Abstract
Whole-genome DNA microarrays were used to evaluate the effect of the redox state of the photosynthetic electron transport chain on gene expression in Synechocystis sp. strain PCC 6803. Two specific inhibitors of electron transport, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), were added to the cultures, and changes in accumulation of transcripts were examined. About 140 genes were highlighted as reproducibly affected by the change in the redox state of the photosynthetic electron transport chain. It was shown that some stress-responsive genes but not photosynthetic genes were under the control of the redox state of the plastoquinone pool in Synechocystis sp. strain PCC 6803.
Photosynthetic organisms must cope with changes in their light environment by various acclimation responses. In order to acclimate to light regimens, organisms need sensors to monitor changing light regimen, signal transduction systems, and output mechanisms for rearrangement of their photosynthetic machinery. The sensing mechanism for changing light conditions and subsequent signal transduction are poorly understood (14). However, it has recently become clear that light-induced electron transport plays a very important role in both transcriptional and posttranscriptional regulations in various photosynthetic organisms. It has been reported that in green algae and higher plants, transcription (9, 17, 25), mRNA stability (2), splicing (8), translation (7, 18), and protein phosphorylation (29) are regulated by the redox state of the photosynthetic electron transport chain. In cyanobacteria, the main targets for redox regulation are transcription and stability of mRNA. mRNA levels of several photosynthesis-related genes, such as psbA, which encodes the reaction center D1 polypeptide of photosystem II (PSII), psaE, which codes for a subunit of photosystem I (PSI), cpcBA, which encodes β and α subunits of phycocyanin, and rbcLS, which encodes subunits of ribulose 1,5-bisphosphate carboxylase, were shown to be affected by the redox state of the photosynthetic electron transport chain (3, 4, 6, 21). Also, the thioredoxin gene (23), genes for fatty acid desaturases (19), some nitrogen-regulated genes (5, 10, 28), and heat shock genes (12) were reported to be under the control of photosynthetic electron transport. This raised the question of how many genes in the whole genome are regulated by photosynthetic electron transport. To answer this question, we investigated the entire profile of accumulated transcripts upon the change in redox state of intersystem electron transfer components in the cyanobacterium Synechocystis sp. strain PCC 6803 with a whole-genome DNA microarray. The microarray was recently used to examine the temporal program of gene expression during acclimation from low to high light intensity (15).
Effects of DCMU and DBMIB on the redox state of the photosynthetic electron transport chain.
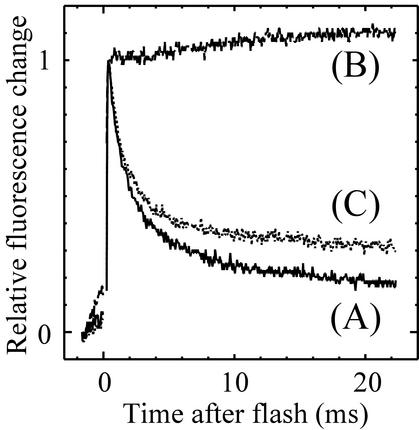
In the present study, we employed two electron transport inhibitors, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), to modulate the redox state of the photosynthetic electron transport chain. DCMU blocks transfer of electrons from PSII to the plastoquinone (PQ) pool, and DBMIB prevents the oxidation of PQ by the cytochrome b6/f complex (31). The two inhibitors have opposite effects on the net redox state of the PQ pool: more PQ is oxidized in the presence of DCMU, and more is reduced in the presence of DBMIB. By comparing the effects of two inhibitors, we aimed at the categorization of open reading frames (ORFs) of Synechocystis sp. strain PCC 6803 according to the patterns of redox regulation. First, the specificity of DBMIB was checked because DBMIB is also known to inhibit electron transfer from the primary electron acceptor in PSII (QA) to the second electron acceptor (QB) at higher concentrations. In the presence of inhibitors, we determined the fluorescence decay kinetics in the millisecond range, which represents electron transfer from QA to QB. The cyanobacterial cells were dark-adapted for 5 min, and then a single-turnover flash (XST; Heinz Waltz, Effeltrich, Germany) was applied. The fluorescence level was monitored with a PAM fluorimeter (PAM 101/103; Heinz Waltz) using the Auto 100-kHz mode with a high-sensitivity measurement unit (ED-101US; Heinz Waltz). Triggering of the single-turnover flash and sampling of the fluorescence signal were done with an A/D converter (AnalogPro II; Canopus, Kobe, Japan) and laboratory-made software with a sampling frequency of 25 kHz and 14-bit resolution. As shown in Fig. 1, addition of 10 μM DCMU totally inhibited the decay of fluorescence, while 10 μM DBMIB only slightly affected the decay kinetics. When the concentration of DBMIB was raised to 100 μM, the rapid phase of the fluorescence decay kinetics in the millisecond range was largely eliminated (data not shown). Thus, 10 μM DBMIB showed only a slight effect, if any, on the electron transfer from QA to QB, under our experimental conditions. Since 10 μM DBMIB totally blocked electron transfer from PQ to a cytochrome b6/f complex, judging from the complete inhibition of overall electron transfer (data not shown), the PQ pool should be highly reduced in the presence of DBMIB at this concentration. Although DBMIB was reported to be relatively unstable (4), we found that 10 μM DBMIB totally blocked oxygen evolution of cultures even after 2 h of incubation under our experimental conditions. At that time, cells were still alive, since oxygen evolution was partially restored after the removal of DBMIB.
FIG. 1.
Decay kinetics of fluorescence in the millisecond range after excitation with a single-turnover flash. The data are normalized with variable fluorescence. Measurement were performed without inhibitors (A), in the presence of 10 μM DCMU (B), and in the presence of 10 μM DBMIB (C).
Growth conditions, RNA isolation, and DNA microarray analyses.
A glucose-tolerant wild-type isolate of Synechocystis sp. strain PCC 6803 was grown at 20 μmol of photons m−2 s−1 to a cell density equivalent to an A730 of 0.5 and then incubated with or without inhibitors. Isolation of total RNA from cultures and successive DNA microarray analysis were performed as previously described (15). For image acquisition and analysis, two scanning and quantification systems were used in this study. Image acquisition with a 418 array scanner (Affymetrix, Santa Clara, Calif.) and data analysis by Imagene version 3.0 software (BioDiscovery, Los Angeles, Calif.) were performed as previously described (15). Image acquisition with a ScanArray 4000 (GSI Lumonics, Watertown, Mass.) was performed with the autobalance-autorange feature. With this feature, the sensitivity of the instrument can be automatically adjusted by changing the laser power and photomultiplier gain settings so that the signal is within 90% of maximum to prevent saturation. The raw data obtained with the ScanArray 4000 were analyzed with QuantArray version 2.0 software (GSI Lumonics). The fluorescence intensity of each spot in both Cy3 and Cy5 images was quantified, and local background fluorescence levels were subtracted. Cy3 and Cy5 images were normalized by adjusting the total signal intensities of the two images (“global normalization”). After normalization, the ratio of the transcript level of each gene with electron transport inhibitors to that without inhibitors was calculated to determine the induction ratio by inhibitors. All induction ratios presented are the averages of three independent experiments yielding duplicate results (n = 6). The raw data are available on the Internet (http://www.genome.ad.jp/kegg/).
The effect of the addition of inhibitors on the overall profile of accumulation of transcripts.
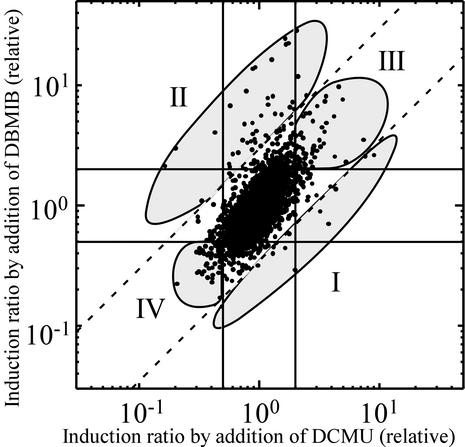
At first, accumulation profiles of transcripts were compared between 15 min and 1 h after the addition of inhibitors. Many genes were affected similarly at both time points. However, the extent of induction or repression was much higher at 1 h than at 15 min (data not shown). Modulation of gene expression in response to the new redox state seemed to be still in progress at 15 min, whereas prominent induction or repression was achieved in many genes at 1 h. Therefore, we chose 1 h as the incubation time for this study to obtain clear and reproducible results with DNA microarrays. Figure 2 shows the effect of the addition of inhibitors on the transcript levels of the whole set of genes in Synechocystis after 1 h of incubation. The majority of the genes were aligned along the diagonal line from the lower left to the upper right, which means that they are similarly affected by both inhibitors. However, most of them did not seem to be redox responsive, since they were located inside two lines representing twofold induction and 50% repression. Only a few genes were located on the diagonal line from the lower right to the upper left, indicating that it is rare for a gene to be oppositely affected by two inhibitors. Genes whose expression was significantly affected by the addition of inhibitors were classified into four groups, as shown by the shaded area in Fig. 2: in group I, induction by DCMU was more than three times larger than that by DBMIB; in group II, induction by DBMIB was more than three times larger than that by DCMU; group III genes were induced by both inhibitors more than twofold; and group IV genes were repressed by both inhibitors to less than half. Representative genes belonging to each group are listed in Table 1.
FIG. 2.
Induction or repression ratio of each gene for the inhibitors DCMU and DBMIB. Twofold induction and 50% repression are shown by solid lines. A >3-fold difference in the effects of the two inhibitors is shown by dashed lines. The four groups categorized by inhibitor effect are indicated.
TABLE 1.
Genes whose expression was significantly affected by the addition of inhibitors of photosynthetic electron transport
| Groupa and ORF (no. and description) | Change (fold) in expression levelb by addition of:
|
Effect of stress conditionc
|
||||
|---|---|---|---|---|---|---|
| DCMU | DBMIB | HL | LT | HS | HO | |
| Group I | ||||||
| sll1647 | 3.23 ± 2.36 | 0.89 ± 0.35 | ||||
| sll1677, spore maturation protein B | 0.98 ± 0.27 | 0.31 ± 0.05 | ||||
| sll1864, chloride channel protein | 3.80 ± 0.46 | 0.70 ± 0.10 | I | |||
| slr0358 | 1.30 ± 0.20 | 0.40 ± 0.14 | ||||
| slr0373 | 9.13 ± 4.74 | 2.61 ± 0.57 | R | |||
| slr0376 | 4.36 ± 1.25 | 1.35 ± 0.25 | R | |||
| slr0772, chlB, protochlorophyllide reductase subunit | 0.49 ± 0.04 | 0.14 ± 0.07 | R | |||
| slr0888 | 1.79 ± 0.30 | 0.58 ± 0.06 | R | |||
| slr1634 | 1.99 ± 0.28 | 0.29 ± 0.05 | R | R | ||
| slr1908, probable porin | 1.66 ± 0.22 | 0.44 ± 0.17 | ||||
| ssl1911, gifA, glutamine synthetase inactivating factor IF7 | 7.70 ± 1.70 | 2.55 ± 0.71 | ||||
| Group II | ||||||
| sll0170, dnaK2 | 0.89 ± 0.19 | 4.83 ± 0.99 | I | I | I | |
| sll0306, sigB, rpoD | 1.90 ± 0.36 | 7.20 ± 3.67 | I | I | ||
| sll0416, groEL-2 | 0.33 ± 0.15 | 1.09 ± 0.52 | I | I | I | |
| sll0430, htpG | 0.43 ± 0.23 | 4.02 ± 0.39 | I | I | ||
| sll0528 | 1.76 ± 0.30 | 22.21 ± 7.85 | I | I | I | |
| sll0549 | 1.81 ± 0.32 | 6.54 ± 2.49 | ||||
| sll0786 | 0.58 ± 0.44 | 1.78 ± 1.85 | ||||
| sll0788 | 2.06 ± 0.35 | 28.42 ± 9.51 | I | I | ||
| sll0789, copR, rre34 | 1.66 ± 0.34 | 24.34 ± 13.32 | I | I | ||
| sll0790, hik31 | 0.93 ± 0.22 | 13.95 ± 5.25 | ||||
| sll0846 | 1.05 ± 0.38 | 5.55 ± 2.37 | I | I | I | |
| sll1159 | 0.73 ± 0.17 | 3.46 ± 1.60 | ||||
| sll1167, pbp, penicillin-binding protein 4 | 0.89 ± 0.37 | 3.92 ± 2.19 | I | |||
| sll1450, nrtA, nitrate transport protein NrtA | 0.67 ± 0.24 | 2.74 ± 0.95 | ||||
| sll1451, nrtB, nitrate transport protein NrtB | 0.93 ± 0.35 | 3.33 ± 0.87 | ||||
| sll1452, nrtC, nitrate transport protein NrtC | 1.20 ± 0.42 | 3.61 ± 1.26 | ||||
| sll1453, nrtD, nitrate transport protein NrtD | 0.92 ± 0.42 | 3.55 ± 0.63 | R | |||
| sll1514, hspA | 1.28 ± 0.31 | 16.14 ± 4.74 | I | I | I | |
| sll1620 | 0.80 ± 0.16 | 3.30 ± 0.45 | ||||
| sll1621 membrane protein | 0.78 ± 0.21 | 8.84 ± 1.95 | R | I | ||
| slr0074, ycf24, ABC transporter subunit | 0.88 ± 0.18 | 2.90 ± 0.42 | ||||
| slr0075, ycf16, ABC transporter subunit | 0.56 ± 0.09 | 2.21 ± 0.13 | ||||
| slr0076 | 0.58 ± 0.04 | 2.09 ± 0.23 | ||||
| slr0093, dnaJ | 1.36 ± 0.33 | 5.76 ± 2.83 | I | I | ||
| slr0095, O-methyltransferase | 1.14 ± 0.19 | 3.80 ± 1.97 | I | |||
| slr0272 | 1.55 ± 0.68 | 5.66 ± 2.86 | R | |||
| slr0898, nirA, ferredoxin-nitrite reductase | 0.57 ± 0.25 | 1.83 ± 0.28 | ||||
| slr1285, hik34 | 0.58 ± 0.09 | 6.72 ± 4.02 | ||||
| slr1291, ndhD2 | 1.68 ± 0.32 | 21.70 ± 11.16 | I | I | ||
| slr1413 | 0.76 ± 0.23 | 4.74 ± 2.33 | ||||
| slr1544 | 2.48 ± 0.80 | 8.16 ± 4.76 | I | I | I | I |
| slr1603 | 2.12 ± 0.44 | 11.72 ± 6.94 | I | I | ||
| slr1641, clpB1 | 1.18 ± 0.27 | 7.75 ± 2.31 | I | I | I | |
| slr1674 | 1.30 ± 0.13 | 8.05 ± 1.77 | I | I | I | |
| slr1675, hypA1 | 0.89 ± 0.39 | 13.59 ± 6.55 | I | I | I | |
| slr1738 | 0.95 ± 0.12 | 4.15 ± 1.28 | I | |||
| slr1963, water-soluble carotenoid protein | 0.61 ± 0.11 | 2.66 ± 0.38 | I | I | I | |
| slr2075, groES | 0.20 ± 0.06 | 2.99 ± 0.83 | I | |||
| slr2076, groEL | 0.17 ± 0.06 | 2.11 ± 0.51 | I | |||
| ssl2542, hliA, scpC | 1.19 ± 0.40 | 6.84 ± 6.70 | I | I | ||
| ssl2971 | 0.86 ± 0.07 | 3.41 ± 1.61 | I | I | ||
| ssr2595, hliB, scpD | 1.98 ± 0.73 | 6.28 ± 6.28 | I | I | ||
| Group III | ||||||
| sll0020, clpC ATP-dependent Clp protease regulatory subunit | 2.01 ± 0.30 | 2.50 ± 0.24 | ||||
| sll0141, putative HlyD family secretion protein | 2.89 ± 0.48 | 3.10 ± 0.25 | ||||
| sll0297 | 3.04 ± 0.50 | 2.84 ± 1.31 | ||||
| sll0749 | 2.02 ± 0.27 | 3.80 ± 1.39 | ||||
| sll0843 | 2.47 ± 0.48 | 5.79 ± 1.16 | ||||
| sll0891, citH, ldh, malate dehydrogenase | 3.20 ± 0.52 | 3.48 ± 0.49 | ||||
| sll0939 | 4.63 ± 1.01 | 9.64 ± 4.87 | I | I | ||
| sll0992, esterase | 2.02 ± 0.70 | 3.77 ± 1.47 | /PICK> | |||
| sll1086 | 3.62 ± 1.14 | 8.01 ± 2.73 | I | |||
| sll1201 | 2.38 ± 0.15 | 2.23 ± 0.75 | ||||
| sll1268 | 2.31 ± 0.42 | 2.93 ± 0.56 | ||||
| sll1432, hypB | 2.92 ± 0.79 | 2.49 ± 0.44 | ||||
| sll1483, transforming growth factor-induced protein | 4.99 ± 2.14 | 7.55 ± 3.58 | I | I | I | |
| sll1515, gifB, glutamine synthetase-inactivating factor IF17 | 7.44 ± 1.67 | 3.50 ± 0.68 | I | |||
| sll1774 | 2.80 ± 0.17 | 2.08 ± 0.85 | ||||
| sll1867, psbA3, PSII D1 protein | 2.17 ± 0.52 | 2.42 ± 0.27 | I | |||
| sll2012, sigD, rpoD | 2.82 ± 0.56 | 5.00 ± 2.40 | I | I | ||
| slr0164, clpP4, clpR, Clp protease proteolytic subunit | 2.07 ± 0.14 | 3.12 ± 1.04 | ||||
| slr0211 | 2.84 ± 0.31 | 2.94 ± 0.15 | ||||
| slr0374, cell division cycle protein | 5.33 ± 1.64 | 2.29 ± 0.24 | R | |||
| slr0397 | 2.12 ± 0.37 | 2.37 ± 0.27 | ||||
| slr0451, ski2, antiviral protein | 2.02 ± 0.59 | 2.90 ± 0.36 | ||||
| slr0551 | 2.83 ± 0.45 | 2.97 ± 1.08 | I | |||
| slr0581 | 2.73 ± 0.30 | 2.27 ± 0.66 | I | I | ||
| slr0599, eukaryotic protein kinase | 2.40 ± 0.18 | 2.42 ± 0.89 | ||||
| slr0600, putative thioredoxin reductase | 2.21 ± 0.19 | 3.94 ± 0.68 | ||||
| slr0839, hemH, scpA, ferrochelatase | 2.18 ± 0.33 | 3.04 ± 0.40 | ||||
| slr0942, aldehyde reductase | 2.20 ± 0.28 | 2.30 ± 0.28 | ||||
| slr0967 | 3.55 ± 0.92 | 7.30 ± 0.85 | I | |||
| slr1113, ABC transporter | 2.77 ± 0.30 | 2.70 ± 0.79 | ||||
| slr1114 | 3.26 ± 0.77 | 3.05 ± 0.63 | ||||
| slr1127 | 3.35 ± 0.31 | 4.43 ± 0.56 | ||||
| slr1128 | 4.21 ± 0.82 | 5.49 ± 1.41 | R | |||
| slr1129, rne, ribonuclease E | 2.33 ± 0.21 | 3.61 ± 0.79 | ||||
| slr1253 | 2.83 ± 0.51 | 2.86 ± 0.54 | ||||
| slr1259 | 4.29 ± 0.80 | 2.53 ± 0.68 | I | |||
| slr1260 | 3.47 ± 0.24 | 3.27 ± 0.63 | ||||
| slr1262 | 3.64 ± 0.20 | 4.82 ± 0.45 | ||||
| slr1604, ftsH, cell division protein FtsH | 2.07 ± 0.17 | 3.01 ± 0.33 | I | I | ||
| slr1687 | 3.07 ± 1.45 | 6.67 ± 3.21 | I | I | ||
| slr1712 | 2.47 ± 0.10 | 2.67 ± 0.51 | ||||
| slr1739, psb28, PSII 13-kDa protein homolog | 2.64 ± 0.93 | 5.82 ± 1.68 | ||||
| slr1751, prc or tsp, carboxyl-terminal protease | 2.45 ± 0.15 | 2.50 ± 0.58 | I | |||
| slr1830, phbC, poly(3-hydroxyalkanoate) synthase | 2.36 ± 0.27 | 2.69 ± 0.97 | ||||
| ssl2501 | 3.53 ± 0.73 | 4.08 ± 1.57 | ||||
| ssl3769 | 2.33 ± 0.50 | 5.47 ± 1.15 | ||||
| Group IV | ||||||
| sll0017, hemL, gsa, glutamate-1-semialdehyde 2,1-aminomutase | 0.49 ± 0.16 | 0.37 ± 0.12 | ||||
| sll0026, ndhF, NADH dehydrogenase subunit 5 | 0.48 ± 0.03 | 0.39 ± 0.05 | ||||
| sll0421, purB, adenylosuccinate lyase | 0.50 ± 0.05 | 0.45 ± 0.02 | ||||
| sll1077, speB, agmatine ureohydrolase | 0.41 ± 0.09 | 0.30 ± 0.06 | ||||
| sll1091, chlP, geranylgeranyl hydrogenase | 0.47 ± 0.02 | 0.34 ± 0.23 | R | R | R | R |
| sll1185, hemF, coproporphyrinogen III oxidase | 0.32 ± 0.03 | 0.28 ± 0.16 | R | R | ||
| sll1212, rfbD, GDP-d-mannose dehydratase | 0.40 ± 0.05 | 0.44 ± 0.22 | ||||
| sll1305 | 0.44 ± 0.03 | 0.46 ± 0.26 | R | R | R | R |
| sll1323, atpG, ATP synthase subunit b′ | 0.35 ± 0.06 | 0.28 ± 0.03 | ||||
| sll1324, atpF, ATP synthase subunit b | 0.30 ± 0.04 | 0.41 ± 0.18 | ||||
| sll1325, atpD, ATP synthase d subunit | 0.32 ± 0.05 | 0.40 ± 0.15 | ||||
| sll1326, atpA, ATP synthase a subunit | 0.42 ± 0.06 | 0.50 ± 0.17 | ||||
| sll1471, cpcG2, phycobilisome rod-core linker polypeptide | 0.21 ± 0.23 | 0.22 ± 0.25 | ||||
| sll1526 | 0.34 ± 0.07 | 0.27 ± 0.08 | ||||
| sll1530 | 0.36 ± 0.08 | 0.46 ± 0.31 | ||||
| sll1531 | 0.37 ± 0.12 | 0.32 ± 0.08 | ||||
| sll1550, probable porin | 0.46 ± 0.12 | 0.27 ± 0.12 | R | |||
| sll1577, cpcB, phycocyanin b subunit | 0.32 ± 0.09 | 0.44 ± 0.40 | R | R | R | |
| sll1579, cpcC, phycocyanin-associated linker protein | 0.41 ± 0.12 | 0.26 ± 0.10 | R | R | ||
| sll1580, cpcC, phycocyanin-associated linker protein | 0.37 ± 0.05 | 0.39 ± 0.28 | R | R | R | R |
| sll1799, rpl3, 50S ribosomal protein L3 | 0.35 ± 0.02 | 0.40 ± 0.10 | I | I | I | |
| sll1800, rpl4, 50S ribosomal protein L4 | 0.38 ± 0.07 | 0.30 ± 0.05 | I | I | ||
| sll1801, rpl23, 50S ribosomal protein L23 | 0.38 ± 0.04 | 0.40 ± 0.04 | I | I | ||
| sll1802, rpl2, 50S ribosomal protein L2 | 0.34 ± 0.05 | 0.42 ± 0.10 | I | |||
| sll1804, rps3, 30S ribosomal protein S3 | 0.40 ± 0.06 | 0.36 ± 0.07 | I | |||
| slr0468 | 0.44 ± 0.15 | 0.38 ± 0.11 | ||||
| slr0676, cysC, adenylylsulfate 3′-phosphotransferase | 0.40 ± 0.05 | 0.40 ± 0.13 | ||||
| slr0750, chlN, protochlorophyllide reductase subunit | 0.47 ± 0.22 | 0.17 ± 0.07 | R | |||
| slr0909 | 0.37 ± 0.07 | 0.38 ± 0.16 | ||||
| slr1056 | 0.49 ± 0.28 | 0.38 ± 0.07 | ||||
| slr1064, rfbU or mtfA, mannosyltransferase B | 0.45 ± 0.10 | 0.38 ± 0.07 | ||||
| slr1161 | 0.41 ± 0.05 | 0.23 ± 0.16 | R | |||
| slr1162 | 0.31 ± 0.09 | 0.27 ± 0.24 | ||||
| slr1329, atpB, ATP synthase b subunit | 0.48 ± 0.08 | 0.36 ± 0.03 | ||||
| slr1396 | 0.38 ± 0.06 | 0.25 ± 0.06 | ||||
| slr1618 | 0.44 ± 0.05 | 0.49 ± 0.09 | ||||
| slr1619 | 0.46 ± 0.04 | 0.41 ± 0.05 | ||||
| slr1854 | 0.37 ± 0.05 | 0.32 ± 0.14 | R | R | R | R |
| slr1855 | 0.41 ± 0.05 | 0.25 ± 0.02 | R | R | R | R |
| slr1859, anti-sigma-F factor antagonist | 0.45 ± 0.08 | 0.38 ± 0.08 | R | R | ||
| slr1860, icfG | 0.46 ± 0.13 | 0.43 ± 0.06 | ||||
In group I, induction by DCMU is more than three times greater than that by DBMIB; in group II, induction by DBMIB is more than three times greater than that by DCMU; group III genes are induced by both inhibitors more than twofold; group IV genes are repressed less than 50% by both inhibitors.
Values are averages±standard deviations for three independent experiments, each in duplicate (n = 6).
Genes differently affected by the two inhibitors.
Genes categorized in groups I and II could be regulated by the redox state of the PQ pool, since DCMU and DBMIB showed quite different effects on the expression of these genes. It is of note that many genes were strongly induced by DBMIB (group II) but not by DCMU (group I). Heat shock genes, such as groESL, groEL-2, dnaK, dnaJ, htpG, clpB, and hspA, genes for high-light-inducible proteins, such as hliA and hliB, and nitrate assimilation-related genes, such as nrtABCD and nirA, were induced by the addition of DBMIB. The reduced state of the PQ pool might be important as the signal for accumulation of transcripts. At any rate, no genes related to photosynthesis were shown to be regulated by the redox state of the PQ pool in this study. Even when inhibitors were added to the high-light-grown cultures, photosynthesis-related genes were not categorized in group I or II (data not shown). This observation is quite unexpected, since the transcriptional regulation of photosynthesis-related genes by the redox state of the PQ pool in green algae and higher plants is well established. For example, transcription of cab genes in Dunaliella tertiolecta (9), the psaAB operon in mustard (25), psaB in pea (32), and petE in tobacco (26) were shown to be oppositely affected by addition of DCMU or DBMIB, indicating the coupling between the redox state of the PQ pool and the rate of transcription. The regulation system for the expression of photosynthetic genes in these organisms seems to be totally different from that in cyanobacteria.
Genes similarly affected by the two inhibitors.
It is assumed that the redox state of the components upstream of QA or downstream of the PQ pool responds similarly to DCMU and DBMIB. Thus, the candidates for the redox sensor responsible for group III and IV genes could be electron transfer components such as PSII, the cytochrome b6/f complex, and the ferredoxin-thioredoxin system. In group III genes that are induced by both inhibitors, we observed genes encoding proteases, housekeeping enzymes, and some PSII-related components, such as psbA and psb28. On the other hand, genes encoding ribosomal proteins, phycocyanin, ATP synthase, and enzymes for biosynthesis of photosynthetic pigments were repressed by both inhibitors and categorized to group IV. Although the extent of repression by both inhibitors was not high enough to be listed in Table 1, some genes reported as redox responsive so far, such as trxA, which encodes thioredoxin (23), desA, -B, and -D, which encode fatty acid desaturases (19), and nitrogen-regulated genes (5, 10, 28), could be also categorized in group IV. It is of interest that genes activated by NtcA, such as glnA, glnB, ntcA, sigE, and amt1 (13), were all down-regulated (data not shown), and genes repressed by NtcA, such as gifA and gifB (11), were up-regulated (Table 1) by both inhibitors. Although nitrate assimilation-related genes, such as nrtABCD and nirA, also have an NtcA-binding motif, their response was quite different from that of other NtcA-regulated genes. In addition to NtcA, NtcB contributes to the regulation of the nitrate assimilation-related genes (1), which may confer special characteristics on the expression profiles of these genes. As for genes encoding subunits of PSI, we observed only slight down-regulation by the addition of inhibitors (data not shown), although several groups reported that PSI genes were largely repressed by inhibitors (3, 20). Some differences in experimental conditions, such as light intensity and/or quality, might be the cause of this discrepancy. We think that PSI genes are redox responsive at least under high-light conditions, since the marked down-regulation of PSI genes under high light (22) was abolished by the addition of inhibitors (data not shown). In summary, genes encoding subunits of phycobilisome, photosystems, ATP synthase, ribosomes, and various enzymes were affected similarly by both inhibitors and therefore supposed to be under the control of the redox state of electron transport components other than the PQ pool. The PQ pool might be used for the sensor of rapid induction of stress-responsive genes, whereas other redox components of the photosynthetic electron transport chain might be important for slower acclimation responses. The Q0 site of the cytochrome b6/f complex is one possible candidate for the redox sensor for the regulation of photosystem genes, as suggested by Bissati and Kirilovsky (6).
Relationship between redox and stress responsiveness.
To date, gene expression in response to high-light (15), low-temperature (30), high-salinity (16), and high-osmolality conditions (16) has been investigated by using DNA microarrays. We noticed that many redox-responsive genes in Table 1 were also listed in these studies. As shown in Table 1, many genes in group II and III were induced and those in group I and IV were repressed under various stress conditions. On the other hand, of the genes listed as stress responsive under high-light, low-temperature, high-salinity, and high-osmolality conditions, 24, 24, 43, and 40% were listed as redox responsive in this study, respectively. From these observations, we concluded that transcript levels of many genes involved in acclimation responses to environmental stresses could be regulated by the redox state of the photosynthetic electron transport chain. The sigma-70 factor genes listed in Table 1, sigB (ORF sll0306) and sigD (ORF sll2012), may work when these genes are induced under stress conditions.
Characterization of multigene families according to redox responsiveness.
There are many genes present in multiple copies in the genome of Synechocystis sp. strain PCC 6803, and the characteristics of each copy were obscure in most cases. However, we found that for some genes, one copy could be clearly distinguished from the others copies by its redox responsiveness. For example, among ndh genes encoding subunits of NAD(P)H dehydrogenase, only one copy, ndhD2, was largely induced by DBMIB and categorized in group II, indicating that this copy has a special role. On the other hand, copies involved in CO2 uptake, such as ndhD3 and ndhF3 (24), were repressed by both inhibitors (data not shown). Most of the other ndh genes did not respond to the addition of inhibitors. Another example is clpP4. There are four copies for the catalytic subunit of Clp protease in Synechocystis sp. strain PCC 6803. Among them, the putative product of only clpP4 lacks the amino acid residues required for the proteolytic activity and is therefore unlikely to function as a true ClpP protease (27). The fact that this gene but not the other clpP genes responded to the addition of the inhibitors suggests the physiological significance of clpP4. It is also noteworthy that psb28 (ORF slr1739) is one of a few photosystem-related genes listed in Table 1. This gene is homologous to psbW (ORF sll1398), which encodes the small subunit of PSII. Although we do not know whether this gene product is also a component of PSII, its redox responsiveness suggests that it has its own physiological role. Information on redox regulation of these genes could be important clues for the elucidation of the physiological function of each gene product.
Conclusion.
In addition to several genes reported to be redox responsive so far (3, 5, 10, 12, 19, 23, 28), more than 100 genes involved in various cellular functions in Synechocystis sp. strain PCC 6803 were shown to be affected by the redox state of the photosynthetic electron transport chain. The photosynthetic electron transport chain seems to play an important role in monitoring environmental changes and in acclimating to them.
Acknowledgments
This work was supported by a Grant-in-Aid for Young Scientists (to Y.H.), by the Genome Frontier Project (to M.K.), and by Grants-in-Aid for Scientific Research (to K.S. and M.I.) from the Ministry of Education, Culture, Sports, Science and Technology.
REFERENCES
- 1.Aichi, M., N. Takatani, and T. Omata. 2001. Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183:5840-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexciev, K., and A. Tullberg. 1997. Regulation of petB mRNA stability in pea chloroplasts by redox poise. Physiol. Plant. 99:477-485. [Google Scholar]
- 3.Alfonso, M., I. Perewoska, S. Constant, and D. Kirilovsky. 1999. Redox control of psbA expression in cyanobacteria Synechocystis strains. J. Photochem. Photobiol. B 48:104-113. [Google Scholar]
- 4.Alfonso, M., I. Perewoska, and D. Kirilovsky. 2000. Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803. Involvement of the cytochrome b6/f complex. Plant Physiol. 122:505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfonso, M., I. Perewoska, and D. Kirilovsky. 2001. Redox control of ntcA gene expression in Synechocystis sp. PCC 6803. Nitrogen availability and electron transport regulate the levels of the NtcA protein. Plant Physiol. 125:969-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissati, K. E., and D. Kirilovsky. 2001. Regulation of psbA and psaE expression by light quality in Synechocystis species PCC 6803. A redox control mechanism. Plant Physiol. 125:1988-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danon, A., and S. P. Mayfield. 1994. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science 266:1717-1719. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande, N. N., Y. Bao, and D. L. Herrin. 1997. Evidence for light/redox-regulated splicing of psbA pre-RNAs in Chlamydomonas chloroplasts. RNA 3:37-48. [PMC free article] [PubMed] [Google Scholar]
- 9.Escoubas, J. M., M. Lomas, J. Laroche, and P. G. Falkowski. 1995. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92:10237-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-Domínguez, M., and F. J. Florencio. 1997. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 35:723-734. [DOI] [PubMed] [Google Scholar]
- 11.García-Domínguez, M., J. C. Reyes, and F. J. Florencio. 2000. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type 1 from Synechocystis sp. PCC 6803. Mol. Microbiol. 35:1192-1201. [DOI] [PubMed] [Google Scholar]
- 12.Glatz, A., I. Horváth, V. Varvasovszki, E. Kovács, Z. Török, and L. Vigh. 1997. Chaperonin genes of the Synechocystis PCC 6803 are differently regulated under light-dark transition during heat stress. Biochem. Biophys. Res. Commun. 239:291-297. [DOI] [PubMed] [Google Scholar]
- 13.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hihara, Y. 1999. The molecular mechanism for acclimation to high light in cyanobacteria. Curr. Top. Plant Biol. 1:37-50. [Google Scholar]
- 15.Hihara, Y., A. Kamei, M. Kanehisa, A. Kaplan, and M. Ikeuchi. 2001. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanesaki, Y., I. Suzuki, S. I. Allakhverdiev, K. Mikami, and N. Murata. 2002. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 290:339-348. [DOI] [PubMed] [Google Scholar]
- 17.Karpinski, S., C. Escobar, B. Karpinska, G. Creissen, and P. M. Mullineaux. 1997. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9:627-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J. M., and S. P. Mayfield. 1997. Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 278:1954-1957. [DOI] [PubMed] [Google Scholar]
- 19.Kis, M., O. Zsiros, T. Farkas, H. Wada, F. Nagy, and Z. Gombos. 1998. Light-induced expression of fatty acid desaturase genes. Proc. Natl. Acad. Sci. USA 95:4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, H., and L. A. Sherman. 2000. A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:4268-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamed, A., and C. Jansson. 1991. Photosynthetic electron transport controls degradation but not production of psbA transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 16:891-897. [DOI] [PubMed] [Google Scholar]
- 22.Muramatsu, M., and Y. Hihara. 2002. Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta. [Online.] http://link.springer.de/link/service/journals/00425/contents/tfirst.htm. DOI 10.1007/s00425-002-0859-5. [DOI] [PubMed]
- 23.Navarro, F., E. Martin-Figueroa, and F. J. Florencio. 2000. Electron transport controls transcription of the thioredoxin gene (trxA) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 43:23-32. [DOI] [PubMed] [Google Scholar]
- 24.Ohkawa, H., G. D. Price, M. R. Badger, and T. Ogawa. 2000. Mutation of ndh genes leads to inhibition of CO2 uptake rather than HCO3− uptake in Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:2591-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfannschmidt, T., A. Nilsson, A. Tullberg, G. Link, and J. F. Allen. 1999. Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life 48:271-276. [DOI] [PubMed] [Google Scholar]
- 26.Pfannschmidt, T., K. Schutze, M. Brost, and R. Oelmuller. 2001. A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J. Biol. Chem. 276:36125-36130. [DOI] [PubMed] [Google Scholar]
- 27.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insight into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 28.Reyes, J. C., and F. J. Florencio. 1995. Electron transport controls transcription of the glutamine synthetase gene (glnA) from the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 27:789-799. [DOI] [PubMed] [Google Scholar]
- 29.Rintamäki, E., P. Martinsuo, S. Pursiheimo, and E.-M. Aro. 2000. Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. USA 97:11644-11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki, I., Y. Kanesaki, K. Mikami, M. Kanehisa, and N. Murata. 2001. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 40:235-244. [DOI] [PubMed] [Google Scholar]
- 31.Trebst, A. 1980. Inhibitors in electron flow: tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 69:675-715. [Google Scholar]
- 32.Tullberg, A., K. Alexciev, T. Pfannschmidt, and J. F. Allen. 2000. Photosynthetic electron flow regulates transcription of the psaB gene in pea (Pisum sativum L.) chloroplasts through the redox state of the plastoquinone pool. Plant Cell Physiol. 41:1045-1054. [DOI] [PubMed] [Google Scholar]