Abstract
Silent Information Regulator 2 (Sir2) enzymes (or sirtuins) are NAD+-dependent deacetylases that modulate gene silencing, aging and energy metabolism. Previous work has implicated several transcription factors as sirtuin targets. Here, we investigated whether mammalian sirtuins could directly control the activity of metabolic enzymes. We demonstrate that mammalian Acetyl-CoA synthetases (AceCSs) are regulated by reversible acetylation and that sirtuins activate AceCSs by deacetylation. Site-specific acetylation of mouse AceCS1 on Lys-661 was identified by using mass spectrometry and a specific anti-acetyl-AceCS antibody. SIRT1 was the only member of seven human Sir2 homologues capable of deacetylating AceCS1 in cellular coexpression experiments. SIRT1 expression also led to a pronounced increase in AceCS1-dependent fatty-acid synthesis from acetate. Using purified enzymes, only SIRT1 and SIRT3 exhibited high catalytic efficiency against acetylated AceCS1. In mammals, two AceCSs have been identified: cytoplasmic AceCS1 and mitochondrial AceCS2. Because SIRT3 is localized to the mitochondria, we investigated whether AceCS2 also might be regulated by acetylation, and specifically deacetylated by mitochondrial SIRT3. AceCS2 was completely inactivated upon acetylation and was rapidly reactivated by SIRT3 deacetylation. Lys-635 of mouse AceCS2 was identified as the targeted residue. Using reversible acetylation to modulate enzyme activity, we propose a model for the control of AceCS1 by SIRT1 and of AceCS2 by SIRT3.
Keywords: acetate, deacetylation, Sir2, acetyltransferase, metabolism
Silent Information Regulator 2 (Sir2) enzymes function in a number of cellular processes such as gene silencing (1, 2), cell cycle regulation (3), fatty acid metabolism (4), lifespan extension (5, 6), and apoptosis (7–9). Sir2 proteins (or sirtuins) catalyze NAD+-dependent protein deacetylation, yielding nicotinamide and the metabolite O-acetyl-ADP-ribose as additional products (10). Among the seven human homologs (SIRT1–7) (11, 12), SIRT1 has been implicated in the regulation of transcription factors p53 (7–9), NFκB (13), FoxO (14–16), Mef2 (17), and PGC-1α (18, 19). PGC-1α is activated by signals that control energy and nutrient homeostasis. Also, PGC-1α controls gene expression that stimulates mitochondrial biogenesis, a thermogenic program in brown fat and metabolic pathways that regulate the fasting response in liver (20, 21). In liver cells, SIRT1 was shown to coordinate with PGC-1α to activate expression of gluconeogenic genes in response to fasting (18, 19). Using beta cell-targeted overexpression, SIRT1 enhanced insulin secretion and blood glucose homeostasis (22). In mitochondria, SIRT3 is implicated in regulating mitochondrial function and thermogenesis in brown adipocytes (23), and variability of the SIRT3 gene is linked to survivorship in the elderly (24, 25). Interestingly, PGC-1α contributes to the regulation of SREBP-1c (21) whose target genes include fatty acid synthase, acetyl-CoA carboxylase, and acetyl-CoA synthetase 1 (AceCS1). Moreover, these enzymes, which control lipogenesis (26–28), are transcriptional targets of insulin action (29). Although the molecular basis for the varied phenotypes remains unanswered, SIRT1 and SIRT3 appear to be regulators that may link transcription and pathways of energy homeostasis.
A Sir2 homologue in Salmonella enterica was shown to regulate the activity of AceCS through deacetylation, permitting bacterial growth on acetate and propionate (4). These observations led to our hypothesis that mammalian sirtuins might control acetyl-CoA synthetases and modulate acetyl-CoA synthesis from acetate. Unlike prokaryotes, mammals have only the AceCS pathway to convert free acetate back to a useable metabolite, acetyl-CoA. AceCS catalyzes the formation of acetyl-CoA from acetate, CoA and ATP. There are two known mammalian AceCSs, AceCS1 and AceCS2 (30). Found throughout the body, AceCS1 is localized to the cytoplasm and is most abundant in liver and kidney (30). Localized to mitochondria and broadly expressed, AceCS2 is found at high levels in kidney and heart muscle (30).
The roles of acetate metabolism and AceCS in mammals remain to be established. The serum levels of acetate are reported to be ≤0.2 mM in humans, although the sources of acetate are varied (ref. 31 and references therein). Acetate can be absorbed in the gut from the diet or from the byproducts of resident enteric bacteria. Acetate can be generated through endogenous metabolic processes, such as ethanol metabolism, acetyl-CoA hydrolases, acetylcholinesterase, and histone deacetylases class I and II (32). After ethanol consumption, acetate levels are elevated by as much as 20-fold (33). Under conditions of prolonged starvation and diabetes, endogenous pathways are the main source of serum acetate (34, 35). Acetate metabolism is impaired in diabetics (ref. 36 and references therein) and as humans age (31). Because mammalian AceCSs are essential in converting free acetate to acetyl-CoA, we explored the molecular mechanism for controlling AcsCS activity at the posttranslational level. Here, we investigated whether mammalian sirtuins regulate AceCS enzymes through reversible acetylation.
Results
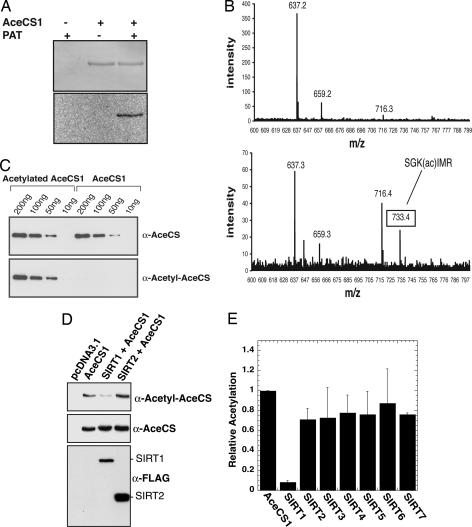
To provide initial evidence that mammalian AceCS proteins may be regulated by reversible acetylation, we used purified recombinant mouse AceCS1 and examined whether the synthetase could be acetylated in vitro by a protein acetyltransferase (PAT) from bacteria (37). We reasoned that, because of the high conservation of AceCSs across diverse species, PAT might acetylate mouse AceCS1. Indeed, PAT was able to catalyze the transfer of the acetyl group from [1-14C]acetyl-CoA to AceCS1, as depicted in Fig. 1A. To identify the site(s) of acetylation, unacetylated and acetylated AceCS1 were enzymatically digested and analyzed by mass spectrometry. Lys-661 was found to be acetylated (Fig. 1B), identifying this residue as the PAT-targeted acetylation site. Lys-661 is a putative catalytic residue, functioning in the ATP-dependent adenylation of acetate to yield acetyl-adenosine monophosphate during the initial step in catalysis (4).
Fig. 1.
AceCS1 is acetylated in vitro and in mammalian cells. (A) AceCS1 is acetylated in vitro by PAT. Recombinant AceCS1 was incubated in the presence or absence of PAT and [1-14C]acetyl-CoA for 1h, resolved by SDS/PAGE and detected by Coomassie (Upper) and autoradiography (Lower). (B) MALDI-TOF confirms Lys-661 is acetylated on AceCS1. (Upper) Tryptic digest of AceCS1. (Lower) tryptic digest of acetylated AceCS1. As expected, the spectra are similar, with Lower showing a new peak corresponding to the acetylated peptide, SGK(ac)IMR. This peptide was further confirmed by MS/MS on the TOF–TOF instrument. (C) Acetylation state of AceCS1 can be detected with an anti-acetyl-AceCS antibody. Recombinant AceCS1 and acetyl-AceCS1 were resolved by SDS/PAGE and detected by Western blotting with anti-AceCS and anti-acetyl-AceCS antibodies, respectively. (D) Acetylation of AceCS1 is decreased upon SIRT1 coexpression. Cos-7 cells cotransfected with a construct expressing AceCS1 and the construct expressing SIRT1 or SIRT2. After 48 h, cell extracts were resolved by SDS/PAGE and detected by Western blot, using anti-acetyl-AceCS, anti-AceCS, and anti-FLAG antibodies. (E) Coexpression of AceCS1 and SIRT1–7 show increased deactylation of AceCS1 by SIRT1. The ratio of acetylated AceCS1 to total AceCS1 from cell lysates was calculated from densitometry of Western blots with anti-AceCS and anti-acetyl-AceCS1 antibodies relative to AceCS1 transfection alone.
To demonstrate that AceCS1 is acetylated on Lys-661 in mammalian cells, we developed an acetylation-specific polyclonal antibody by using the AceCS1 peptide sequence LPKTRSGK(ac)IMRR. The specificity and cross-reactivity of the immunopurified antibody (α-acetyl-AceCS) were examined by using purified recombinant mouse AceCS1 that was either acetylated by PAT or left unmodified (Fig. 1C). Western blot analysis with the α-acetyl-AceCS antibody showed high reactivity with the acetylated protein but negligible reactivity with the unmodified protein (Fig. 1C Lower). A polyclonal antibody (α-AceCS) that recognizes both acetylated and unmodified AceCS1 was used as a control (Fig. 1C Upper). These data clearly demonstrate that PAT acetylates Lys-661 of mouse AceCS1.
Having established the utility of the α-acetyl-AceCS antibody, next we determined whether AceCS1 is acetylated in mammalian cells by an endogenous mammalian acetyltransferase. Transfection of Cos-7 cells was carried out with DNA plasmids encoding AceCS1 or an empty vector (pcDNA3.1). To detect the acetylation state of AceCS1, extracts from these cells were analyzed by Western blotting using both α-acetyl-AceCS and α-AceCS (Fig. 1D). Immunoreactivity with the α-acetyl-AceCS antibody was prominent in cells overexpressing the AceCS1 protein. To verify that the immunoreactivity was due to acetylation, cotransfection of FLAG-tagged SIRT1 and SIRT2 (two sirtuins that display cytoplasmic localization; refs. 22 and 38) with AceCS1 was performed. SIRT1 coexpression led to a dramatic decrease in the acetyl-specific reactivity with α-acetyl-AceCS, whereas SIRT2 coexpression yielded no net change in the acetylation status of AceCS1 (Fig. 1D). These data provide evidence that AceCS1 is acetylated by an endogenous protein acetyltransferase and that sirtuins, at least SIRT1, are capable of rendering deacetylated AceCS1.
To examine whether sirtuins SIRT3 through SIRT7 were able to induce the deacetylation of AceCS1 in this system, coexpression studies were extended to all sirtuins (Fig. 1E). AceCS1 was coexpressed with either a vector control or a vector encoding one of the seven human homologs SIRT1–7, and the acetylation state was determined relative to total AceCS1 protein (Fig. 1E), using densitometry of the α-acetyl-AceCS and α-AceCS Western blots, as in Fig. 1D. The average values of five separate experiments with error bars showing one standard deviation are depicted in Fig. 1E. Strikingly, only SIRT1 induced the dramatic loss of acetylation on AceCS1.
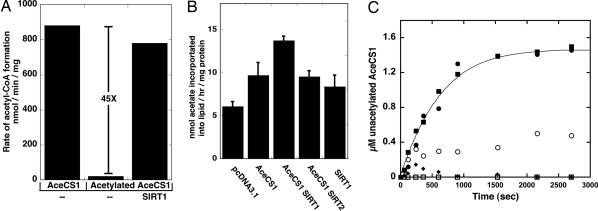
To establish the functional consequence of mammalian AceCS1 acetylation, we measured in vitro AceCS1's activity before and after acetylation by PAT (Fig. 2A). Using a TLC-based assay to monitor acetyl-CoA synthesis, we found that purified recombinant AceCS1 displayed robust activity (880 nmol/min per mg), which was similar to that reported for AceCS from Salmonella enterica (4). In dramatic contrast, acetylation rendered the enzyme essential inactive (≤20 nmol/min per mg). To examine whether acetylated AceCS1 could be reactivated by direct deacetylation, SIRT1 was incubated with acetylated AceCS1 and activity assays were performed. AceCS1 was completely reactivated (780 nmol/min per mg or >45-fold) upon deacetylation (Fig. 2A).
Fig. 2.
AceCS1 activity is regulated by sirtuin-catalyzed deacetylation. (A) AceCS1 activity is regulated by reversible acetylation. AceCS1 or acetylated AceCS1 (5 μM) was incubated for 1 h with 2 μM SIRT1 at 37°C. AceCS1 activity was determined by measuring the formation of acetyl-CoA over time from [14C]acetate, and reported as nmol of acetyl-CoA per min per mg of protein. (B) 1,2-[14C]acetate incorporation into lipids through AceCS1 is enhanced upon SIRT1 coexpression. Cos-7 cells were cotransfected with a vector encoding AceCS1 and/or SIRT1, SIRT2, or empty vector where indicated. Lipids were extracted as described in Materials and Methods, and [14C]acetate incorporation was analyzed by scintillation counting and normalized to protein content. ANOVA statistical analysis was performed, indicating significant differences (P < 0.05) between SIRT1 and AceCS1 coexpression and AceCS1 expression alone. (C) SIRT1 and SIRT3 show high catalytic activity against acetylated AceCS1. Catalytic amounts of SIRT1 (filled circles), SIRT2 (open circles), SIRT3 (filled squares), SIRT5 (filled diamonds), and SIRT6 (open squares) (50 nM) were incubated with 1.5 μM acetylated AceCS1 in 1 mM NAD+, 1 mM DTT, and 50 mM Tris (pH 7.5) at 37°C for the indicated time and deacetylation of AceCS1 was measured by AceCS1 activity assays. Data from SIRT1 and SIRT3 were fitted to the integrated Michaelis–Menten equation (45).
Previously, AceCS1 overexpression in mammalian cells was shown to yield enhanced rates of fatty acid synthesis through acetate (27). To provide cellular evidence that AceCS1 can be regulated by SIRT1, we coexpressed AceCS1 with either control vector, SIRT1, or SIRT2, and measured the incorporation of [14C]acetate into total lipids using chloroform–methanol extraction (39). Compared to vector alone, AceCS1 expression led to an overall 60% increase of acetate incorporation into lipids (Fig. 2B). Coexpression of AceCS1 with SIRT1, but not SIRT2, enhanced acetate incorporation by 130% compared to vector alone (Fig. 2B). Fig. 2B represents the averages of three separate experiments and the error bars indicate one standard deviation. Although acetate incorporation with SIRT1 expression alone was slightly higher than that for the vector control, this particular difference was not statistically significant. Because DNA transfection efficiency was typically <90%, the observed lipogenesis enhanced by SIRT1/AceCS1 overexpression (Fig. 2B) may be a lower limit of the actual acetate flux into lipids. The synergistic effect of SIRT1 and AceCS1 expression on lipogenesis provides a functional link between AceCS1 and SIRT1.
To provide direct evidence for the deacetylation of AceCS1 by SIRT1, we performed quantitative deacetylation assays with catalytic amounts (50 nM) of various sirtuins. For this analysis, we compared the efficiency of recombinant, purified SIRT1, SIRT2, SIRT3, SIRT5, and SIRT6 to deacetylate and activate acetylated AceCS1 (1.5 μM) (Fig. 2C). Progress curves of AceCS1 deacetylation were monitored over 40 min, and where possible, the data were fitted to the integrated form of the Michaelis–Menten equation to obtain the specificity constant Vmax/Km. SIRT5 and SIRT6 displayed no significant activity against acetylated AceCS1. In stark contrast, SIRT1 and SIRT3 displayed highly efficient deacetylation and activation of AceCS1, with identical Vmax/Km values of 3.2 × 104 M−1·s−1 (Fig. 2C). SIRT2 showed a considerably lower rate of deacetylation that could not be fitted accurately. The ability of SIRT1 and SIRT3 to reactivate AceCS1 with nearly identical kinetics suggested that SIRT3 might also function as an AceCS deacetylase. However, AceCS1 would not likely be the in vivo target of SIRT3, because SIRT3 is localized to mitochondria (40, 41). The lack of SIRT3 induced deacetylation of cytoplasmic AceCS1 in cell expression studies (Fig. 1E) supports this assertion. Interestingly, AceCS2 is localized exclusively to mitochondria (30). Because both SIRT3 (40, 41) and AceCS2 are found in mitochondria, we reasoned that AceCS2 might be regulated by reversible acetylation and specifically controlled by SIRT3.
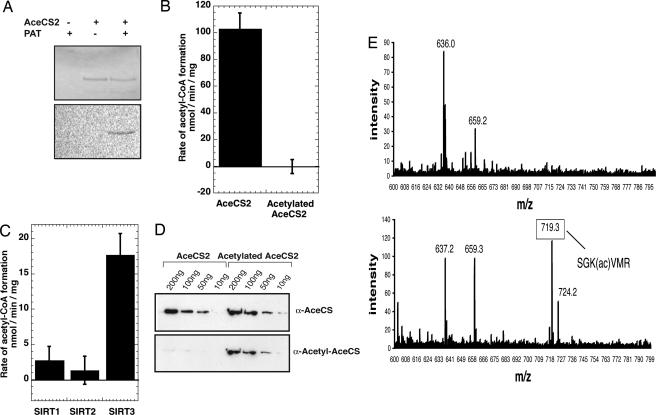
To explore this idea, we cloned, expressed, and purified AceCS2 and subjected this enzyme to a similar analysis to that for AceCS1. Full-length AceCS2 (amino acids 1–682) was inactive, whereas a truncated version (amino acids 39–682), which is the putative mitochondrial form, displayed enzymatic activity (100 nmol/min per mg). To demonstrate that AceCS2 is regulated by reversible acetylation, we show that PAT readily acetylated AceCS2 (Fig. 3A), yielding completely inactive enzyme (Fig. 3B). Deacetylation by SIRT3 re-activated AceCS2 at a rate that was ≈5-fold higher than SIRT1 (Fig. 3C), whereas SIRT2 displayed no significant ability to deacetylate AceCS2 under the assay conditions. To verify that Lys-635 of AceCS2 (homologous to Lys-661 of AceCS1) is the regulatory acetylation site, both Western blot analyses and mass spectrometry were performed. Indeed, the α-acetyl-AceCS antibody detected acetylation of AceCS2 (Fig. 3D), and mass spectrometry confirmed a single acetylation at Lys-635 (Fig. 3E). These data provide direct evidence that AceCS2 is inactivated by acetylation on Lys-635, and suggest that SIRT3 would function as the regulatory deacetylase in mitochondria.
Fig. 3.
AceCS2 activity is regulated by SIRT3-catalyzed deacetylation. (A) AceCS2 is acetylated in vitro by PAT. Recombinant AceCS2 was incubated in the presence or absence of PAT and [14C]acetyl-CoA for 1 h, resolved by SDS/PAGE, and detected by Coomassie (Upper) and autoradiography (Lower). (B) AceCS2 activity is regulated by reversible acetylation. AceCS2 or acetylated AceCS2 (2 μM) activity was determined by measuring the formation of acetyl-CoA over time from [14C]acetate, and reported as nmol of acetyl-CoA per min per mg of protein. (C) AceCS2 is preferentially deacetylated by SIRT3. Catalytic amounts of SIRT1, SIRT2, and SIRT3 (50 nM) were incubated with 1.0 μM acetylated AceCS2 in 1 mM NAD+, 1 mM DTT, and 50 mM Tris (pH 7.5) at 37°C with time points taken over 40 min. The ability of sirtuins to deacetylate and activate acetylated AceCS2 was followed by measuring the rate acetyl-CoA formation (nmol of acetyl-CoA per min per mg of protein). (D) Acetylation state of AceCS2 can be detected with an anti-acetyl-AceCS antibody. Recombinant AceCS2 and acetyl-AceCS2 were resolved by SDS/PAGE and detected by Western blotting with anti-AceCS and anti-acetyl-AceCS antibodies, respectively. (E) MALDI-TOF confirms Lys-635 is acetylated on AceCS2. (Upper) Tryptic digest of AceCS2. (Lower) Tryptic digest of acetylated AceCS2. As expected, the spectra are similar, with Lower showing a new peak corresponding to the acetylated peptide, SGK(ac)VMR. This peptide was further confirmed by MS/MS on the TOF-TOF instrument.
Discussion
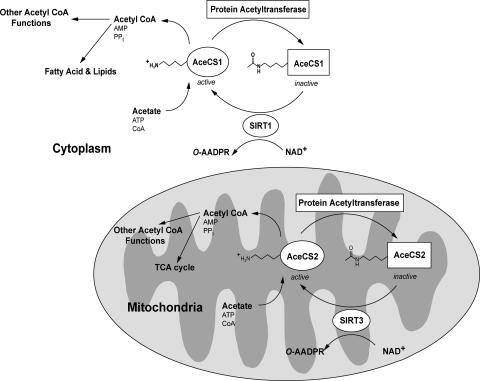
Based on the data presented, we propose the following working model where mammalian metabolic enzymes AceCS1 and AceCS2 are controlled by reversible acetylation (Fig. 4). Amazingly, this regulatory mechanism appears to be evolutionarily conserved, as Salmonella enterica harbor a similar regulatory circuit (37). This regulatory “on–off” switch for controlling AceCS activity is modulated by SIRT1 acting on cytoplasmic AceCS1, and by SIRT3 acting on AceCS2 in the mitochrondria (Fig. 4). Acetylation of the conserved lysine residue inactivates the two AceCSs, whereas sirtuin-catalyzed deacetylation completely reactivates the synthetases. We show that AceCS1 is acetylated in cells, and that specific deacetylation by SIRT1 leads to enhanced fatty acid synthesis through acetate incorporation (Fig. 2B). Furthermore, we provide direct evidence that mitochondrial AceCS2 is activated by SIRT3-catalyzed deacetylation at Lys-635. Expression of AceCS2 (J.M.D. and W.C.H. unpublished data, and ref. 30) causes a dramatic increase in CO2 output (tricarboxylic acid cycle) in mitochondria. Expression of AceCS2 led to a 10-fold increase in CO2, whereas AceCS1 expression led to no change in CO2 liberated (J.M.D. and W.C.H. unpublished data), indicating that the acetyl-CoA pools are distinct. Together, these results suggest that SIRT3 may regulate acetyl-CoA levels through the modulation of AceCS2 activity in mitochondria (Fig. 4).
Fig. 4.
Proposed model for the regulation of mammalian AceCSs AceCS1 and AceCS2 by sirtuins SIRT1 and SIRT3. AceCSs are inactivated by protein acetyltransferases (PATs), which acetylate an important catalytic lysine residue. The PAT(s) responsible for acetylating AceCSs remain to be determined. In the cytoplasm, SIRT1 catalyzes the deacetylation of AceCS1 on Lys-661 (murine). In the mitochondria, SIRT3 catalyzes the deacetylation of AceCS2 on Lys-635 (murine). Free acetate, generated from endogenous cellular reactions or absorbed from the gut, is converted to acetyl-CoA that can be used in metabolic pathways, such as fatty acid synthesis (cytoplasm) or the tricarboxylic acid cycle in mitochondria. Other acetyl-CoA requiring enzymes/pathways also may be regulated by sirtuin-controlled AceCS activity.
Caloric restriction in mammals is known to increase lifespan. In recent reports, SIRT1 and SIRT3 have been implicated in organism longevity. With calorie-restricted mice, SIRT1 was required for the induction of an increased-activity phenotype (42). Variability of the SIRT3 gene was linked to survivorship in the elderly (24, 25). Moreover, SIRT3 was implicated in regulating mitochondrial function and thermogenesis in brown adipocytes (23). The molecular basis for these phenotypes is unclear. It will be interesting to examine whether the control of AceCSs by SIRT1 and SIRT3 are involved in these processes, and whether additional metabolic enzymes are regulated by reversible acetylation.
Although previous reports (and the current study) show that the acetyl-CoA generated from AceCS1 and AceCS2 can be used in fatty acid synthesis and the tricarboxylic acid cycle, respectively, the acetyl-CoA generated by AceCSs may feed directly into other acetyl-CoA-requiring pathways, such as those dependent on protein acetyltransferases. In addition to future studies toward understanding the role of acetate metabolism and AceCSs in human physiology, it will be critical to identify the mammalian AceCS PAT(s) and elucidate the signaling pathways that modulate SIRT1, SIRT3, and PAT(s) activity.
Materials and Methods
Materials.
The plasmids pcDNA3.1 encoding SIRT1–7 with a FLAG tag, were obtained from E. Verdin (University of California, San Francisco). The plasmids pCD-SRα encoding AceCS1 and AceCS2 was obtained from T. Fujino (Tohoku University, Sendai, Japan). The anti-AceCS antibody was obtained from A. J. Wolfe (Loyola University Chicago, Maywood, IL). The PAT was obtained from J. C. Escalante-Semerena (University of Wisconsin, Madison). A rabbit polyclonal antibody specific for acetylated AceCS was raised against the AceCS1 peptide LPKTRSGK(ac)IMRR, conjugated to keyhole limpet hemocyanin (KLH), and was affinity purified against acetylated peptide (Invitrogen). Additionally anti-FLAG (M2 Sigma) was used.
Cell Culture and Transfection.
Cos-7 cells were cultured as a monolayer in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cotransfection of Cos-7 cells was performed by using 4 μg of total DNA with lipofectamine 2000 (Invitrogen) according to manufacturer's protocol and as described in the figure legends. Soluble cell extracts were made in RIPA buffer (Sigma) with 5 mM nicotinamide, 100 μM TSA, and HALT protease inhibitor (Pierce). Protein concentration was quantified by Bradford assay (Bio-Rad).
Expression and Purification of Recombinant AceCS1, AceCS2, SIRT1, SIRT2, SIRT3, SIRT5, and SIRT6.
AceCS1, AceCS2 full length, AceCS2 truncated, SIRT1, SIRT3 SIRT5, and SIRT6 from mammalian expression vectors were cloned into the bacterial expression vector pQE-80 (Qiagen). AceCS2 full length contains amino acids 1–682, and AceCS2 truncated contains amino acids 39–682. His-tagged SIRT2 was developed as described (38). The plasmids were transformed into Escherichia coli BL21DE3. The transformed bacteria were induced with isopropyl-d-thiogalactopyranoside (IPTG), lysed and purified by nickel affinity chromatography. AceCS1, AceCS2 full length, and truncated AceCS2 were purified further on a phenyl-Sepharose column (detailed primer sequence, expression, and purification are described in Supporting Text, which is published as supporting information on the PNAS web site).
Acetylation and Purification of AceCS1 and AceCS2.
AceCS1 and truncated AceCS2 (5–100 μg) were acetylated by PAT (1–10 μg) in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) (pH 7.5), 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and 20 μM acetyl-CoA or 20 μM 1-[14C]acetyl-CoA (10 mCi/mmol; 1 Ci = 37 GBq) where indicated (37). Acetylated AceCS1 and acetylated AceCS2 were separated from PAT through nickel affinity chromatography.
AceCS Activity Assay.
AceCS1, AceCS2, acetylated AceCS1 and acetylated AceCS2 activities were measured in 100 μl of 50 mM Hepes (pH 7.5), 200 μM TCEP, 5 mM ATP, 5 mM MgCl2, 5 mM CoA, 250 μM 1-[14C]acetate (5 mCi/mmol), and 5 mM nicotinamide. Reactions were quenched with trifluoroacetic acid (TFA) and separated by thin layer chromatography (TLC) in 3:2 chloroform/methanol. The amount of [14C]acetyl-CoA formed over time was detected and quantified by PhosphorImaging (43). Assays were carried out under saturating levels of acetate, CoA, and ATP. Rates were determined where product formed was linear with time.
Deacetylation Assay.
Deacetylation of acetylated AceCS1 and AceCS2 by sirtuins was performed in 100 μl of Tris (pH 7.5), 1 mM NAD+, and 1 mM DTT at 37°C (44). Specific enzyme concentrations and times are indicated in figure legends. For the deacetylation progress curves, catalytic amounts of SIRT1, SIRT2, SIRT3, SIRT5, and SIRT6 (50 nM) were incubated with 1.5 μM acetylated AceCS1. Aliquots were taken over time and the extent of AceCS1 deacetylation was quantified from the rate of acetyl-CoA formation. The fractional activation of maximal AceCS1 activity was multiplied by the initial acetylated AceCS1 concentration to obtain the amount of deacetylated AceCS1. Data from SIRT1 and SIRT3 were fitted to the integrated Michaelis–Menten equation (45).
[14C]Acetate Incorporation into Lipids.
Two days after transfection, Cos-7 cells expressing AceCS1 and the indicated sirtuins were incubated in Hanks' balanced salt solution with 25 mM glucose and 1 mM 1,2-[14C]acetate (1.9 mCi/mmol) at 37°C for 2 h. The lipids were then extracted as described by Folch et al. (39). The cells were collected by centrifugation, quenched with 25 μl of 10% TFA, and extracted twice with 2:1 chloroform/methanol. The extracted lipids were then added to liquid scintillation fluid and the radioactivity was quantified by using liquid scintillation counting.
Mass Spectrometry.
The MALDI-TOF analysis of tryptic digests of AceCS1, AceCS2, acetylated AceCS1, and acetylated AceCS2 identified peptides from the respective proteins with only the acetylated peptides, SGK(ac)IMR and SGK(ac)VMR, present in the acetylated AceCS1 and acetylated AceCS2 samples, respectively. Nanospray LC-MS/MS analyses for both protein sets did not identify other acetylation sites. See Supporting Text and Table 1, which is published as supporting information on the PNAS web site, for detailed methods and data.
Supplementary Material
Acknowledgments
We thank Terri Kowieski for assistance with the cloning and expression of the mammalian Sir2 homologs and Olivera Grubisha for assistance in editing. This work supported by National Institutes of Health Grants GM065386 and GM059785.
Abbreviations
- AceCS
acetyl-CoA synthetase
- PAT
protein acetyltransferase
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Rusche L. N., Kirchmaier A. L., Rine J. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 2.Gasser S. M., Cockell M. M. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- 3.Dryden S. C., Nahhas F. A., Nowak J. E., Goustin A. S., Tainsky M. A. Mol. Cell. Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 5.Rogina B., Helfand S. L. Proc. Natl. Acad. Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tissenbaum H. A., Guarente L. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 7.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 8.Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 9.Langley E., Pearson M., Faretta M., Bauer U. M., Frye R. A., Minucci S., Pelicci P. G., Kouzarides T. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denu J. M. Curr. Opin. Chem. Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Frye R. A. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 12.Frye R. A. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 13.Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., et al. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 15.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 16.Daitoku H., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A. Proc. Natl. Acad. Sci. USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X., Sternsdorf T., Bolger T. A., Evans R. M., Yao T. P. Mol. Cell. Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 19.Nemoto S., Fergusson M. M., Finkel T. J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 20.Puigserver P. Int. J. Obes. (London) 2005;29(Suppl. 1):S5–S9. doi: 10.1038/sj.ijo.0802905. [DOI] [PubMed] [Google Scholar]
- 21.Lin J., Handschin C., Spiegelman B. M. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Moynihan K. A., Grimm A. A., Plueger M. M., Bernal-Mizrachi E., Ford E., Cras-Meneur C., Permutt M. A., Imai S. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Shi T., Wang F., Stieren E., Tong Q. J. Biol. Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 24.Rose G., Dato S., Altomare K., Bellizzi D., Garasto S., Greco V., Passarino G., Feraco E., Mari V., Barbi C., et al. Exp. Gerontol. 2003;38:1065–1070. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 25.Bellizzi D., Rose G., Cavalcante P., Covello G., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., et al. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda Y., Yamamoto J., Okamura M., Fujino T., Takahashi S., Takeuchi K., Osborne T. F., Yamamoto T. T., Ito S., Sakai J. J. Biol. Chem. 2001;276:34259–34269. doi: 10.1074/jbc.M103848200. [DOI] [PubMed] [Google Scholar]
- 27.Luong A., Hannah V. C., Brown M. S., Goldstein J. L. J. Biol. Chem. 2000;275:26458–26466. doi: 10.1074/jbc.M004160200. [DOI] [PubMed] [Google Scholar]
- 28.Sone H., Shimano H., Sakakura Y., Inoue N., Amemiya-Kudo M., Yahagi N., Osawa M., Suzuki H., Yokoo T., Takahashi A., et al. Am. J. Physiol. Endocrinol. Metab. 2002;282:E222–30. doi: 10.1152/ajpendo.00189.2001. [DOI] [PubMed] [Google Scholar]
- 29.Horton J. D., Goldstein J. L., Brown M. S. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujino T., Kondo J., Ishikawa M., Morikawa K., Yamamoto T. T. J. Biol. Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 31.Skutches C. L., Holroyde C. P., Myers R. N., Paul P., Reichard G. A. J. Clin. Invest. 1979;64:708–713. doi: 10.1172/JCI109513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfe A. J. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundquist F., Tygstrup N., Winkler K., Mellemgaard K., Munck-Petersen S. J. Clin. Invest. 1962;41:955–961. doi: 10.1172/JCI104574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheppach W., Pomare E. W., Elia M., Cummings J. H. Clin. Sci. (London) 1991;80:177–182. doi: 10.1042/cs0800177. [DOI] [PubMed] [Google Scholar]
- 35.Akanji A. O., Humphreys S., Thursfield V., Hockaday T. D. Clin. Chim. Acta. 1989;185:25–34. doi: 10.1016/0009-8981(89)90127-7. [DOI] [PubMed] [Google Scholar]
- 36.Akanji A. O., Hockaday T. D. Am. J. Clin. Nutr. 1990;51:112–118. doi: 10.1093/ajcn/51.1.112. [DOI] [PubMed] [Google Scholar]
- 37.Starai V. J., Escalante-Semerena J. C. J. Mol. Biol. 2004;340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 38.North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 39.Folch J., Lees M., Sloane Stanley G. H. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 40.Onyango P., Celic I., McCaffery J. M., Boeke J. D., Feinberg A. P. Proc. Natl. Acad. Sci. USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwer B., North B. J., Frye R. A., Ott M., Verdin E. J. Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen D., Steele A. D., Lindquist S., Guarente L. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 43.Starai V. J., Takahashi H., Boeke J. D., Escalante-Semerena J. C. Genetics. 2003;163:545–555. doi: 10.1093/genetics/163.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borra M. T., Denu J. M. Methods Enzymol. 2004;376:171–187. doi: 10.1016/S0076-6879(03)76011-X. [DOI] [PubMed] [Google Scholar]
- 45.Berndsen C. E., Denu J. M. Methods. 2005;36:321–331. doi: 10.1016/j.ymeth.2005.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.