Abstract
We have identified an NiFe-hydrogenase exclusively localized in the cytoplasm of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 (T. kodakaraensis hydrogenase). A gene cluster encoding T. kodakaraensis hydrogenase was composed of four open reading frames (hyhBGSLTk), where the hyhSTk and hyhLTk gene products corresponded to the small and the large subunits of NiFe-hydrogenase, respectively. A putative open reading frame for hydrogenase-specific maturation endopeptidase (hybDTk) was found downstream of the cluster. Polyclonal antibodies raised against recombinant HyhLTk were used for immunoaffinity purification of T. kodakaraensis hydrogenase, leading to a 259-fold concentration of hydrogenase activity. The purified T. kodakaraensis hydrogenase was composed of four subunits (β, γ, δ, and α), corresponding to the products of hyhBGSLTk, respectively. Each αβγδ unit contained 0.8 mol of Ni, 22.3 mol of Fe, 21.1 mol of acid-labile sulfide, and 1.01 mol of flavin adenine dinucleotide. The optimal temperature for the T. kodakaraensis hydrogenase was 95°C for H2 uptake and 90°C for H2 production with methyl viologen as the electron carrier. We found that NADP+ and NADPH promoted high levels of uptake and evolution of H2, respectively, suggesting that the molecule is the electron carrier for the T. kodakaraensis hydrogenase.
Hydrogenase catalyzes the reversible activation of molecular hydrogen (H2). The enzyme is widely distributed among bacteria and archaea as well as some unicellular eukaryotes (41, 45). Hydrogenase enables an organism either to utilize H2 as a source of reducing power or to use protons as a terminal electron acceptor, evolving H2. Due to its ability to produce H2, hydrogenase is an enzyme of great biotechnological interest and has been the target of extensive research (43, 44).
Hydrogenases can be generally classified into two classes by their metal contents, NiFe-hydrogenases and Fe-only hydrogenases (41), with an exceptional metal-free hydrogenase isolated from a methanogen (6, 12, 46). NiFe-hydrogenase is usually a heterodimeric complex with a large Ni-containing subunit and a small subunit bearing one or more Fe-S clusters. In 1995, the first structure of an NiFe-hydrogenase, from Desulfovibrio gigas, was determined (42). The crystal structure revealed that the catalytic center located in the large subunit was binuclear and composed of Ni and Fe atoms. The Ni atom is coordinated by four cysteine residues, two of which also bridge to the Fe atom.
Infrared spectroscopic analysis of the D. gigas NiFe-hydrogenase revealed that the Fe atom is also coordinated by three diatomic ligands, one CO and two CN (27). On the other hand, Fe-only hydrogenases have no sequence similarity with the NiFe-hydrogenases and harbor a unique catalytic center comprised of six Fe atoms (designated the H cluster) (26). However, the presence of Fe-only hydrogenases is limited to certain anaerobic bacteria and some anaerobic eukaryotes (41).
Hyperthermophiles are microorganisms that optimally grow at temperatures above 80°C (36), most of them belonging to the domain Archaea. Enzymes produced by hyperthermophiles have attracted much attention as thermostable biocatalysts for industrial use (1). This is particularly the case for hydrogenase, as a soluble, thermostable hydrogenase could potentially provide a stable biocatalyst for hydrogen production (43). Accordingly, cytosolic hydrogenases have been identified and studied from various hyperthermophiles, including the archaea Pyrococcus furiosus (19), Thermococcus litoralis (29), Thermococcus stetteri (47), and Thermococcus celer (3) and the bacterium Thermotoga maritima (40). The archaeal enzymes have all been found to be NiFe-hydrogenases, while the enzyme from T. maritima was an Fe-only hydrogenase.
Among the thermostable NiFe hydrogenases, the enzymes from P. furiosus are by far the best characterized compared to the enzymes from the Thermococcus species (Table 1). P. furiosus harbors two cytoplasmic hydrogenases (5, 21) and one membrane-bound hydrogenase (33, 35), all of them NiFe-hydrogenases. The two cytosolic enzymes (hydrogenase I and hydrogenase II) are similar in structure and enzymatic characteristics except that hydrogenase II has a much lower specific activity than hydrogenase I and is responsible for only 10% of the total H2 evolution activity in the cytoplasmic fraction (21). These cytoplasmic hydrogenases are responsible for the production of not only H2 but also H2S and are therefore sometimes referred to as sulfhydrogenases (20, 21).
TABLE 1.
Comparison of cytosolic hydrogenases from hyperthermophilic archaea
| Property | T. kodakaraensis hydrogenase | T. litoralis hydrogenase | T. stetteri hydrogenase | T. celer hydrogenase |
P. furiosus
|
|
|---|---|---|---|---|---|---|
| Hydrogenase I | Hydrogenase II | |||||
| Subunit composition | αβγδ | αβγδ | Not determined | αβ | αβγδ | αβγδ |
| Gene isolation | Yes | Yes | Not performed | Not performed | Yes | Yes |
| Sequence identity with T. kodakaraensis hydrogenase | 100% | 81.7% | Not determined | Not determined | 83.7% | 44.2% |
| Holoenzyme | (αβγδ)na | αβγδ | Not determined | (αβ)2 | αβγδ | (αβγδ)2 |
| Ni content | 0.8 ± 0.1b | 0.9 ± 0.1 | 1d | 1.9-2.3 | 0.8 ± 0.1e | 0.9 ± 0.1b |
| Fe content | 22.3 ± 0.7b | 22.0 ± 1.0 | 13d | 24-30 | 26 ± 2.5e | 21 ± 1.6b |
| Acid-labile sulfide | 21.1 ± 0.7b | 22.0 ± 2.0 | Not determined | 24-36 | 20 ± 3.2e | 19 ± 2.1b |
| Flavin cofactor species | FAD | Not determined | Flavin mononucleotide | Not determined | FAD | FAD |
| Flavin cofactor content | 1b | Not determined | Not determined | Not determined | 1 | 1b |
| H2 uptake (U/mg at 80°C) | ||||||
| NADP+ (1 mM) | 48.7 | Not determined | 0 | Not determined | 75 | 0.3 |
| Methyl viologen (1 mM) | 319 | 110c | 20,000 (98°C) | 12,900 (88°C) | 250 | 11 |
| H2 evolution (U/mg at 80°C) | ||||||
| NADPH (1 mM) | 2.72 | Not determined | Not determined | Not determined | 10 | 0.2 |
| Methyl viologen (1 mM) | 283 | 33c | Not determined | 9,160 (88°C) | 290 | 40 |
| Reference(s) | This work | 29 | 47 | 3 | 5,19 | 19, 21 |
n ≥ 3.
per αβγδ.
Vmax value.
13 mol of Fe per mol of Ni.
Original data (5) were recalculated according to the molecular mass determined from the sequence.
Thermococcus kodakaraensis KOD1 (previously classified as a Pyrococcus sp.) is an anaerobic, sulfur-reducing hyperthermophilic archaeon that was isolated from a geothermal spring in the coastal area of Kodakara Island, Kagoshima, Japan (24). We have been focusing on the application of several enzymes from this hyperthermophile, such as KOD DNA polymerase (13, 37). Here, we report the purification and a detailed biochemical characterization of a cytoplasmic NiFe-hydrogenase from T. kodakaraensis KOD1.
Hydrogenase activity in T. kodakaraensis KOD1.
T. kodakaraensis KOD1 grows heterotrophically on amino acids with elemental sulfur as a terminal electron acceptor, producing hydrogen sulfide (24). We have also observed fermentative growth on pyruvate and starch, with the evolution of H2. In all cases, H2-producing activity could be observed in cell extracts of T. kodakaraensis KOD1 with reduced methyl viologen as the electron donor, indicating the presence of a cytosolic hydrogenase (T. kodakaraensis hydrogenase). However, high oxygen sensitivity and the low concentration of the enzyme hampered its purification by traditional methods. Therefore, we set out to develop a rapid purification procedure, mainly based on immunoaffinity chromatography under anaerobic conditions.
Cloning of T. kodakaraensis hydrogenase gene.
Cytosolic sulfhydrogenase genes have been isolated from Pyrococcus furiosus (21, 25). Highly similar orthologues are present on the genomes of Pyrococcus abyssi and Pyrococcus horikoshii (PAB0634-41, PAB1784-7, and PH1290-4). As these archaeal strains, along with T. kodakaraensis KOD1, belong to the same order, Thermococcales, we designed primers from these sequences, Hy-1 (5′-GGAATTCTCGTCAATCAGGTCTATCGC-3′) and Hy-2 (5′-GGGAATTCAGGTATGTTAAGTTA-CCCAAGGAAAACA-3′), to amplify the hydrogenase gene from strain KOD1. The amplified DNA fragment was used to isolate the entire T. kodakaraensis hydrogenase gene from a genomic DNA library. The isolated DNA fragment was subcloned into a plasmid vector (pUC19) with Escherichia coli strain DH5α as the host (30).
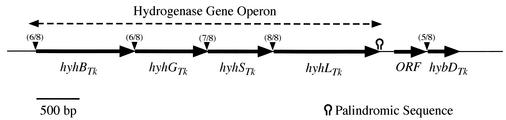
Sequence analysis of T. kodakarensis hydrogenase gene. Sequencing analysis with the ABI Prism kit and model 310 capillary DNA sequencer (Applied Biosystems, Foster City, Calif.) revealed the presence of a gene cluster composed of four open reading frames (hyhBGSL; Fig. 1), which resembled the genes encoding the four subunits (β, γ, δ, and α, respectively) of the cytosolic sulfhydrogenase from P. furiosus. The four genes used ATG as the initiation codon except for hyhBTk, in which the rare TTG codon was present. hyhBTk overlapped hyhGTk, and hyhSTk overlapped hyhLTk. Each gene had a typical archaeal Shine-Dalgarno sequence about 5 to 7 bases upstream of their initiation codons. The calculated molecular mass of each subunit was 43,366 Da (β), 33,216 Da (γ), 30,002 Da (δ), and 48,286 Da (α). Each protein displayed 87.5%, 93.2%, 82.0%, and 86.4% identity, respectively, to the corresponding subunits of P. furiosus hydrogenase I and 51.8%, 34.2%, 46.4%, and 39.3% identity, respectively, with P. furiosus hydrogenase II. Near the stop codon of the last gene, hyhLTk, there was an archaeal transcription termination region, with a set of 6-bp palindromic sequences predicting the formation of a hairpin-loop structure (GAGGCT-N2-AGCCTC) followed by a pyrimidine-rich region (4). These data strongly suggest that the gene cluster constitutes an operon, terminating after hyhLTk.
FIG. 1.
Structure of T. kodakaraensis hydrogenase gene operon. Arrowheads indicate the presence of putative Shine-Dalgarno sequences. The ideal Shine-Dalgarno sequence (8 bp) interpreted from the 3′-terminal nucleotide sequence of the 16S rRNA of strain KOD1 is 5′-GGAGGTGA-3′ (22). Numbers in parentheses above each arrowhead indicate the number of nucleotides matching the ideal Shine-Dalgarno sequence. A putative transcription termination sequence is also illustrated with a hairpin.
Downstream of the hyhBGSLTk operon, two adjacent open reading frames were found. The first open reading frame, located 239 bp downstream of hyhBGSLTk, did not display significant sequence similarity with other proteins, whereas the second open reading frame (hybDTk) resembled previously identified hydrogenase-specific maturation endopeptidases (see below).
Structural motifs in T. kodakaraensis hydrogenase and HybDTk.
The α subunit of T. kodakaraensis hydrogenase displayed similarity with the large subunit of various NiFe-hydrogenase heterodimers and contained a set of motifs that presumably formed the catalytic center (25, 45). The δ subunit of T. kodakaraensis hydrogenase corresponded to the small subunit of NiFe-hydrogenases. Three [4Fe-4S] cluster motifs usually located within the δ subunit of archaeal sulfhydrogenases (34) were also present in T. kodakaraensis hydrogenase. The β and γ subunits were homologous to AsrA and AsrB, respectively, both of which are the subunits of Salmonella enterica serovar Typhimurium anaerobic sulfite reductase (14). The β subunit of T. kodakaraensis hydrogenase contained two typical CxxCxxCxxxCP motifs (residues 233 to 244 and 312 to 323), corresponding to two [4Fe-4S] clusters (25, 34).
The γ subunit of T. kodakaraensis hydrogenase contained a plant ferredoxin-type [2Fe-2S] cluster consisting of the motifs CxxGxCxxC (residues 255 to 263) and CxxxP (residues 275 to 279) (34). Within the γ subunit of archaeal sulfhydrogenases, Rákhely et al. proposed that an additional four sequence motifs were involved in the binding of flavin nucleotides and NADP(H). These four motifs were completely conserved in the γ subunit of T. kodakaraensis hydrogenase (residues 52 to 62, 99 to 112, 125 to 139, and 222 to 227).
The large subunit of NiFe-hydrogenase undergoes a series of posttranslational modifications, which is called maturation (7, 9, 18). The last step of this maturation is the cleavage of a region following a carboxy-terminal DPCxxCxxH motif by a specific endopeptidase (maturation endopeptidase) (31, 32, 38, 39). This motif was completely conserved in the α subunit of T. kodakaraensis hydrogenase, followed by a four-amino-acid stretch (VARL) leading to the carboxy terminus. While the region processed in the hydrogenases from E. coli and other bacteria is 15 or more amino acids in length (32), the short sequence found in T. kodakaraensis hydrogenase was similar to those of P. furiosus hydrogenase-I (VVRL), P. furiosus hydrogenase-II (FVKL), and T. litoralis hydrogenase (VVRL).
As mentioned above, an open reading frame, designated hybDTk, was found downstream of the hyhBGSLTk operon (Fig. 1). HybDTk displayed similarity to E. coli HybD (26.9% identity), the maturation endopeptidase of E. coli hydrogenase 2 (11, 23), and the P. furiosus FrxA protein (56.8%) (16). According to the crystal structure of E. coli HybD, it was proposed that the maturation endopeptidase recognizes the Ni-bound large subunit prior to proteolytic cleavage. Amino acid residues of E. coli HybD involved in the recognition of Ni (Glu16, Asp62, and His93) and those located in close proximity to the metal ion-binding region (Gly9, Gly19, Ala63, and Gly118) (11) were all conserved in HybDTk except for Glu16, which was replaced with Asp.
Immunoaffinity purification of T. kodakaraensis hydrogenase.
To purify T. kodakaraensis hydrogenase from cell extracts of T. kodakaraensis, immunoaffinity chromatography was performed. For this purpose, purified recombinant HyhLTk was prepared as an antigen for antibody production. With the primers EXP-DF (5′-GGTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAGCGAGAAGAAGATAAGGATTGGG-3′) and EXP-AR (5′-GGGGATCCACCACTGGAAAATAGGCTTG-3′), a DNA fragment containing hyhSLTk was amplified and inserted into pET21a(+) (Novagen, Madison, Wis.). Recombinant E. coli BL21(DE3) (Stratagene, La Jolla, Calif.) cells were cultivated until the optical density at 660 nm reached 0.5, and gene expression was induced with isopropyl-β-d-thiogalactopyranoside (0.1 mM). Cell extracts were heat precipitated for 30 min at 85°C. Samples were centrifuged (17,000 × g, 10 min), and the resulting supernatant was applied to a ResourceQ column (Amersham Biosciences, Buckinghamshire, United Kingdom). Recombinant HyhLTk was eluted with Tris-HCl buffer (50 mM, pH 8.0) at a salt concentration between 0.3 and 0.4 M NaCl. A fraction containing HyhLTk was applied to a Superdex 200HR 10/30 column in 50 mM Tris-HCl buffer (pH 8.0) with 0.15 M NaCl.
The purified HyhLTk was used to prepare rabbit polyclonal antibodies. The total immunoglobulin G fraction containing anti-HyhLTk antibody was purified and applied to a column immobilized with recombinant HyhLTk. In order to obtain an immunoglobulin G fraction with moderate affinity for HyhLTk, antibodies retained in the column were eluted with 1.5 M KCl-containing ethylene glycol buffer (50%, vol/vol). The eluted antibodies were then immobilized on a matrix to prepare the affinity column for purification of T. kodakaraensis hydrogenase.
All the purification steps hereafter were performed under a strictly anaerobic atmosphere in anaerobic chambers (Tabai Espec, Osaka, Japan). First, T. kodakaraensis KOD1 (24) was grown at 85°C for 17 h in screw-cap bottles with MA medium, containing, per liter, 4.8 g of Marine Art SF A salt and 26.4 g of Marine Art SF B salt (Senju Seiyaku, Osaka, Japan) as artificial sea salts, 5 g of yeast extract, and 5 g of tryptone (Nacalai Tesque, Kyoto, Japan). After autoclaving the MA medium, 1 g of elemental sulfur was added per liter. T. kodakaraensis cells after cultivation were centrifuged and suspended in buffer containing 50 mM N-(2-hydroxyethyl)-piperazine-N′-(3-propanesulfonic acid) (EPPS) and lysozyme (1 mg/ml), DNase (0.1 mg/ml), and a protease inhibitor tablet (Complete; Roche Diagnostics, Basel, Switzerland). Cells were disrupted by stirring the solution for 60 min. After centrifugation (15,000 × g, 10 min), the supernatant was applied to the antibody-immobilized column.
Proteins attached to the column were eluted with ethylene glycol buffer (50%, vol/vol) containing 1.5 M KCl, and the eluted fraction was dialyzed with phosphate-buffered saline (1× PBS, pH 7.4). These purification steps were repeated to remove nonspecific bound proteins. Homogeneity of the purified hydrogenase was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the method of Laemmli (17). The protein concentration was determined by the Bradford method with the Bio-Rad protein assay system (Bio-Rad, Hercules, Calif.), with bovine serum albumin as a calibration standard.
The protein sample obtained after immunoaffinity chromatography was subjected to nondenaturing polyacrylamide gel electrophoresis followed by Western blot analysis with anti-HyhLTk antibodies. A single positive signal was detected in the purified fraction with the same electrophoretic mobility as that observed with the cell extract (data not shown). The result indicated that the immunoaffinity-purified fraction contained active T. kodakaraensis hydrogenase with the same subunit composition as that of the original enzyme present in the cell extract. Hydrogenase activity was then examined by measuring H2 evolution at 80°C in a stopper-sealed bottle (total volume = 13.5 ml). The assay mixture (2 ml) contained 1 mM methyl viologen and 20 mM sodium dithionite in 50 mM EPPS buffer, pH 8.4 (5), and generation of H2 was quantified by gas chromatography (GC-14A; Shimadzu, Kyoto, Japan). One unit of hydrogenase activity was defined as the production of 1 μmol of H2 per minute. The purified fraction displayed a 259-fold increase in hydrogenase activity compared with the cell extracts.
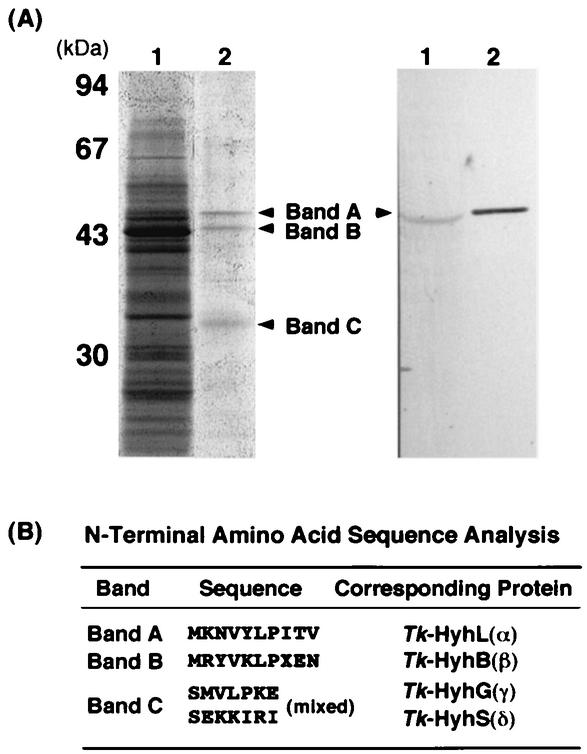
SDS-PAGE analysis of the active fraction revealed the presence of three bands with similar intensities (Fig. 2A, left, bands A, B, and C). Western blot analysis with anti-HyhLTk antibody revealed that band A corresponded to HyhLTk (Fig. 2A, right). With a protein sequencer (model 491 cLC; Applied Biosystems), the amino-terminal amino acid sequence of each band was determined to be as follows: band A, MKNVYLPITV; band B, MRYVKLPXEN; and band C, S(M/E)(V/K)(L/K)(P/I)(K/R)(E/I) (Fig. 2B). The amino acid sequences of bands A and B were in good accordance with those of HyhLTk and HyhBTk, respectively. On the other hand, band C was a mixture of two proteins, and their amino acid sequences agreed with those of a mixture of HyhGTk (SMVLPKE) and HyhSTk (SEKKIRI). The initial Met residues of both proteins were processed. These results indicated that the T. kodakaraensis hydrogenase is composed of the protein products of the hyhBGSLTk operon, and the relative intensities of the bands implied an αβγδ composition. Examination by gel filtration with a Superdex 200 HR10/30 column (Amersham Biosciences) revealed that the holoenzyme displayed a molecular mass higher than that of ferritin (440 kDa). As the sum of the deduced molecular mass of αβγδ was predicted to be approximately 155 kDa, the quaternary structure of T. kodakaraensis hydrogenase holoenzyme was suggested to be (αβγδ)n (n ≥ 3).
FIG. 2.
(A) SDS-PAGE (left) and Western blot analysis (right) of purified T. kodakaraensis hydrogenase. Lane 1, cell extract of T. kodakaraensis KOD1 (30 μg); lane 2, purified T. kodakaraensis hydrogenase (1 μg). Anti-HyhLTk antibodies were used in the Western blot analysis. (B) Amino-terminal amino acid sequences of bands A to C.
Metal and cofactor content of T. kodakaraensis hydrogenase.
Metal content of the purified T. kodakaraensis hydrogenase was determined by plasma emission spectroscopy with an ICPS-7000 (Shimadzu). The result indicated that T. kodakaraensis hydrogenase contained 0.8 ± 0.1 mol of Ni and 22.3 ± 0.7 mol of Fe per mol of αβγδ. The content of acid-labile sulfide was also examined with methods described in the literature (2, 8) and estimated to be 21.1 ± 0.7 mol per mol of αβγδ. The presence of flavin cofactor was also determined by releasing the flavin group through treatment with 10% trichloroacetic acid, followed by analysis by thin-layer chromatography (10). Under UV light, a spot showing the same mobility as flavin adenine dinucleotide (FAD) was detected. FAD content was then estimated by measuring the fluorescence intensity of the neutralized trichloroacetic acid supernatant. Commercially available FAD (Nacalai Tesque) was used as the standard for quantification. These procedures showed that T. kodakaraensis hydrogenase contained 1.01 mol of FAD per mol of αβγδ.
Enzymatic properties of T. kodakaraensis hydrogenase.
Optimal temperature, pH, and thermostability of T. kodakaraensis hydrogenase were determined with methyl viologen as an electron carrier. Methods to measure H2 evolution are described above. H2 uptake activity of T. kodakaraensis hydrogenase was determined by measuring H2-dependent reduction of methyl viologen. The assay mixture containing the purified enzyme and 1 mM methyl viologen in 50 mM EPPS buffer (pH 8.4) was applied in a stopper-sealed cuvette, and the headspace was purged with H2. The mixture was then maintained at 80°C or the desired temperature, and reduction of methyl viologen was followed with a spectrophotometer.
The optimum temperature of the T. kodakaraensis hydrogenase was 90°C for the H2 evolution reaction and 95°C for the H2 uptake reaction. The optimum pH value was determined to be 8.0 for the H2 evolution reaction. As for thermostability, the time required for 50% loss of activity was 203 min at 80°C and 18 min at 90°C for the H2 evolution reaction.
With oxidized methyl viologen, T. kodakaraensis hydrogenase showed an H2 uptake activity of 319 U/mg at 80°C (Table 2). Under the same conditions, we examined H2 uptake activity with various electron carriers. Artificial electron carriers such as 2,6-dichlorophenol indophenol, methylene blue, and benzyl viologen could function as electron acceptors (Table 2). Among the natural electron acceptors, T. kodakaraensis hydrogenase utilized NADP+ efficiently, and a specific activity of 48.7 U/mg was obtained at 1 mM NADP+. NAD+ could also be utilized as an electron acceptor, but specific activity at 1 mM NAD+ was 150 times lower than that with 1 mM NADP+. On the other hand, FAD and riboflavin 5′-phosphate (flavin mononucleotide) could not be used as electron acceptors. The specific activities with methylene blue, benzyl viologen, methyl viologen, and NADP+ represent maximum values, as no significant increase in activity was observed at higher concentrations of these compounds.
TABLE 2.
Electron acceptor specificity of T. kodakaraensis hydrogenase (H2 uptake)
| Electron acceptora | E°′ (mV) | Concn (mM) | Sp act (U/mg) |
|---|---|---|---|
| DCIP | +217 | 0.1 | 18.5 |
| Methylene blue | +11 | 0.2 | 435 |
| FAD | −219 | 0.3 | 0 |
| FMN | −219 | 0.3 | 0 |
| NAD+ | −320 | 1 | 0.2 |
| NADP+ | −320 | 1 | 48.7 |
| Benzyl viologen | −360 | 1 | 1,120 |
| Methyl viologen | −446 | 1 | 319 |
DCIP, 2,6-dichlorophenol indophenol; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide.
The electron carrier specificity was also determined for the H2 evolution reaction (Table 3). The reduced form of benzyl viologen as well as that of methyl viologen could function as an electron donor. As for natural electron donors, when NADPH was utilized at a 1 mM concentration, a specific activity of 2.72 U/mg was observed. The specific activities with benzyl viologen, methyl viologen, and NADPH represent maximum values. H2 evolution was also examined with low concentrations of NADP+ in an NADPH regeneration system. Instead of NADPH, 0.01 mM NADP+ was added to a reaction mixture along with the purified NADP-dependent glutamate dehydrogenase of T. kodakaraensis (1.7 μg) (28) and 10 mM l-glutamate. With this system, evolution of H2 was detected at 85°C with a specific activity of 4.12 U/mg. When any one of these components were omitted, no H2 evolution was detected (data not shown). Furthermore, T. kodakaraensis hydrogenase showed NADPH-dependent reduction of methyl viologen (diaphorase activity; data not shown). These results indicated that T. kodakaraensis hydrogenase had the ability to utilize NADPH as an electron donor.
TABLE 3.
Electron donor specificity of T. kodakaraensis hydrogenase (H2 evolution)
| Electron donora | E°′ (mV) | Concn (mM) | Sp act (U/mg) |
|---|---|---|---|
| Methylene blue | +11 | 0.1 | 0 |
| FAD | −219 | 0.3 | 0 |
| FMN | −219 | 0.3 | 0 |
| NADH | −320 | 1 | 0 |
| NADPH | −320 | 1 | 2.72 |
| NADP+ + GDH + l-Glu | −320 | 0.01 | 4.12 |
| Benzyl viologen | −360 | 1 | 87.3 |
| Methyl viologen | −446 | 1 | 283 |
GDH, glutamate dehydrogenase of T. kodakaraensis KOD1 (1.7 μg); l-Glu, l-glutamate (final, 10 mM); sodium dithionite (final, 20 mM) was added as a reducing reagent except for NADH, NADPH, and NADP+ + glutamate dehydrogenase + l-glutamate. Also see Table 2, footnote a.
Localization of T. kodakaraensis hydrogenase.
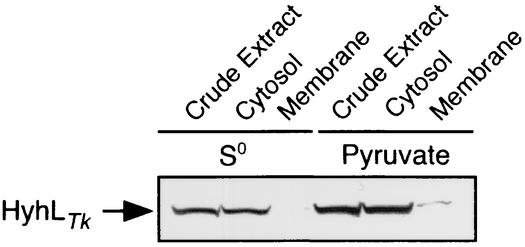
With the anti-HyhLTk antibody, we determined the subcellular localization of T. kodakaraensis hydrogenase. Total cell extracts and the cytosolic and membrane fractions separated by ultracentrifugation were subjected to SDS-PAGE and Western blot analysis. The results clearly indicated that T. kodakaraensis hydrogenase was localized exclusively in the cytoplasmic fraction (Fig. 3).
FIG. 3.
Western blot analysis of HyhLTk in cell extract and cytosol and membrane fractions of T. kodakaraensis KOD1. Cells were grown on MA medium with 0.1% elemental sulfur (S0) or 0.5% sodium pyruvate (pyruvate). The crude extract was ultracentrifuged (110,000 × g) for 2 h to separate the cytosol and membrane fractions. The resulting fractions (40 μg) were subjected to SDS-PAGE, followed by Western blot analysis with anti-HyhLTk antibody.
Diversity and function of cytosolic hydrogenase in Thermococcales.
In this manuscript, we described the gene analysis, purification, and characterization of a cytosolic NiFe-hydrogenase from T. kodakaraensis KOD1. So far, there have been a number of reports on the purification and characterization of cytosolic hydrogenases from hyperthermophilic archaea belonging to the order Thermococcales (3, 5, 21, 29, 47). A comparison of their properties with those of the T. kodakaraensis hydrogenase is shown in Table 1. Among these hydrogenases, the T. stetteri and T. celer enzymes are less well characterized, and sequence information is not available at present. However, the unusually high activity of the two enzymes suggests that these hydrogenases may not be homologous to T. kodakaraensis hydrogenase or P. furiosus hydrogenase I and imply a divergence among hydrogenases from the Thermococcales.
Although some properties of the T. litoralis enzyme are yet to be revealed, several characteristics are commonly observed among the enzymes from T. kodakaraensis, T. litoralis, and P. furiosus (I and II). All were composed of four different subunits (αβγδ), and each heterotetramer contained 1 Ni and 19 to 26 Fe atoms as well as 21 to 22 atoms of acid-labile sulfide. Their amino acid sequences showed significant similarity, although P. furiosus hydrogenase II exhibited relatively low similarity among these enzymes. According to a recent sequence classification of NiFe-hydrogenases, these tetrameric (sulf)hydrogenases are classified into group 3b (41). In terms of activity, T. kodakaraensis hydrogenase and P. furiosus hydrogenase I exhibited similar levels of H2-evolving activity with methyl viologen, one order higher than that of those displayed by the T. litoralis enzyme and P. furiosus hydrogenase II.
Besides these properties, T. kodakaraensis hydrogenase utilized NADP(H) as an electron acceptor and donor and contained 1 mol of FAD per mol of heterotetramer; both of which are also characteristics of P. furiosus hydrogenases I and II. As for the T. litoralis hydrogenase, a ferredoxin-dependent hydrogenase activity has been observed. However, on the basis of sequence analysis, it has been proposed that a flavin cofactor and NADP(H) may also be utilized in this enzyme (29).
From our results and other studies on heterotetrameric hydrogenases (29, 34), the electron transfer pathway in T. kodakaraensis hydrogenase can be proposed as follows. NADPH, after binding to the γ subunit, releases two electrons that reduce the FAD cofactor. The reduced FAD then transfers electrons to the multiple Fe-S clusters present in the γβδ subunits. T. kodakaraensis hydrogenase contains one [2Fe-2S] cluster in the γ subunit, two [4Fe-4S] clusters in the β subunit, and three [4Fe-4S] clusters in the δ subunit. The electrons are finally translocated to the NiFe metal center in the α subunit, where H2 is produced from H+.
With methyl viologen as an electron carrier, T. kodakaraensis hydrogenase could catalyze both H2 uptake and H2 evolution reactions at equivalent rates (ratio of H2 evolution to H2 uptake was 0.89 at 1 mM methyl viologen). Recent kinetic analyses of H2 production and catabolism in P. furiosus have indicated that the generation rate of intracellular reducing equivalents is much higher than the NADPH-dependent evolution rate of H2 (35). This suggested that soluble NADPH-dependent sulfhydrogenases could not account for the total electron transfer of the reducing equivalents. A model was proposed in which a transmembrane hydrogenase was responsible for proton respiration coupled to energy transduction. In this case, the soluble sulfhydrogenases contribute as safety valves for the disposal of excessive reducing equivalents and/or the retrieval of the respiration product, H2. This may also be the case in T. kodakaraensis KOD1. A large portion of the hydrogenase activity in the cell extract was not attached to the immunoaffinity column, suggesting the presence of a second hydrogenase. Corresponding with the data, we have identified a putative membrane-bound hydrogenase gene with 87% similarity to the enzyme from P. furiosus (unpublished data).
Enzyme maturation process of NiFe-hydrogenase in hyperthermophilic archaea.
After polypeptide synthesis, the large subunit of NiFe-hydrogenase encounters a series of posttranslational modifications by specific accessory proteins (7, 9, 18). Although their order is still unknown, this maturation process includes the following events: binding to a specific chaperone, insertion of Ni and Fe cations, formation of diatomic ligands, and proteolytic cleavage of the carboxy-terminal region. Several accessory proteins required for this process have been identified, such as the HypABCDEF proteins and hydrogenase-specific maturation endopeptidases. At present, this maturation process has been studied almost exclusively with hydrogenases of bacterial origin, and little is known concerning the maturation of archaeal hydrogenases.
In this study, we found a gene (hybDTk) downstream of the hyhBGSLTk operon which encodes a protein similar to the hydrogenase-specific maturation endopeptidase. Sequence comparison of the encoded protein (HybDTk) with the E. coli maturation endopeptidase HybD revealed that the amino acid residues that coordinate Ni or are present around the Ni recognition site were essentially conserved. These residues are also preserved among HybDTk homologs identified among other strains of the Thermococcales. Using purified recombinant proteins, we could not detect the carboxy-terminal cleavage of HyhLTk by HybDTk in vitro (data not shown). This is not surprising, as proteolysis of the large subunit is presumed to occur only after Ni incorporation (15, 32), whereas recombinant HyhLTk did not contain Ni (data not shown). As we have also identified hypACDEF orthologs on the genome of T. kodakaraensis (unpublished data), it is likely that a maturation mechanism similar to that for bacterial enzymes also exists in the case of the T. kodakaraensis hydrogenase.
Nucleotide sequence accession number.
Nucleotide sequence data for the T. kodakaraensis hydrogenase will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession number AB075598.
REFERENCES
- 1.Adams, M. W., and R. M. Kelly. 1998. Finding and with hyperthermophilic enzymes. Trends Biotechnol. 16:329-332. [DOI] [PubMed] [Google Scholar]
- 2.Beinert, H. 1983. Semimicro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131:373-378. [DOI] [PubMed] [Google Scholar]
- 3.Blamey, J. M., M. Chiong, and E. T. Smith. 2001. Purification and characterization of an iron-nickel hydrogenase from Thermococcus celer. J. Biol. Inorg. Chem. 6:517-522. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. W., C. J. Daniels, and J. N. Reeve. 1989. Gene structure, organization, and expression in archaebacteria. Crit. Rev. Microbiol. 16:287-338. [DOI] [PubMed] [Google Scholar]
- 5.Bryant, F. O., and M. W. Adams. 1989. Characterization of hydrogenase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J. Biol. Chem. 264:5070-5079. [PubMed] [Google Scholar]
- 6.Buurman, G., S. Shima, and R. K. Thauer. 2000. The metal-free hydrogenase from methanogenic archaea: evidence for a bound cofactor. FEBS Lett. 485:200-204. [DOI] [PubMed] [Google Scholar]
- 7.Casalot, L., and M. Rousset. 2001. Maturation of the [NiFe] hydrogenases. Trends Microbiol. 9:228-237. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. S., and L. E. Mortenson. 1977. Inhibition of methylene blue formation during determination of the acid-labile sulfide of iron-sulfur protein samples containing dithionite. Anal. Biochem. 79:157-165. [DOI] [PubMed] [Google Scholar]
- 9.Dernedde, J., T. Eitinger, N. Patenge, and B. Friedrich. 1996. hyp gene products in Alcaligenes eutrophus are part of a hydrogenase-maturation system. Eur. J. Biochem. 235:351-358. [DOI] [PubMed] [Google Scholar]
- 10.Fazekas, A. G., and K. Kokai. 1971. Extraction, purification, and separation of tissue flavin for spectrophotometric determination. Methods Enzymol. 18:385-398. [Google Scholar]
- 11.Fritsche, E., A. Paschos, H. G. Beisel, A. Böck, and R. Huber. 1999. Crystal structure of the hydrogenase maturating endopeptidase HybD from Escherichia coli. J. Mol. Biol. 288:989-998. [DOI] [PubMed] [Google Scholar]
- 12.Hartmann, G. C., A. R. Klein, M. Linder, and R. K. Thauer. 1996. Purification, properties and primary structure of H2-forming N5,N10-methylenetetrahydromethanopterin dehydrogenase from Methanococcus thermolithotrophicus. Arch. Microbiol. 165:187-193. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto, H., M. Nishioka, S. Fujiwara, M. Takagi, T. Imanaka, T. Inoue, and Y. Kai. 2001. Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J. Mol. Biol. 306:469-477. [DOI] [PubMed] [Google Scholar]
- 14.Huang, C. J., and E. L. Barrett. 1991. Sequence analysis and expression of the Salmonella typhimurium asr operon encoding production of hydrogen sulfide from sulfite. J. Bacteriol. 173:1544-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobi, A., R. Rossmann, and A. Böck. 1992. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch. Microbiol. 158:444-451. [DOI] [PubMed] [Google Scholar]
- 16.Kletzin, A., and M. W. Adams. 1996. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 178:248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lutz, S., A. Jacobi, V. Schlensog, R. Böhm, G. Sawers, and A. Böck. 1991. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol. Microbiol. 5:123-135. [DOI] [PubMed] [Google Scholar]
- 19.Ma, K., and M. W. Adams. 2001. hydrogenases I and II from Pyrococcus furiosus. Methods Enzymol. 331:208-216. [DOI] [PubMed] [Google Scholar]
- 20.Ma, K., R. N. Schicho, R. M. Kelly, and M. W. Adams. 1993. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc. Natl. Acad. Sci. USA 90:5341-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, K., R. Weiss, and M. W. Adams. 2000. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182:1864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuda, T., M. Morikawa, M. Haruki, H. Higashibata, T. Imanaka, and S. Kanaya. 1999. Isolation of TBP-interacting protein (TIP) from a hyperthermophilic archaeon that inhibits the binding of TBP to TATA-DNA. FEBS Lett. 457:38-42. [DOI] [PubMed] [Google Scholar]
- 23.Menon, N. K., C. Y. Chatelus, M. Dervartanian, J. C. Wendt, K. T. Shanmugam, H. D. Peck, Jr., and A. E. Przybyla. 1994. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J. Bacteriol. 176:4416-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedroni, P., A. Della Volpe, G. Galli, G. M. Mura, C. Pratesi, and G. Grandi. 1995. Characterization of the locus encoding the [Ni-Fe] sulfhydrogenase from the archaeon Pyrococcus furiosus: evidence for a relationship to bacterial sulfite reductases. Microbiology 141:449-458. [DOI] [PubMed] [Google Scholar]
- 26.Peters, J. W., W. N. Lanzilotta, B. J. Lemon, and L. C. Seefeldt. 1998. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Å resolution. Science 282:1853-1858. [DOI] [PubMed] [Google Scholar]
- 27.Pierik, A. J., W. Roseboom, R. P. Happe, K. A. Bagley, and S. P. Albracht. 1999. Carbon monoxide and cyanide as intrinsic ligands to iron in the active site of [NiFe]-hydrogenases. NiFe(CN)2CO, biology's way to activate H2. J. Biol. Chem. 274:3331-3337. [DOI] [PubMed] [Google Scholar]
- 28.Rahman, R. N., S. Fujiwara, M. Takagi, and T. Imanaka. 1998. Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and comparison of the enzymatic characteristics of native and recombinant GDHs. Mol. Gen. Genet. 257:338-347. [DOI] [PubMed] [Google Scholar]
- 29.Rákhely, G., Z. H. Zhou, M. W. Adams, and K. L. Kovács. 1999. Biochemical and molecular characterization of the [NiFe] hydrogenase from the hyperthermophilic archaeon Thermococcus litoralis. Eur. J. Biochem. 266:1158-1165. [DOI] [PubMed] [Google Scholar]
- 30.Raleigh, E. A., K. Lech, and R. Brent. 1994. E. coli, plasmids, and bacteriophages. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 31.Rossmann, R., T. Maier, F. Lottspeich, and A. Böck. 1995. Characterisation of a protease from Escherichia coli involved in hydrogenase maturation. Eur. J. Biochem. 227:545-550. [DOI] [PubMed] [Google Scholar]
- 32.Rossmann, R., M. Sauter, F. Lottspeich, and A. Böck. 1994. Maturation of the large subunit (HycE) of Escherichia coli hydrogenase 3 requires nickel incorporation followed by C-terminal processing at Arg537. Eur. J. Biochem. 220:377-384. [DOI] [PubMed] [Google Scholar]
- 33.Sapra, R., M. F. Verhagen, and M. W. Adams. 2000. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 182:3423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva, P. J., B. de Castro, and W. R. Hagen. 1999. On the prosthetic groups of the NiFe sulfhydrogenase from Pyrococcus furiosus: topology, structure, and temperature-dependent redox chemistry. J. Biol. Inorg. Chem. 4:284-291. [DOI] [PubMed] [Google Scholar]
- 35.Silva, P. J., E. C. van den Ban, H. Wassink, H. Haaker, B. de Castro, F. T. Robb, and W. R. Hagen. 2000. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267:6541-6551. [DOI] [PubMed] [Google Scholar]
- 36.Stetter, K. O. 1999. Extremophiles and their adaptation to hot environments. FEBS Lett. 452:22-25. [DOI] [PubMed] [Google Scholar]
- 37.Takagi, M., M. Nishioka, H. Kakihara, M. Kitabayashi, H. Inoue, B. Kawakami, M. Oka, and T. Imanaka. 1997. Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl. Environ. Microbiol. 63:4504-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theodoratou, E., A. Paschos, A. Magalon, E. Fritsche, R. Huber, and A. Böck. 2000. Nickel serves as a substrate recognition motif for the endopeptidase involved in hydrogenase maturation. Eur. J. Biochem. 267:1995-1999. [DOI] [PubMed] [Google Scholar]
- 39.Theodoratou, E., A. Paschos, W. Mintz, and A. Böck. 2000. Analysis of the cleavage site specificity of the endopeptidase involved in the maturation of the large subunit of hydrogenase 3 from Escherichia coli. Arch. Microbiol. 173:110-116. [DOI] [PubMed] [Google Scholar]
- 40.Verhagen, M. F., and M. W. Adams. 2001. Fe-only hydrogenase from Thermotoga maritima. Methods Enzymol. 331:216-226. [DOI] [PubMed] [Google Scholar]
- 41.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 42.Volbeda, A., M. H. Charon, C. Piras, E. C. Hatchikian, M. Frey, and J. C. Fontecilla-Camps. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580-587. [DOI] [PubMed] [Google Scholar]
- 43.Woodward, J., S. M. Mattingly, M. Danson, D. Hough, N. Ward, and M. Adams. 1996. In vitro hydrogen production by glucose dehydrogenase and hydrogenase. Nat. Biotechnol. 14:872-874. [DOI] [PubMed] [Google Scholar]
- 44.Woodward, J., M. Orr, K. Cordray, and E. Greenbaum. 2000. Enzymatic production of biohydrogen. Nature 405:1014-1015. [DOI] [PubMed] [Google Scholar]
- 45.Wu, L. F., and M. A. Mandrand. 1993. Microbial hydrogenases: primary structure, classification, signatures and phylogeny. FEMS Microbiol. Rev. 10:243-269. [DOI] [PubMed] [Google Scholar]
- 46.Zirngibl, C., W. Van Dongen, B. Schworer, R. Von Bunau, M. Richter, A. Klein, and R. K. Thauer. 1992. H2-forming methylenetetrahydromethanopterin dehydrogenase, a novel type of hydrogenase without iron-sulfur clusters in methanogenic archaea. Eur. J. Biochem. 208:511-520. [DOI] [PubMed] [Google Scholar]
- 47.Zorin, N. A., M. Medina, M. A. Pusheva, I. N. Gogotov, and R. Cammack. 1996. Hydrogenase from the thermophilic bacterium Thermococcus stetteri: isolation and characterisation of EPR-detectable redox centres. FEMS Microbiol. Lett. 142:71-76. [Google Scholar]