7ΔB | HB101 | ND | |
| p42d (pSym) | CFNX689 | traI::pSUPΩSp, pd::Tn5GDYN1 | CFN2001(p42a− pSym−) | ND |
| CFNX690 | traR::loxPSp, pd::Tn5GDYN1 | CFN2001(p42a− pSym−) | ND | |
| CFNX691 | cinR::loxPSp, pd::Tn5GDYN1 | CFN2001(p42a− pSym−) | ND | |
| CFNX692 | pC-13, pd::Tn5GDYN1 | CFN2001(p42a− pSym−) | 3 × 10−6 |
All crosses were repeated at least twice.
Expressed as transconjugants per donor.
ND, not detected
The conjugative ability of pC-13 from different backgrounds was tested. This cosmid showed the same transfer frequency as the complete p42a, either from R. etli derivatives lacking one or more plasmids or from A. tumefaciens GMI9023 (Table 3). These experiments show that pC-13 carries all of the genes necessary for p42a transfer. As mentioned above, we have previously shown that p42a is able to mobilize the pSym by a cointegration-mediated mechanism (5). In this work we tested whether pC-13 could also do that. Our results showed that pC-13 is also able to mobilize pSym (Table 3). Analysis of the transconjugants indicated that the pC-13-mediated transfer of pSym in all cases involved a cointegration event, similar to what happens with p42a. In contrast, we did not detect transfer of pC-13 from E. coli HB101 even though several different recipients were utilized. This suggests that the transfer genes contained in pC-13 are not efficiently expressed in an E. coli background.
Sequence analysis of the tra genes contained in pC-13.
The complete DNA sequence of pC-13 was determined. It revealed 21 open reading frames (ORFs) whose predicted products are similar to known transfer proteins of A. tumefaciens pTiC58 and related plasmids (Fig. 1). The gene arrangement in pC-13 was similar to that found in pTiC58 (11, 33), except that no opine catabolism-related genes were identified. The transfer origin (oriT) was located between traCDG and traAFB, which are transcribed divergently from this site. The oriT was identical to the oriT of pNGR234a and also to the oriT of pTiC58.
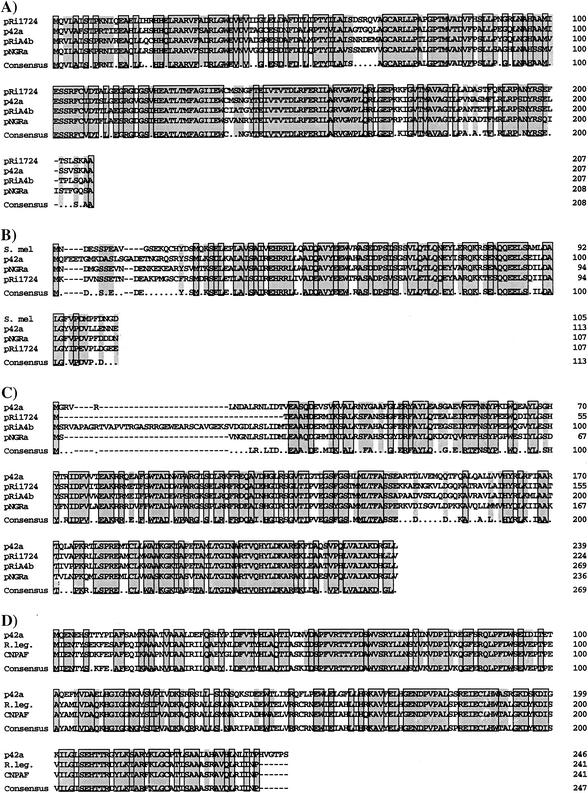
Four ORFs encoding regulator proteins were identified in pC-13. Homologous proteins for three of them have been previously characterized for the pTiC58 transfer system. Their products, TraR, TraM, and TraI, are highly conserved with those from other related plasmids (Fig. 2). However, the identity of the pC-13 genes to those from pTiC58 was low (31% for TraR, 29% for TraM, and 50% for TraI). This could explain why we were not able to detect signals for traR and traM genes in our hybridization experiments (see above). TraR of pC-13 is a 239-aa protein, showing 60% identity to the LuxR family quorum-sensing transcriptional factor of pRiA4b (a plasmid of A. tumefaciens) (AB050904), 62% identity to the TraR protein of pRi1724 of Rhizobium rhizogenes (NC_002575), and 58% identity to TraR of pNGR234a of Rhizobium sp. strain NGR234 (AE00069).
FIG. 2.
Alignment of p42a transfer regulator proteins with the corresponding proteins of related systems. S. mel., S. meliloti; pNGRa, plasmid pNGR234a; R. leg., R. leguminosarum. Identical amino acids are boxed, and conserved amino acids are shown in the consensus. (A) TraI; (B) TraM; (C) TraR; (D) CinR.
We found another regulator gene, cinR. CinR from pC-13 is a 246-aa protein that is 59% identical to CinR of R. leguminosarum bv. Viciae (AF210630) and 57% identical to CinR of R. etli CNPAF512 (AF393621). Both CinR and TraR are clearly proteins belonging to the LuxR family of transcriptional regulators.
TraI of pC-13 is a 207-aa protein with 75% identity to TraI of pRi1724 (NC_002575), 74% identity to an autoinducer synthase of pRiA4b (AB050904), and 70% identity to TraI of pNGR234a (AE000068). Therefore, TraI is evidently an acylated HSL synthase member of the LuxI family proteins. In the intergenic region between traI and repA, a tra box with the sequence GTTGTAGAATCCTACAAG was found 59 bp upstream of traI, suggesting that the transcription of traI is dependent on TraR. Another tra box was localized upstream of traC.
The fourth regulator, TraM, is a 113-aa protein homologous to an antiactivator that inhibits TraR activity through interaction with this protein in domains located at their respective C termini. It shows 69% identity to TraM of pNGR234a (AE000069), 65% identity to TraM of pRiA4b (AB050904), and 57% identity to TraM of pRi1724 (NC_002575). All of these proteins show low identity to transfer regulator proteins of pTiC58 (NC_003065).
ORFs similar to trbB, -C, -D, -E, -J, -K, -L, -F, -G, -H, and -I, involved in mating-pair formation (33), were also found between traI and cinR; all of them are transcribed in the same direction as traI. Replication genes repABC are located upstream of traI (Fig. 1).
Characterization of oriT.
The oriT of pC-13, identified through similarity with transfer origins of pNGR234a and pTiC58, was localized between traA and traC. The sequence analysis revealed a motif identical to that present in the oriT region of pTiC58, which is common to the region where nick sites are located in pTF1 and RSF1010. However, the inverted repeats near this motif (8) seem to be absent. To test the functionality of this transfer origin, a fragment of 143 bp containing the oriT was cloned in pBBR1MCS5 (pCT11) (Fig. 1) and introduced by conjugation into different strains. pCT11 could be mobilized from wild-type CFN42 but not from CFNX182, a strain lacking p42a (Table 3). As a control we tested the mobilization ability of the vector pBBR1MCS5. As can be seen in Table 3, pBBR1MCS5 could not be transferred. These results indicate that the fragment cloned in pCT11 carries a functional oriT dependent upon p42a-encoded proteins. Similar results were found with plasmid pDCKE.7ΔB, which contains the oriT of pTiC58 (Table 3); it can be mobilized from a strain containing p42a but not from a strain lacking this plasmid.
traI is responsible for the production of an autoinducer in R. etli CFN42.
In pTiC58, TraI is responsible for the production of a conjugal factor or autoinducer, which is a member of a family of substituted HSLs (acyl-HSLs) (25). To determine if an acyl-HSL is involved in the transfer of p42a, the production of these compounds by R. etli CFN42 and some derivatives was determined by using the TLC system described by Shaw et al. (48).
Acyl-HSLs can differ with respect to the length of the acyl side chain, the presence or absence of one or more unsaturations, and the nature of the substitution at the carbon 3 position. In some acyl-HSLs, carbon 3 is hydroxylated (3-hydroxyl); it also may carry a carbonyl group (3-oxo) or a fully reduced methylene. The 3-oxo derivatives characteristically produce tailing spots with diffuse edges, whereas the 3-unsubstituted forms produce circular spots with sharp edges (48). To determine whether the traI gene found in pC-13 is responsible for production of one of the autoinducers, we analyzed the autoinducer production in strain CFNX669 (traI::pSUPΩSp). The results showed that there is one tailing reactive spot which comigrates with a 3-oxo-C8-HSL. This 3-oxo-C8-HSL is detected in the wild-type strain but disappears in the traI mutant (Fig. 3, lanes 2 and 6) and in the p42a-cured derivative (Fig. 3, lane 3). There is another reactive spot that comigrates with the 3-oxo-C8-HSL, and its circular form indicates that it is a 3-OH-C8-HSL. This spot is present in strain CFNX182 (cured of p42a) (Fig. 3, lane 3), indicating that it is encoded in the chromosome or in a plasmid different from p42a. Two unidentified spots were detected in the wild type at low levels; they may be contaminants, similar to those previously detected with this technique (7).
FIG. 3.
Thin-layer chromatogram of the acyl-HSL produced by R. etli CFN42 and derivatives. Lane 1, A. tumefaciens NT1(pTiC58ΔaccR); lane 2, CFN42; lane 3, CFNX182 (cured of p42a); lane 4, CFNX671 (cinR::loxPSp); lane 5, CFNX670 (traR::loxPSp); lane 6, CFNX669 (traI::pSUPΩSp); lane 7, CFNX668 (traM::pSUPΩSp); lane 8, 3-oxo-C8-HSL; lane 9, YM medium control.
In order to determine the role of the 3-oxo-C8-HSL in conjugative transfer, the mobilization of p42a containing the interrupted traI was tested and found not to be detectable (Table 3). Complementation with the wild-type gene (CFNX669/pCT12) restored the transfer ability of the strain (Table 3). Also, when the traI mutant CFNX699 was grown in a conditioned medium consisting of the supernatant of a CFN42 culture grown to stationary phase or in a medium supplemented with 5 μl of 3-oxo-C8-HSL, the transfer frequency of p42a traI::pSUPΩSp was similar to that of the wild-type strain (1.7 × 10−2 and 0.77 × 10−2, respectively). Our conclusion from these experiments is that TraI is a positive regulator of the transfer process through the synthesis of a 3-oxo-C8-HSL. On the other hand, in spite of the fact that CFN42 produces at least two acyl-HSLs at readily detectable levels, only the one encoded by the traI localized on p42a seems to be involved in conjugative transfer; no other product can substitute for its function, indicating very specific regulation.
Mobilization of p42a, and consequently of pSym, depends on quorum sensing.
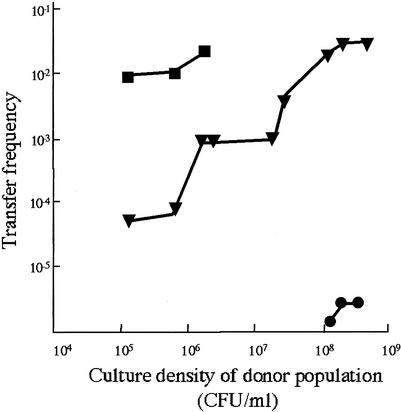
Acyl-HSLs are molecules involved in quorum-sensing processes, processes that are cell density and growth phase dependent. We have shown that one of these compounds is necessary for p42a mobilization. We also know that the transfer of pSym depends on the presence of p42a (5). Thus, to test if transfer of R. etli plasmids is a quorum-sensing-dependent process, we constructed an R. etli derivative harboring both p42a and p42d (pSym) labeled with different markers (CFNX688). We determined the transfer frequencies of both plasmids at different growth stages of the donor strain (Fig. 4). The results showed that transfer of p42a initiates when the donor population density is approximately 105 and the frequency increases as the donor population augments its density. Furthermore, when the donor strain is pregrown for 6 h in a conditioned medium, transfer is induced at an earlier stage (Fig. 4). These results clearly indicate that the transfer of p42a is dependent on quorum sensing.
FIG. 4.
Effect of culture density on the transfer of p42a and pSym from CFNX688. Transfer frequencies of p42a (triangles), of p42a with conditioned medium added before mating (squares), and of pSym (circles) are shown. Strain CFN2001 (cured of p42a and pSym) was always used as the recipient.
The transfer of pSym initiates only after the p42a transfer frequency reaches its highest level (at a population density of 108 to 109), and it also increases in accordance with the donor population (Fig. 4). Therefore, the transfer of pSym is also dependent on quorum sensing, albeit indirectly.
Two regulators of the LuxR family are activators of p42a transfer.
To analyze the role of cinR and traR in p42a mobilization, we tested the transfer ability of p42a in derivatives containing mutations in these genes (Table 3). Transfer of p42a was completely abolished in derivatives containing mutations in traI, cinR, or traR and was restored when complemented with the respective wild-type genes (CFNX669/pCT12, CFNX670/pCT13, and CFNX671/pCT14) (Table 3). Therefore, the three genes are necessary and sufficient for p42a transfer. Since mobilization of pSym depends on the p42a transfer system, we confirmed that these mutations also eliminate pSym transfer (Table 3).
To further understand the regulation of the p42a transfer genes, we constructed derivatives containing fusions of each of the regulatory genes to a reporter gene: pCT7 (traI::uidA), pCT8 (traR::uidA), and pCT10 (cinR::uidA). Expression of these fusions was tested in the wild-type and traI, traR, and cinR mutant backgrounds. The analysis (Table 4) showed that traI expression is reduced when traR is mutated (CFNX680). A more dramatic effect was observed in cinR or traI mutants (CFNX684 and CFNX676), where traI expression was completely abolished. These results led us to predict that production of the traI-dependent acyl-HSL should be affected in these mutants. TLC detection analysis demonstrated that in CFNX669, CFNX670, and CFNX671, the reactive-spot profile was similar to the profile of strain CFNX182 (cured of p42a), while in CFNX668 (traM mutant), the acyl-HSL profile was identical to that of the wild type (Fig. 3). Regarding cinR, its expression was reduced only in a traI mutant background (CFNX679) (Table 4), while the other mutations had no effect on its expression. Finally, traR expression was similar in all of the different backgrounds (Table 4), suggesting that it is constitutively expressed.
TABLE 4.
Expression of traI::uidA, traR::uidA, and cinR::uidA gene fusions in R. etli wild-type and mutant backgrounds
| Strain | Strain genotype | Mean β-glucuronidase sp act ± SDa |
|---|---|---|
| CFNX672 | Wild type, traI::uidA | 116.53 ± 16.62 |
| CFNX676 | traI::ΩSp traI::uidA | 13 ± 3.03 |
| CFNX680 | traR::loxSp traI::uidA | 19.15 ± 3.26 |
| CFNX684 | cinR::loxSp traI::uidA | NDb |
| CFNX673 | Wild type, traR::uidA | 15.39 ± 5.15 |
| CFNX677 | traI::ΩSp traR::uidA | 15.035 ± 1.9 |
| CFNX681 | traR::loxSp traR::uidA | 20.69 ± 4.91 |
| CFNX685 | cinR::loxSp traR::uidA | 18.98 ± 2.79 |
| CFNX675 | Wild type, cinR::uidA | 20.59 ± 2.073 |
| CFNX679 | traI::ΩSp cinR::uidA | NDb |
| CFNX683 | traR::loxSp cinR::uidA | 24.54 ± 2.35 |
| CFNX687 | cinR::loxSp cinR::uidA | 20.94 ± 3.57 |
| CFNX182/pCT7+pCT13 | p42a−, pRK7813-traR, traI::uidA | NDb |
| CFNX182/pCT7+pCT14 | p42a−, pRK7813-cinR, traI::uidA | NDb |
Values are expressed as nanomoles per minute per milligram of protein and are means from two independent experiments performed in duplicate.
ND, not detected.
Since cinR and traR mutants affect traI expression and this gene is responsible for 3-oxo-C8-HSL production, we performed an experiment to determine whether 3-oxo-C8-HSL allows cinR and traR mutants to perform conjugative transfer. The results showed that transfer ability was recovered only at a low frequency in the traR mutant (1.2 × 10−6), and in the cinR mutant (3.7 × 10−5). These results indicate that in addition to their role in traI expression, both regulators participate directly in regulation of tra genes expression.
The traI fusion (pCT7) was introduced into a derivative of the p42a-cured strain CFNX182 carrying the clone containing the traR gene (pCT13) or the cinR gene (pCT14). The β-glucuronidase activity in cultures of these strains grown in conditioned medium was determined. It was found that traI is not expressed in these backgrounds (Table 4). These results suggest that both genes are required for traI expression and that they function in parallel. This is in agreement with previous results (see above), since the wild-type strain carrying mutations in either traR or cinR shows a decrease in traI expression and transfer, in spite of the fact that traR has no effect on cinR expression and vice versa.
TraM does not repress conjugative transfer in p42a.
TraM has been shown to be a negative regulator of pTiC58 conjugative transfer through interaction with TraR in domains located at their respective C termini (26, 27, 41). To verify whether the traM is an antiactivator in p42a, we constructed a derivative (CFNX668) containing a mutation in traM (see Materials and Methods) and tested its conjugative ability. Surprisingly, the transfer frequency was not affected (Table 3). Since we knew by sequence analysis that the C terminus of TraR was conserved with those of other homologous proteins of related pTi plasmids, we questioned whether it could be inactivated by an heterologous TraM repressor. To this end, pCF424 (Table 1), a pBBR1MCS5 derivative containing traM of pNGR234a, was introduced into CFNX187 by conjugation. In this strain, we found that the p42a transfer frequency decreased by 2 orders of magnitude (Table 3). This indicates that traR of p42a is able to interact with TraM but, for some unknown reason, this interaction is not carried out with the endogenous TraM. To further explore this issue, a transcriptional fusion of traM with the β-glucuronidase reporter (pCT9) was constructed. We did not detect expression in any of the backgrounds tested, i.e., wild-type CFN42 or the mutants CFNX669 (traI::pSUPΩsp), CFNX670 (traR::loxPSp), or CFNX671 (cinR::loxPSp) (data not shown). We concluded that TraR in CFN42 is not inactivated because the traM gene is not expressed, and thus the antiactivator is not produced, at least under the experimental conditions tested.
DISCUSSION
In the last few years, it has been found that quorum-sensing-dependent processes are widespread among gram-negative bacteria, including plant-associated bacteria (52). Production of multiple autoinducers and complex regulatory networks has been reported for R. etli, R. leguminosarum, and S. meliloti (19, 36, 45, 53, 54). R. etli CNPAF512 produces seven different acyl-HSLs (45). Three of these autoinducers have been shown to be produced inside the nodules by bacteroids; mutations in one of them result in a substantial decrease in acetylene reduction activity, indicating that they are involved in nodulation and nitrogen fixation (9). In R. etli CFN42 we detected the production of only two acyl-HSLs. Strains CNPAF512 and CFN42 also differ in other plasmid-encoded features, such as the presence of a FixJ homolog in CNPAF512, which is absent in CFN42, and the presence of a novel FixL in the latter strain (18). The organization of these genes was analyzed in 24 R. etli strains from different geographic origins. Seventeen (70%) were similar to CFN42 (6). All of these data support the suggestion that in R. etli there is intraspecies variability regarding different plasmid-encoded traits.
In R. leguminosarum, acyl-HSLs have been related to inhibition of bacterial growth through the production of a small bacteriocin (19, 53, 54). An intricate regulatory cascade involving various luxI-luxR-type regulatory genes (cinR, bisR, and triR) has been shown to participate in regulation of the conjugative transfer of a symbiotic plasmid in R. leguminosarum (34, 53). In this cascade, TriR positively regulates conjugative transfer genes in response to the 3-oxo-C8-HSL synthesized by traI. triR expression is positively controlled by BisR; this induction requires 3OH-C14:1-HSL synthesized by the chromosomally encoded CinI. Also, BisR plays a role in repression of cinI expression. Although a trb operon was identified, localization of the transfer origin, oriT, and other transfer genes (traAFB and traCDG), as well as analysis of traI, will surely contribute to complete the regulatory scheme for transfer of this plasmid. In addition, a recently described traR-traM locus from S. meliloti has been proposed to be involved in transfer (36); however, experimental confirmation is still required.
In this work, we performed a comprehensive analysis of the conjugative transfer mechanism of p42a of R. etli and distinctly showed that it is quorum sensing dependent. Interestingly, as pSym transfer depends on p42a, we find that the transfer of the symbiotic plasmid also depends on cell density, in an even more restricted manner than for p42a.
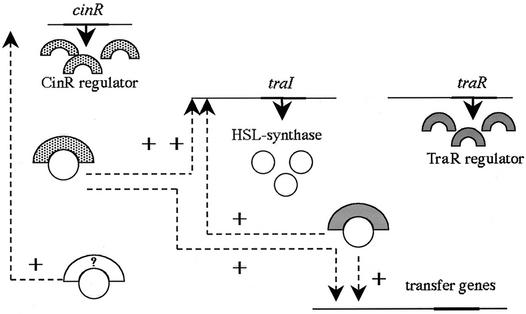
Based on the expression assays of regulatory gene fusions in different backgrounds, we propose a regulatory model for p42a transfer (Fig. 5). Analysis of β-glucuronidase activity demonstrated that traI is positively autoregulated and is upregulated by TraR and CinR. According to the proposed model, a complex should be formed by TraR and the 3-oxo-C8-HSL synthesized by TraI. This complex would be able to bind to the promoters of transfer genes where tra boxes have been identified. This complex would also mediate the autoregulatory control of traI, where a tra box was also found. A mutation in traR still allows a low level of traI expression, but TraI activity is not high enough to synthesize the autoinducer required to promote transfer of p42a, or even to be detected by TLC analysis. Also, CinR was found to be a positive regulator of traI independently of TraR, because a mutation in cinR abolishes traI expression but has no effect on traR. As mentioned above (see Results), the transfer frequency of pC-13 is similar to that of the complete plasmid p42a. This indicates that no other regulators are involved in transfer of p42a. Therefore, we predict that the 3-oxo-C8-HSL produced by TraI is able to form a complex with CinR and to upregulate traI (Fig. 5).
FIG. 5.
Model for the regulation of transfer gene expression in p42a. +, positive regulation; ?, unknown regulatory protein from the LuxR family.
Regarding cinR, its expression is upregulated by TraI and decreases when traI is mutated. As CinR is a positive regulator for traI, we would expect a decrease of cinR expression in a cinR mutant. However, the results show that cinR expression remains unaltered in a cinR mutant. We propose that traI is transcribed at a basal level independent of TraR and CinR, allowing the accumulation of enough 3-oxo-C8-HSL to activate cinR. In the traI mutant, this basal level produces an inactive TraI, completely abolishing autoinducer production. The transcriptional activator for cinR has not been defined. This hypothetical activator could be either TraR or CinR, acting separately and being able to replace each other, or an unknown and yet-to-be-described regulator. Finally, traR expression was found to be constitutive under all conditions tested.
Regulation of p42a transfer resembles that of pTiC58 in many aspects. Nevertheless, they differ in some important aspects. In pTiC58, opines secreted by the plant are the first signal for transfer. In contrast, p42a mobilization does not require its symbiotic host. Another important difference among the transfer systems of these plasmids concerns TraM activity. According to Piper and Farrand (41), in pTiC58 TraM serves to inactivate the small amount of TraR which is constitutively produced in the absence of the conjugal opines. As a consequence, conjugal transfer of this plasmid requires not only the accumulation of autoinducer but also expression of traR at levels that allow it to overcome the activity of TraM. In contrast, we found no expression of TraM in p42a. As opines are not required to initiate conjugal transfer, we believe that traM might be a relic of an acquired transfer machine in R. etli. The absence of the inhibitory effect of TraM, as well as the fact that p42a is able to initiate transfer at a lower culture density than pTiC58, may explain the high frequency of p42a transfer (compare Fig. 4 with Fig. 2 of reference 41). However, we cannot rule out the possibility that TraM might be expressed and have a regulatory role under other experimental conditions.
From the results presented in this paper, it is evident that there is a regulatory cascade controlling conjugative transfer of p42a and an even more complex regulation for pSym transfer. To complete the regulatory scheme, it will be necessary to identify other controlling elements, such as the transcriptional regulator of cinR.
We have previously shown both that pSym transfer depends on the presence of p42a and the feasibility of cointegration among both plasmids (5). Here, we show the existence of another level for pSym transfer regulation, indirectly linked to the environmental conditions, which may have an effect on the cellular density. Although p42a transfer is not restricted to the presence of the symbiotic host (see above), we expect that the cellular population is more likely to achieve the level required for plasmid transfer in a closed environment, such as the nodule or infection thread, than in soil.
Acknowledgments
We are grateful to Laura Cervantes, Javier Rivera, and Rosa Gómez Barreto for technical assistance and to Stephen K. Farrand for his help in the detection of acyl-HSL and for providing tra probes. We also thank Paul Gaytán and Eugenio López for the synthesis of oligonucleotides.
This work was partially supported by grants IN202599 (DGAPA-UNAM) and 102375 and 102366 (DGEP-UNAM).
REFERENCES
- 1.Beck von Bodman, S., J. E. McCutchan, and S. K. Farrand. 1989. Characterization of conjugal transfer functions of Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 171:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck von Bodman, S., G. T. Hayman, and S. K. Farrand. 1992. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl. Acad. Sci. USA 89:643-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bittinger, M. A., J. A. Gross, J. Widom, J. Clardy, and J. Handelsman. 2000. Rhizobium etli CE3 carries vir gene homologs on a self-transmissible plasmid. Mol. Plant-Microbe Interact. 13:1019-1021. [DOI] [PubMed] [Google Scholar]
- 4.Brom, S., A. García de los Santos, T. Stepkowsky, M. Flores, G. Dávila, D. Romero, and R. Palacios. 1992. Different plasmids of Rhizobium leguminosarum bv. Phaseoli are required for optimal symbiotic performance. J. Bacteriol. 174:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brom, S., A. García de los Santos, L. Cervantes, R. Palacios, and D. Romero. 2000. In Rhizobium etli symbiotic plasmid transfer, nodulation competitivity and cellular growth require interaction among different replicons. Plasmid 44:34-43. [DOI] [PubMed] [Google Scholar]
- 6.Brom, S., L. Girard, A. García-de los Santos, J. M. Sanjuán-Pinilla, J. Olivares, and J. Sanjuán. 2002. Conservation of plasmid-encoded traits among bean-nodulating Rhizobium species. Appl. Environ. Microbiol. 68:2555-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha, C., P. Gao, Y. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 8.Cook, D. M., and S. K. Farrand. 1992. The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T-region borders. J. Bacteriol. 174:6238-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels, R., D. E. De Vos, J. Desair, G. Raedschelders, E. Luyten, V. Rosemeyer, C. Verreth, E. Schoeters, J. Vanderleyden, and J. Michiels. 2002. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J. Biol. Chem. 277:462-468. [DOI] [PubMed] [Google Scholar]
- 10.DeJong, T. M., N. J. Brewin, W. B. Johnston, and D. A. Phillips. 1982. Improvement of symbiotic properties in Rhizobium leguminosarum by plasmid transfer. J. Gen. Microbiol. 128:1829-1838. [Google Scholar]
- 11.Farrand, S. K., I. Hwang, and D. M. Cook. 1996. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 178:4233-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores, M., S. Brom, T. Stepkowski, L. Girard, G. Dávila, D. Romero, and R. Palacios. 1993. Gene amplification in Rhizobium: identification and in vivo cloning of discrete amplifiable DNA region (amplicons) from Rhizobium leguminosarum bv. phaseoli. Proc. Natl. Acad. Sci. USA 90:4932-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 14.García de los Santos, A., S. Brom, and D. Romero. 1996. Rhizobium plasmids in bacteria-legume interactions. World J. Microbiol. Biotechnol. 12:119-125. [DOI] [PubMed] [Google Scholar]
- 15.García de los Santos, A., and S. Brom. 1997. Characterization of two plasmid-borne lpsβ loci of Rhizobium etli required for lipopolysaccharide synthesis and for optimal interaction with plants. Mol. Plant-Microbe Interact. 10:891-902. [DOI] [PubMed] [Google Scholar]
- 16.Geniaux, E., G. Laguerre, and N. Amarger. 1993. Comparison of geographically distant populations of Rhizobium isolated from root nodules of Phaseolus vulgaris. Mol. Ecol. 2:1-8. [Google Scholar]
- 17.Girard, M. L., M. Flores, S. Brom, D. Romero, R. Palacios, and G. Dávila. 1991. Structural complexity of the symbiotic plasmid of Rhizobium leguminosarum bv. Phaseoli. J. Bacteriol. 173:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girard, L., S. Brom, A. Dávalos, O. López, M. Soberón, and D. Romero. 2000. Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol. Plant-Microbe Interact. 13:1283-1292. [DOI] [PubMed] [Google Scholar]
- 19.Gray, K. M., J. P. Pearson, J. A. Downie, B. E. A. Boboye, and E. P. Greenberg. 1996. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J. Bacteriol. 178:372-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green, E. A., and P. C. Zambrysky. 1993. Agrobacteria mate in opine dens. Curr. Biol. 3:507-509. [DOI] [PubMed] [Google Scholar]
- 21.Grunstein, M., and D. G. Hogness. 1975. Colony hybridization. A method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl. Acad. Sci. USA 72:3961-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrera-Cervera, J. A., J. Caballero-Mellado, E. Romero-Martínez, J. Olivares, and J. Sanjuán. 1999. At least five rhizobial species nodulate Phaseolus vulgaris in Spanish soil. FEMS Microbiol. Ecol. 30:87-97. [Google Scholar]
- 23.Hirsch, P. R., M. Van Montagu, A. W. B. Johnston, N. J. Brewin, and J. Schell. 1980. Physical identification of bacteriocinogenic, nodulation and other plasmids in strains of R. leguminosarum. J. Gen. Microbiol. 120:403-412. [Google Scholar]
- 24.Huerta-Zepeda, A., L. Ortuño, G. Du Pont, S. Durán, A. Lloret, H. Merchant-Larios, and J. Calderón. 1997. Isolation and characterization of Rhizobium etli mutants altered in degradation of asparagine. J. Bacteriol. 179:2068-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang, I., P. L. Li, L. Zhang, K. R. Piper, D. M. Cook, M. E. Tate, and S. K. Farrand. 1994. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acyl homoserine lactone autoinducer. Proc. Natl. Acad. Sci. USA 91:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang, I., D. M. Cook, and S. K. Farrand. 1995. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, I., A. J. Smyth, Z.-Q. Lou, and S. K. Farrand. 1999. Modulating quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol. Microbiol. 34:282-294. [DOI] [PubMed] [Google Scholar]
- 28.Johnston, A. W. B., J. L. Beynon, A. V. Buchanan-Wollanston, S. M. Setchell, P. R. Hirsch, and J. E. Beringer. 1978. High frequency transfer of nodulating ability between strains and species of Rhizobium. Nature 276:634-636. [Google Scholar]
- 29.Jones, J. D. G., and N. Gutterson. 1987. An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens HV37a. Gene 61:299-306. [DOI] [PubMed] [Google Scholar]
- 30.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 31.Laguerre, G., S. M. Nour, V. Macheret, J. Sanjuan, P. Drouin, and N. Amarger. 2001. Classification of Rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981-993. [DOI] [PubMed] [Google Scholar]
- 32.Leemans, J., G. Soberón, M. A. Cevallos, L. Fernández, M. A. Pardo, H. de la Vega, M. Flores, C. Quinto, and R. Palacios. 1984. General organization in R. phaseoli nif plasmids, p. 710. In C. Veeger and W. E. Newton (ed.), Advances in nitrogen fixation research. Nijhoff-Junk-Pudoc, The Hague, The Netherlands.
- 33.Li, P.-L., M. D. Everhart, and S. K. Farrand. 1998. Genetic and sequence analysis of the pTiC58 trb locus, encoding a mating-pair formation system related to members of the type IV secretion family. J. Bacteriol. 180:6164-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lithgow, J. K., A. Wilkinson, A. Hardman, B. Rodelas, F. Wisniewski-Dyé, P. Williams, and J. A. Downie. 2000. The regulatory locus cinR in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol. Microbiol. 37:81-97. [DOI] [PubMed] [Google Scholar]
- 35.Lou, Z.-Q., Y. Qin, and S. K. Farrand. 2000. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J. Biol. Chem. 275:7713-7722. [DOI] [PubMed] [Google Scholar]
- 36.Marketon, M. M., and J. E. González. 2002. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J. Bacteriol. 184:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez-Salazar, J. M., and D. Romero. 2000. Role of ruvB gene in homologous and homeologous recombination in Rhizobium etli. Gene 243:125-131. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Noel, K. D., A. Sánchez, L. Fernández, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 41.Piper, K. R., and S. K. Farrand. 2000. Quorum sensing but not autoinduction of pTi plasmid conjugal transfer requires control by opine regulon and the antiactivator TraM. J. Bacteriol. 182:1080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 43.Quinto, C., H. de la Vega, M. Flores, L. Fernández, T. Ballado, G. Soberón, and R. Palacios. 1982. Reiteration of nitrogen fixation gene sequences in Rhizobium phaseoli. Nature 299:724-728. [Google Scholar]
- 44.Quinto, C., H. de la Vega, M. Flores, J. Leemans, M. A. Cevallos, M. A. Pardo, R. Azpiroz, M. L. Girard, E. Calva, and R. Palacios. 1985. Nitrogenase reductase: a functional multigene family in Rhizobium phaseoli. Proc. Natl. Acad. Sci. USA 82:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosemeyer, V., J. Michiels, C. Verreth, and J. Vanderleyden. 1998. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J. Bacteriol. 180:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg, C., and T. Hughet. 1984. The pTiC58 plasmid of Agrobacterium tumefaciens is not essential for tumor induction. Mol. Gen. Genet. 196:533-536. [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Shaw, P. D., P. Gao, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehard, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 50.Southern, E. M. 1975. Detection of sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 51.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. Blackwell Scientific Publications, Oxford, United Kingdom.
- 52.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 53.Wilkinson, A., V. Danino, F. Wisniewski-Dyé, J. K. Lithgow, and J. A. Downie. 2002. N-Acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer. J. Bacteriol. 184:4510-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wisniewski-Dyé, F., J. Jones, S. R. Chhabra, and J. A. Downie. 2002. raiIR genes are part of a quorum-sensing network controlled by cinI and cinR in Rhizobium leguminosarum. J. Bacteriol. 184:1597-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]