Abstract
A major role of the methyl-directed mismatch repair (MMR) system of Escherichia coli is to repair postreplicative errors. In this report, we provide evidence that MMR also acts on oxidized DNA, preventing mutagenesis. When cells deficient in MMR are grown anaerobically, spontaneous mutation frequencies are reduced compared with those of the same cells grown aerobically. In addition, we show that a dam mutant has an increased sensitivity to hydrogen peroxide treatment that can be suppressed by mutations that inactivate MMR. In a dam mutant, MMR is not targeted to newly replicated DNA strands and therefore mismatches are converted to single- and double-strand DNA breaks. Thus, base pairs containing oxidized bases will be converted to strand breaks if they are repaired by MMR. This is demonstrated by the increased peroxide sensitivity of a dam mutant and the finding that the sensitivity can be suppressed by mutations inactivating MMR. We demonstrate further that this repair activity results from MMR recognition of base pairs containing 8-oxoguanine (8-oxoG) based on the finding that overexpression of the MutM oxidative repair protein, which repairs 8-oxoG, can suppress the mutH-dependent increase in transversion mutations. These findings demonstrate that MMR has the ability to prevent oxidative mutagenesis either by removing 8-oxoG directly or by removing adenine misincorporated opposite 8-oxoG or both.
In Escherichia coli, the methyl-directed mismatch repair (MMR) system is initiated after replication and one of its primary functions is to remove base-base mismatches or small insertion-deletion loops generated by misincorporation or strand slippage during replication of DNA (10). MMR has been conserved from prokaryotes to eukaryotes and has been shown to function in homologous and homeologous recombination and transcription-coupled repair and to act on base pairs containing lesions (5, 6, 8, 15, 20, 28). In humans, defects in MMR result in elevated spontaneous mutation rates and microsatellite instability, leading to an increased predisposition to certain cancers (19).
Reactive oxygen species are considered to be a major threat to the integrity of DNA, as well as that of proteins, lipids, and carbohydrates (1, 14). In aerobically growing cells, reactive oxygen species are produced as by-products of normal metabolic pathways and have been shown to contribute to human diseases including cancer, cardiovascular disease, immune system decline, brain dysfunction, and cataracts (1). Some of these by-products include singlet oxygen (1O2), peroxide radicals (·O2), hydrogen peroxide (H2O2), and hydroxyl radicals (·OH) (1, 7). Although H2O2 is relatively stable, it can rapidly react with Fe2+ to produce highly reactive ·OH radicals in a process described by the Fenton reaction (11, 12). This ·OH radical can then react with DNA to produce a variety of DNA lesions. Reactions with guanine lead to 7,8-dihydro-8-oxoguanine (8-oxoG), which is the most common lesion produced (27).
In E. coli, several enzymes are involved in processing oxidative DNA damage due to 8-oxoG. One enzyme is the MutM glycosylase, or formamidopyrimidine-DNA glycosylase, encoded by the mutM gene (17). This enzyme functions to remove 8-oxoG lesions found in DNA (22). If MutM removes the lesion prior to replication, then the base excision repair pathway can restore the original G·C base pair (18). If the lesion is not removed prior to replication, then this will result in either another C·8-oxoG pair, which is subject to another attempt at repair by MutM, or misincorporation of adenine opposite the 8-oxoG lesion, leading to GC → TA transversions (3, 17). Another enzyme involved in removal of oxidative damage due to 8-oxoG is MutY, encoded by the mutY gene (18). Whereas the MutM protein removes 8-oxoG lesions from DNA, the MutY protein removes the adenine base from the A·8-oxoG mispair (16). Once the misincorporated adenine is removed, MutM can then make another attempt at repair. Together, these enzymes function to reduce the GC → TA transversions most commonly associated with 8-oxoG.
Recently, mismatch correction has been implicated in the repair of oxidatively damaged bases, possibly due to 8-oxoG. In a recent report by DeWeese et al. (5), mouse embryonic stem cells deficient in MMR were shown to display increased levels of 8-oxoG after exposure to low-level radiation compared with those in wild-type cells. Earley and Crouse (6) have determined that Saccharomyces cerevisiae cells deficient in MMR that are grown anaerobically display a reduction in reversion rates, presumably due to 8-oxoG, and Ni et al. (20) have shown that the MutS homologs MSH2 and MSH6 function to remove adenine misincorporated opposite 8-oxoG. In E. coli, the overexpression of MutS protein was shown to reduce GC → TA transversions, suggesting the ability to correct A·8-oxoG mismatches (28), and human homologs of the MutS protein have been shown to bind to mismatched 8-oxoG lesions (15). In this study, we show that the MMR system of E. coli acts on DNA containing oxidative damage. In addition, we show that overexpression of MutM in a MutH-deficient strain reduces the rate of the GC → TA transversions most commonly associated with 8-oxoG. Therefore, the results presented here, along with previously reported results, indicate that the MMR system of E. coli functions to recognize and repair oxidative damage due to 8-oxoG.
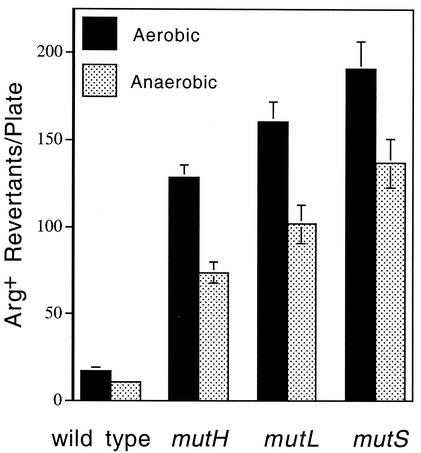
Reduction of spontaneous mutagenesis in anaerobically growing MMR-deficient strains.
Since oxidative mutagenesis results from the production of reactive oxygen species, which are by-products of normal metabolic pathways in aerobically growing cells, we investigated whether growing cells anaerobically would reduce the mutation frequency of MMR-deficient strains. Figure 1 presents a comparison of mutagenesis in wild-type strains and MMR-deficient strains grown aerobically or anaerobically. The strains used in this experiment (Table 1) are Arg− and grow to a constant level until the limited amount of arginine is exhausted, forming a lawn of approximately 5 × 109 cells per plate. Arg+ revertants continue to grow, forming colonies on the lawn of Arg− mutants, and are counted after incubation (25). Strains carrying a mutation in mutH, mutL, or mutS that are grown aerobically show an increased reversion frequency compared with that of the wild type. When these strains are grown anaerobically, the spontaneous mutation rate is decreased by roughly 30 to 40%. This indicates that some of the increase in mutations seen in mutH, mutL, or mutS strains can be reduced by limiting oxygen levels and implies that MMR may function to prevent oxidative mutagenesis. These results are similar to those of Earley and Crouse (6), who examined the effect of anaerobiosis on mutagenesis in MMR-deficient strains of S. cerevisiae, but must be taken with caution as factors other than oxidative damage may affect mutagenesis when cells grown with and without oxygen are compared.
FIG. 1.
Reduction of spontaneous mutagenesis in anaerobically growing MMR-deficient strains. Overnight cultures grown in Luria broth were plated onto semienriched (ESEM) plates (24) for selection of Arg+ revertants and incubated aerobically at 37°C for 2 days and anaerobically at 37°C for 5 days. The Arg+ revertants were counted. Data shown are averages of 19 independent experiments; error bars indicate the standard errors of the means.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype | Description | Source or reference |
|---|---|---|---|
| AB1157 and its derivatives | |||
| AB1157 | Wild type | argE3 hisG4 leu-6 proA2 thr-1 ara14 galK2 lacY1 mtl-1 xyl-5 rpsL31 supE44 tsx-6 | 2 |
| GM3819 | dam-16::kan | dam-16::kan derivative of AB1157 | M. Marinusa |
| GM5556 | dam-16 mutH464 | dam-16::kan mutH464::Tn10 derivative of AB1157 | M. Marinusa |
| GM7650 | dam-16 mutS215 | dam-16::kan mutS215::Tn10 derivative of AB1157 | M. Marinusa |
| MV1161 and its derivatives | |||
| MV1161 | Wild type | rfa-550 derivative of AB1157 | 26 |
| MV4478 | mutH472::Tn10 | mutH transductant of MV1161 | This study |
| MV4479 | mutL218::Tn10 | mutL transductant of MV1161 | This study |
| MV4480 | mutS215::Tn10 | mutS transductant of MV1161 | This study |
| CC104 and its derivatives | |||
| CC104 | Wild type | lacZ CC104 | 4 |
| MV6015 | Wild type/pTrc99a | pTrc99a transformant of CC104 | This study |
| MV6016 | Wild type/pmutM | pmutM transformant of CC104 | This study |
| MV4511 | mutH472::Tn10 | mutH transductant of CC104 | This study |
| MV6017 | mutH472::Tn10/pTrc99a | pTrc99a transformant of MV4511 | This study |
| MV6018 | mutH472::Tn10/pmutM | pmutM transformant of MV4511 | This study |
| Plasmids | |||
| pTrc99a | Vector | Pharmacia | |
| pmutM | mutM+ cloned into pTrc99a | Lab stock |
Full descriptions of these strains are available at the following website: http://users.umassmed.edu/martin.marinus/dstrains.html.
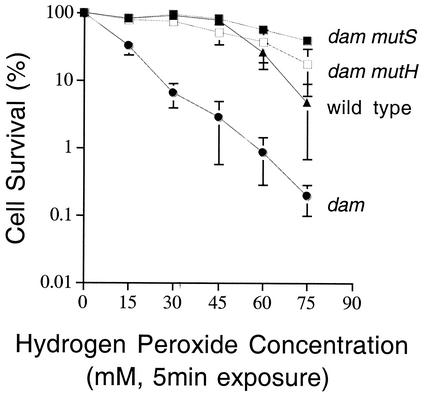
dam mutation results in increased sensitivity to oxidative DNA damage.
To determine whether MMR repairs mismatches of bases containing oxidative lesions, a dam mutant, which lacks the ability to methylate DNA, was examined for increased sensitivity. A dam mutant strain retains the ability to carry out MMR; however, without methylation of DNA, MutH cannot discriminate between parental and newly synthesized DNA strands and will therefore nick both strands at d(GATC) sequences in a mutS-dependent reaction (13). The predominant oxidative lesion, 8-oxoG, is readily bypassed by DNA polymerases and has little effect on lethality (9). In a dam mutant strain, base pairs containing 8-oxoG will be converted to single- and double-strand breaks if MMR recognizes base pairs containing oxidative lesions, which will increase sensitivity to hydrogen peroxide treatment. The data in Fig. 2 show this to be the case; the dam mutant strain is more sensitive to hydrogen peroxide treatment than is a wild-type strain. This result strongly suggests that MMR recognizes base pairs containing oxidative lesions.
FIG. 2.
MutHLS suppression of dam-mediated peroxide sensitivity. Frozen stocks were streaked onto Luria broth (LB) plates containing, when necessary, kanamycin (75 μg/ml) and/or tetracycline (15 μg/ml) and grown overnight at 30°C. Single colonies were then inoculated into LB medium containing the appropriate antibiotics along with 1× MOPS (morpholinepropanesulfonic acid) buffer and 2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when necessary. Cultures were grown with aeration to mid-log phase and then stored at 4°C overnight. Cultures were then diluted 1:50 in fresh Luria-Bertani medium containing the appropriate antibiotics, MOPS buffer, and IPTG when necessary and grown with aeration to mid-log phase. Cells were harvested and resuspended in 5 ml of 1× E salts (23). Samples were taken and treated with H2O2 at the indicated concentrations for 5 min at 37°C. After treatment, cells were diluted in 1× E salts, plated on LB plates containing the appropriate antibiotics, and incubated at 37°C overnight. Colonies were counted to determine the percentage of surviving cells. Data shown are averages of three independent experiments; error bars indicate the standard errors of the means.
dam-mediated peroxide sensitivity requires MMR.
If dam-mediated peroxide sensitivity is due to conversion of oxidative lesions to strand breaks by MMR, then inactivation of MMR should prevent strand break formation and restore peroxide resistance in a dam mutant. Figure 2 demonstrates that dam mutH and dam mutS strains are much more resistant to hydrogen peroxide than the dam single mutant and are even slightly more resistant to hydrogen peroxide than is a wild-type strain. These results indicate that the peroxide sensitivity of the dam mutant is due to the action of MMR on DNA containing oxidized bases, and that dam-mediated peroxide sensitivity requires the action of mutH and mutS. Based on these results, we conclude that mutS recognizes base pairs containing oxidative lesions and triggers a mutH-dependent cleavage at GATC sites. The small but reproducible increase in resistance seen when the mutH dam and mutS dam strains are compared with the wild type suggests that conversion of nonlethal oxidative lesions to lethal strand breaks may also occur in the wild type, minimizing mutagenesis but increasing lethality.
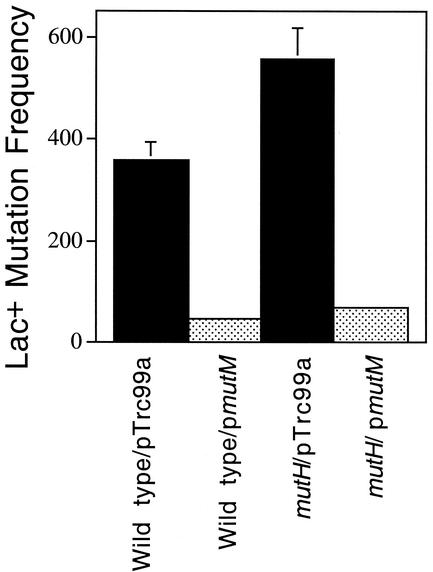
Reduction of GC → TA transversions by overexpression of mutM in a mutH mutant strain.
Since 8-oxoG lesions in DNA result in GC → TA transversions, we determined whether MMR acts specifically on 8-oxoG-containing base pairs by using the bacterial strain CC104. This strain carries a point mutation at the glutamic acid codon 461 located in the active site of the β-galactosidase enzyme that reverts only by GC → TA transversion (4). Therefore, if MMR prevents 8-oxoG-mediated mutagenesis, then CC104 strains deficient in MMR should show an elevated frequency of GC → TA transversions. Moreover, if MutM levels in wild-type strains are limiting, then it should be possible to reduce GC → TA transversions by increasing MutM protein levels. Figure 3 shows that overexpression of MutM from a strong promoter on a high-copy plasmid decreases GC → TA transversions in a wild-type strain. This indicates that most spontaneous GC → TA transversions result from oxidative damage that has escaped repair by the MutM system and that increasing levels of MutM can repair most of the residual damage, preventing mutagenesis. When GC → TA transversions are measured in a mutH strain, there is a substantial increase relative to those seen in the wild type. mutM overexpression in this strain reduces the transversion frequency to a level similar to that of the wild-type strain. This indicates that the mutH-dependent increase in GC → TA transversions is due to lesions that can be repaired by MutM.
FIG. 3.
Reduction of GC → TA transversions by overexpression of mutM in a mutH mutant strain. Lac+ revertants were determined by using the strain CC104 developed by Cupples and Miller (4). Strains were grown overnight at 37°C with aeration in Luria-Bertani medium containing 200 μg of ampicillin/ml and then spread on Lac mutagenesis assay plates by using the plating medium of Cupples and Miller (4) with the addition of 50 μg of carbenicillin/ml to maintain plasmids. IPTG was used when necessary to induce expression of mutM on the plasmid pTrc99a. The plates were incubated at 37°C for 4 days, and then the Lac+ revertants were counted. Mutation frequencies are expressed as lacZ revertants per 109 viable cells. Data shown are averages of 10 independent experiments; error bars indicate the standard errors of the means.
A role for MMR in the repair of oxidative DNA damage was first identified in yeast (6), and it was suggested that the ability of this repair system to perform this function may have evolved because yeast lack MutY and MutT homologs. Organisms such as E. coli and mammals, which have both MutY and MutT, may not need to repair oxidized bases by the MMR system (15). Since we found that the action of MMR on oxidative DNA damage is influenced by the dam mutation, and the hemimethylated state required for MMR exists only transiently after replication in E. coli (21), then it is possible that the role of MMR is to immediately repair the products of misreplication past oxidative lesions. Furthermore, if it is found that MMR can also repair 8-oxoG itself, then MMR may also function to remove 8-oxoG incorporated by the replication machinery. Other repair systems acting on oxidized bases may function to repair the bulk of the oxidative lesions not closely associated with replication forks, repairing oxidized bases in fully methylated DNA.
Acknowledgments
This work was supported by grant no. GM56420 from the National Institutes of Health.
We thank Jen-Yeu Wang for construction of the mutM-expressing plasmid and Martin Marinus, Department of Biochemistry and Molecular Pharmacology, University of Massachusetts, for critical reading of the manuscript.
REFERENCES
- 1.Ames, B. N., M. K. Shigenaga, and T. M. Hagen. 1993. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA 90:7915-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann, B. J. 1987. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 1190-1219. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C.
- 3.Cabrera, M., Y. Nghiem, and J. H. Miller. 1988. mutM, a second mutator locus in Escherichia coli that generates GC → TA transversions. J. Bacteriol. 170:5405-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeWeese, T. L., J. M. Shipman, N. A. Larrier, N. M. Bickley, L. R. Kidd, J. D. Groopman, R. G. Cutler, H. te Riele, and W. G. Nelson. 1998. Mouse embryonic stem cells carrying one or two defective Msh2 alleles respond abnormally to oxidative stress inflicted by low-level radiation. Proc. Natl. Acad. Sci. USA 95:11915-11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earley, M. C., and G. F. Crouse. 1998. The role of mismatch repair in the prevention of base mutations in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:15487-15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 8.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 9.Henderson, P. T., J. C. Delaney, F. Gu, S. R. Tannenbaum, and J. M. Essigmann. 2002. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry 41:914-921. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh, P. 2001. Molecular mechanisms of DNA mismatch repair. Mutat. Res. 486:71-87. [DOI] [PubMed] [Google Scholar]
- 11.Imlay, J. A., S. M. Chin, and S. Linn. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640-642. [DOI] [PubMed] [Google Scholar]
- 12.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 13.Marinus, M. G., and N. R. Morris. 1974. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J. Mol. Biol. 85:309-322. [DOI] [PubMed] [Google Scholar]
- 14.Marnett, L. J. 2000. Oxyradicals and DNA damage. Carcinogenesis 21:361-370. [DOI] [PubMed] [Google Scholar]
- 15.Mazurek, A., M. Berardini, and R. Fishel. 2002. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J. Biol. Chem. 277:8260-8266. [DOI] [PubMed] [Google Scholar]
- 16.Michaels, M. L., C. Cruz, A. P. Grollman, and J. H. Miller. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 89:7022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaels, M. L., L. Pham, C. Cruz, and J. H. Miller. 1991. MutM, a protein that prevents GC → TA transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 19:3629-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michaels, M. L., J. Tchou, A. P. Grollman, and J. H. Miller. 1992. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 31:10964-10968. [DOI] [PubMed] [Google Scholar]
- 19.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65:101-133. [DOI] [PubMed] [Google Scholar]
- 20.Ni, T. T., G. T. Marsischky, and R. D. Kolodner. 1999. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell 4:439-444. [DOI] [PubMed] [Google Scholar]
- 21.Palmer, B. R., and M. G. Marinus. 1994. The dam and dcm strains of Escherichia coli—a review. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 22.Tchou, J., H. Kasai, S. Shibutani, M. Chung, J. Laval, A. P. Grollman, and S. Nishimura. 1991. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. USA 88:4690-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification of some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 24.Volkert, M. R. 1989. Altered induction of the adaptive response to alkylation damage in Escherichia coli recF mutants. J. Bacteriol. 171:99-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkert, M. R., N. A. Elliott, and D. E. Housman. 2000. Functional genomics reveals a family of eukaryotic oxidation protection genes. Proc. Natl. Acad. Sci. USA 97:14530-14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volkert, M. R., and D. C. Nguyen. 1984. Induction of specific Escherichia coli genes by sublethal treatments with alkylating agents. Proc. Natl. Acad. Sci. USA 81:4110-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiseman, H., and B. Halliwell. 1996. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem. J. 313:17-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao, J., and M. E. Winkler. 2000. Reduction of GC → TA transversion mutation by overexpression of MutS in Escherichia coli K-12. J. Bacteriol. 182:5025-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]