Abstract
Plant cell walls are comprised of cellulose and hemicellulose and other polymers that are intertwined, and this complex structure presents a barrier to degradation by pure cellulases or hemicellulases. In this study, we determined the synergistic effects on corn cell wall degradation by the action of cellulosomal xylanase XynA and cellulosomal cellulases from Clostridium cellulovorans. XynA minicellulosomes and cellulase minicellulosomes were found to degrade corn cell walls synergistically but not purified substrates such as xylan and crystalline cellulose. The mixture of XynA and cellulases at a molar ratio of 1:2 showed the highest synergistic effect of 1.6 on corn cell wall degradation. The amounts both of xylooligosaccharides and cellooligosaccharides liberated from corn cell walls were increased by the synergistic action of XynA and cellulases. Although synergistic effects on corn cell wall degradation were found in simultaneous reactions with XynA and cellulases, no synergistic effects were observed in sequential reactions. The possible mechanism of synergism between XynA and cellulases is discussed.
Plant cell walls are the most abundant and renewable source of fermentable sugars on earth (7). The cell wall of plants is comprised of cellulose, hemicellulose, pectin, lignin, and other components. Cellulose and hemicellulose are the major components and are tightly connected and intertwined; this hampers biomass degradation by pure cellulases or pure hemicellulases. The main focus of the present study was to determine whether there is any synergistic effect of cellulases and xylanase when a natural substance such as corn fiber is used as their substrate.
Clostridium cellulovorans produces an enzyme complex (cellulosome) containing a variety of glycosyl hydrolytic subunits attached to the nonenzymatic scaffolding component termed CbpA (3). We have cloned and sequenced cellulosomal xylanase gene xynA (11), mannanase gene manA (21), and pectate lyase gene pelA (22), as well as eight cellulosomal cellulase genes (engB [4], engE [20], engH [23], engK [23], engL [21], engM [23], engY [22], and exgS [13]). Therefore, the existence of hemicellulase genes and pectate lyase gene, as well as cellulase genes, suggests that C. cellulovorans enzymes can degrade plant cell walls effectively. In fact, it has been shown recently that C. cellulovorans cellulosomes liberated reducing sugars from corn cell walls (16) and released protoplasts from cultured tobacco and Arabidopsis cells (24).
One of our goals is the preparation of “designer” cellulosomes that can degrade plant cell walls efficiently to fermentable sugars. Therefore, by biologically converting plant cell walls to fermentable sugars for fuel (e.g., ethanol), we could obtain not only economic but also environmental benefits, such as the reduction of greenhouse gas emission (14). Recently, we assembled recombinant cellulosomes in vitro (15, 17) and optimized the cellulase composition with three cellulosomal cellulases from C. cellulovorans (EngE, EngH, and ExgS) to increase the cellulase activity (17). This development was our first step to prepare designer cellulosomes that can degrade plant cell walls efficiently.
To determine the mechanism of corn cell wall degradation by C. cellulovorans cellulosomes, we have purified cellulosome fractions, which degrade corn cell walls (16). In the purified cellulosome fractions, xylanase XynA and mannanase ManA, as well as cellulases, have been identified as enzymatic subunits. In the case of corn, which is a potential substrate for biomass conversion to obtain fermentable sugars (9), arabinoxylans are known to be major hemicelluloses (2). The arabinoxylan in corn cell walls is thought to cross-link several cellulose microfibrils to generate a network structure in cell walls (2). Therefore, cleavage of xylan cross-linkages is considered to be one of the key reactions to degrade corn cell walls. The subunit composition of cellulosomes and the cell wall structure suggest that xylanases may play an important role in corn cell wall degradation.
To determine the enzyme composition for designer cellulosomes that can degrade corn cell walls effectively, it is important to analyze the role of xylanase. Although synergistic effects between cellulases on cellulose degradation are well known (5, 6, 8, 18, 26, 28), there are few studies concerning synergistic effects between xylanases and cellulosomal cellulases on plant cell wall degradation. In the present study, we used synthetic complexes of xylanase A and miniscaffolding proteins (XynA cellulosomes) and of cellulases and miniscaffolding proteins (cellulase cellulosomes) to form minicellulosomes with specific enzymatic functions. We used these XynA cellulosomes and cellulase cellulosomes to determine whether they had any synergistic effects on corn cell wall degradation. We found that XynA cellulosomes and cellulase cellulosomes did degrade corn cell walls synergistically. The results presented here may help us not only to understand the mechanism of plant cell wall degradation but also to design better enzyme compositions for effective degradation of various plant cell walls.
MATERIALS AND METHODS
Materials.
Avicel (crystalline cellulose) was purchased from FMC Corporation. Xylan from oat spelts was purchased from Sigma. The powder of the water-insoluble fraction of corn stem (corn cell walls) was supplied by Meiji Seika Kaisha, Ltd.
Bacterial strains and media.
Escherichia coli BL21(DE3) (Novagen) was used as an expression host for mini-CbpA, ExgS, EngE, and XynA production with pET-22b-mini-CbpA (15), pET-22b-ExgS (17), pENGE (10), and pEXYNA29 (11). E. coli TOP10 (Invitrogen) was used as an expression host for EngH production with pBAD/Thio-EngH (17). Recombinant strains were cultivated in Luria-Bertani medium supplemented with ampicillin (50 μg/ml) for expression of mini-CbpA, EngE, ExgS, and EngH or kanamycin (20 μg/ml) for expression of XynA.
Expression of recombinant proteins.
For production of recombinant mini-CbpA, ExgS, EngE, and XynA, E. coli BL21(DE3) harboring pET-22b-mini-CbpA, pET-22b-ExgS, pENGE, or pEXYNA29 was grown and recombinant proteins were induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) as an inducer. The E. coli cells were grown in 1 liter of medium at 37°C to an optical density at 600 nm of 0.9. After the culture broth was cooled on ice for 30 min, IPTG was added to a final concentration of 0.4 mM for mini-CbpA and XynA production or to a final concentration of 0.04 mM for ExgS or EngE production. The culture was then grown at 18°C for 16 h. For production of the recombinant EngH, E. coli TOP 10 harboring pBAD/Thio-EngH was grown, and recombinant proteins were induced by adding l-arabinose as an inducer. The E. coli cells were grown in 1 liter of medium at 37°C to an optical density at 600 nm of 0.9. After the culture broth was cooled on ice for 30 min, l-arabinose was added to a final concentration of 0.1%. The culture was then grown at 18°C for 16 h.
Purification of recombinant proteins.
The recombinant mini-CbpA, ExgS, EngE, EngH, and XynA were purified in the same manner as follows. After the E. coli cells, grown as described above, were collected by centrifugation, the cells were resuspended in 30 ml of the lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 1 mg of lysozyme/ml; pH 8.0). The mixture was incubated on ice for 30 min and then the cells were disrupted by sonication to extract soluble proteins. After clarification by centrifugation, the clarified extract was applied to 4 ml of nickel-nitrilotriacetic acid agarose resin (Qiagen), and the proteins bound to the resin were purified and pooled according to the product manual. The pooled solution was desalted and concentrated into 1 ml of 20 mM Tris-HCl buffer (pH 8.0) by use of the Ultrafree 10-kDa membrane (Millipore). The concentrated solution was applied to an anion-exchange column Mono Q HR5/5 (Amersham Pharmacia Biotech AB) preequilibrated with 20 mM Tris-HCl buffer (pH 8.0). After the column was washed with 5 ml of the same buffer, the proteins were eluted with a linear-gradient from 20 mM Tris-HCl buffer (pH 8.0) to 1 M NaCl in 20 mM Tris-HCl buffer (pH 8.0). The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12), and the fractions containing recombinant proteins were collected and dialyzed against 50 mM sodium acetic acid buffer (pH 6.0).
Protein determination.
Protein was measured by the method of Bradford (1) with a protein assay kit from Bio-Rad by using bovine serum albumin as a standard. The molar amount of each recombinant protein was calculated by use of the theoretical molecular weight of each protein.
Assembly of recombinant cellulosomes.
For the cellulases, the purified mini-CbpA (5.0 nmol) and the recombinant cellulosomal cellulases (2.5 nmol of ExgS, 2.5 nmol of EngE and 5 nmol of EngH) were mixed in 100 μl of the binding buffer (25 mM sodium acetate buffer [pH 6.0], 15 mM CaCl2) and kept for 1 h at 4°C. For XynA, the purified mini-CbpA (5.0 nmol) and the purified XynA (10 nmol) were mixed in 100 μl of the binding buffer and kept for 1 h at 4°C. The assembly of recombinant cellulosomes was confirmed by native PAGE analysis by using a 4 to 15% ready-made gel (Bio-Rad) as described previously (15, 17). For clarity, we call the minicellulosomes containing mini-CbpA and cellulases as cellulase cellulosomes and the minicellulosomes containing mini-CbpA and XynA as XynA cellulosomes.
Determination of enzymatic activities.
The enzymatic activities were assayed in 100 μl of the reaction mixtures (0.5% substrate, 50 mM sodium acetate buffer; pH 6.0) at 37°C by measuring the liberated reducing sugars, as d-glucose equivalents, by the Somogyi-Nelson assay method (25). The substrates were corn cell walls for cellwallase, Avicel for cellulase, and xylan for xylanase. Activities were expressed as units with 1 U defined as the amount of enzyme releasing 1 μmol of reducing sugar per min.
Determination of glucose.
The amounts of glucose were determined by use of a glucose assay kit (Sigma). To convert cellooligosaccharides to glucoses for glucose detection, 100 μl of the reaction solution for cellwallase activity determination were incubated with 5 U of β-glucosidase from almond (Sigma) in 125 mM sodium acetate buffer (pH 5) at 37°C for 2 h.
Determination of sugar composition by TLC.
The products formed upon degradation of plant cell wall powder were analyzed by thin-layer chromatography (TLC). The hydrolysis products were applied to a silica gel plate (Merck). For development, a solvent mixture of butanol, acetic acid, and water (3:3:1 [vol/vol/vol]) was used. The sugars on the plate were visualized by spraying the plate with 0.1% methanolic orcinol in 10% sulfuric acid, followed by heating at 110°C.
RESULTS
Preparation of recombinant cellulosomes.
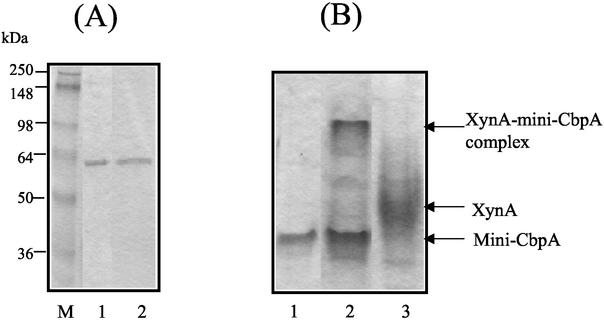
The xylanase A (XynA), which is a family 11 glycoside hydrolase with a dockerin domain and an NodB domain, was expressed by E. coli with a C-terminal His tag from the vector pET-29b as described recently (11) and purified by combination of nickel-affinity chromatography and anion-exchange chromatography. As a scaffolding protein, the mini-CbpA contained one cellulose-binding domain, one hydrophilic domain, and two cohesin domains of CbpA as described previously (15). The mini-CbpA was also expressed by E. coli and purified by combination of nickel-affinity chromatography and anion-exchange chromatography. SDS-PAGE analysis indicated that both XynA and mini-CbpA were purified almost to homogeneity (Fig. 1A).
FIG. 1.
Purification of XynA and mini-CbpA and assembly of XynA cellulosomes. (A) SDS-PAGE gel of the purified mini-CbpA and the purified XynA. Lane M, molecular weight markers; lane 1, purified mini-CbpA; lane 2, purified XynA. (B) Native PAGE analysis of purified mini-CbpA, purified XynA and the XynA-mini-CbpA complex (XynA cellulosome). Lane 1, purified mini-CbpA; lane 2, mixture of mini-CbpA and XynA; lane 3, purified XynA.
Since one cohesin domain binds one cellulosomal enzyme, one mini-CbpA could bind at most two cellulosomal enzymes. Therefore, XynA was mixed with mini-CbpA at a molar ratio of 2:1 to assemble the recombinant cellulosomes containing XynA and mini-CbpA. The assembly of the mini-CbpA and the XynA was confirmed by native PAGE analysis (Fig. 1B). The XynA or the mini-CbpA showed one major band on the native PAGE gel. The mixture of XynA and mini-CbpA showed one new band, and the XynA band had disappeared. The new band was considered to be the band for the “XynA cellulosome.” Although XynA was mixed with mini-CbpA at molar ratio of 2:1, the band for mini-CbpA still appeared in the mixture of XynA and mini-CbpA. This result indicated that the molar amount of mini-CbpA was underestimated or XynA was overestimated by Bradford method. Native PAGE analysis indicated that most of the XynA was bound with mini-CbpA to generate XynA cellulosomes.
Recently, we determined the synergistic effects on cellulase activities between three cellulosomal cellulases (EngE [EC 3.2.1.4/Swiss Prot Q9XD99], EngH [EC 3.2.1.4/Swiss Prot O65987], and ExgS [EC 3.2.1.29/Swiss Prot O65986]) (17). Among the compositions, we determined that the mixture of EngE, EngH, and ExgS at a molar ratio of 1:2:1 with mini-CbpA had the highest cellulase activity. Therefore, in the present study, we used this cellulase cellulosome mixture as the “cellulase cellulosomes” (Table 1). All three cellulosomal cellulases were purified almost to homogeneity and confirmed to bind with mini-CbpA to generate cellulase cellulosomes by native PAGE analysis as described recently (17).
TABLE 1.
Substrate specificities of recombinant cellulosomes of XynA and cellulases
| Cellulosome | Compositiona | Content (U/μmol of enzyme)
|
Products from cell wallsb | ||
|---|---|---|---|---|---|
| Cellwallase | Cellulase | Xylanase | |||
| XynA | XynA (2), mini-CbpA (1) | 0.103 | NDc | 741 | X2 (X3) |
| Cellulase | EngE (1), EngH (1), ExgS (1), and mini-CbpA (2) | 0.274 | 0.540 | 41 | G2 (G3, G4) |
The values in parentheses indicate relative molar amounts.
X2, xylobiose; X3, xylotriose; G2, cellobiose; G3, cellotriose; G4, cellotetraose. The sugars in parentheses indicate sugars liberated in trace amounts.
ND, not detected.
Characterization of recombinant cellulosomes.
The hydrolytic properties of the recombinant cellulosomes are summarized in Table 1. The XynA cellulosomes showed xylanase activity but not cellulase activity. The major hydrolytic product generated from corn cell walls by XynA cellulosomes was xylobiose with xylotriose as a minor product. On the other hand, the cellulase cellulosomes had both cellulase and xylanase activities. Since purified EngE has been shown to possess xylanase activity (20), the xylanase activity of cellulase cellulosomes was considered to come from EngE. The major hydrolytic product generated from corn cell walls by the cellulase cellulosomes was cellobiose, with trace amounts of cellotriose and cellotetraose. Despite the presence of xylanase activity in cellulase cellulosomes, xylooligosaccharides were not detected in hydrolytic products from corn cell walls by TLC analysis. These results with corn cell walls indicated that XynA cellulosomes degraded xylan and that the cellulase cellulosomes degraded cellulose in cell walls.
Synergistic effect on corn cell wall degradation activity (cellwallase) between XynA and cellulases.
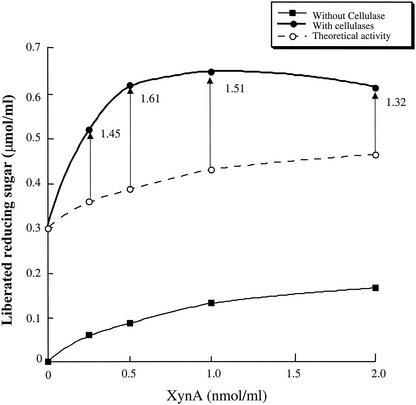
We determined the synergistic effects on corn cell wall degradation activity (cellwallase [16]) between XynA cellulosomes and cellulase cellulosomes. Corn cell walls were degraded by various amounts of XynA cellulosomes (0 to 2 nmol/ml) with or without a constant amount of cellulase cellulosomes (1 nmol/ml). The amounts of liberated reducing sugars were determined after reaction by the cellulosomes for 15 h. The activity of the mixtures of XynA cellulosomes and cellulase cellulosomes was compared with the theoretical activity (sum of the activities of XynA cellulosomes and cellulase cellulosomes). The results are shown in Fig. 2. The activity of the mixture of XynA cellulosomes and cellulase cellulosomes was higher than the corresponding theoretical activity. These results strongly suggested that XynA cellulosomes and cellulase cellulosomes degraded corn cell walls synergistically.
FIG. 2.
Synergistic effects between XynA cellulosomes and cellulase cellulosomes on corn cell wall degradation. The amounts of reducing sugars liberated from corn cell walls were determined. Corn cell walls were degraded by XynA cellulosomes with or without 1 nmol of cellulase cellulosomes/ml at 37°C for 15 h. The x axis indicates the concentrations of added XynA cellulosome. The theoretical activity indicates the sum of individual activities of XynA cellulosomes and cellulase cellulosomes. The synergy degrees (the activity of XynA cellulosome and cellulase cellulosome mixtures was divided by the corresponding theoretical activities) are indicated by arrow bars.
The synergy effects of cellulase and xylanase activities between XynA and cellulase cellulosomes were also determined. The synergy degree (the activities of XynA and cellulase cellulosome mixtures divided by the corresponding theoretical activities) of cellulase was 1.0, and that of xylanase was 0.96. These results indicated that XynA cellulosomes and cellulase cellulosomes degraded crystalline cellulose or xylan independently. These results also suggested that the synergy effect of cellwallase (cell wall degrading activity) activity between XynA cellulosomes and cellulase cellulosomes did not come from synergistic degradation of cellulose or xylan in corn cell walls.
Since the mixture of 0.5 nmol of XynA and 1 nmol of cellulase cellulosomes/ml showed the highest synergy degree of cellwallase (1.61), this cellulosome mixture was used for further determinations.
Sugar composition of hydrolytic products from corn cell walls.
The sugar composition of hydrolytic products from corn cell wall by the mixture of XynA and cellulase cellulosomes was determined.
TLC analysis showed that the major hydrolytic product from corn cell walls was cellobiose with xylobiose, cellotriose, and cellotetraose as minor products. As described above, XynA cellulosome liberated only xylooligosaccharides from corn cell walls and cellulase cellulosomes only cellooligosaccharides (Table 1). Therefore, xylobiose by XynA cellulosomes from corn cell walls and cellooligosaccharides was considered to be produced by cellulase cellulosomes.
The TLC pattern of liberated cellooligosaccharides by mixtures of XynA cellulosomes and cellulase cellulosomes was similar to that caused by cellulase cellulosomes alone (data not shown). These results suggested that the existence of XynA cellulosomes did not affect the degradation manner of cellulase cellulosomes against cellulose in corn cell walls.
To determine the amount of cellooligosaccharides present in the hydrolytic product, the total amount of glucose in the reaction mixtures was determined. The total amount of glucose was quantified after β-glucosidase treatment, which converted all cellooligosaccharides to glucose. The total amount of glucose produced by cellulase cellulosomes was also determined. The results are shown in Table 2. In the case of hydrolytic products produced by cellulase cellulosomes, the total amount of glucose was 0.714 μmol/ml, and the calculated average degree of polymerization of liberated cellooligosaccharides was 2.38. On the other hand, the total amount of glucose in the hydrolytic product produced by the mixture of XynA cellulosomes and cellulase cellulosomes was 1.095 μmol/ml and 1.53-fold higher than that produced by cellulase cellulosomes. These results indicated the cellwallase activity of cellulase cellulosomes was increased by a synergistic effect between XynA cellulosomes and cellulase cellulosomes.
TABLE 2.
Determination of sugar composition of plant cell wall degradation products
| Cellulosome | Mean amt (μmol/ml) of reducing sugara ± SD | Mean total glucosea (μmol/ml) ± SD | Avg DPb of Gnc | Calculated amt (μmol/ml) of:
|
|
|---|---|---|---|---|---|
| Gnc | Xylobiose | ||||
| XynA | 0.087 ± 0.004 | NDg | 0.000 | 0.087 | |
| Cellulase | 0.300 ± 0.003 | 0.714 ± 0.017 | 2.38 | 0.300 | 0.000 |
| XynA + cellulase | 0.622 ± 0.014 | 1.095 ± 0.056 | (2.38)d | 0.460e | 0.162f |
The values are the average of three determinations. Corn cell walls were degraded by 0.5 nmol of XynA cellulosome (XynA), 1 nmol of cellulase cellulosomes (cellulase), or 1 nmol of cellulase cellulosome plus 0.5 nmol of XynA cellulosome (XynA + cellulase/ml) at 37°C for 15 h.
DP, degree of polymerization.
Gn, cellooligosaccharides.
The degree of polymerization of cellooligosaccharides liberated by XynA plus cellulase was assumed to be equal to that of cellulase alone based on the similarity of the TLC patterns of the cellooligosaccharides.
This value was calculated by the amount of glucose at 1.095 μmol/ml divided by the assumed average degree of polymerization (2.38).
This value was calculated by taking the amount of reducing sugar (0.622 μmol/ml) and subtracting the calculated amount of cellooligosaccharide (0.460 μmol/ml).
ND, not detected.
The approximate amount of xylobiose in the hydrolytic products was also estimated. The approximate amount of xylobiose was calculated by subtracting the calculated amount of cellooligosaccharides from the total amount of reducing sugar (Table 2). The approximate amount of xylobiose liberated by the mixture of XynA cellulosomes and cellulase cellulosomes was calculated to be 0.162 μmol/ml and 1.86-fold higher than that liberated by XynA cellulosome alone. These results suggested that the cellwallase activity of XynA cellulosomes was also increased by a synergistic effect between XynA cellulosomes and cellulase cellulosomes.
Determination of the mechanism of synergistic effects on cellwallase between XynA and cellulases.
To determine the mechanism of synergistic effects on cellwallase activity between XynA cellulosomes and cellulase cellulosomes, time course experiments and sequential reactions were carried out.
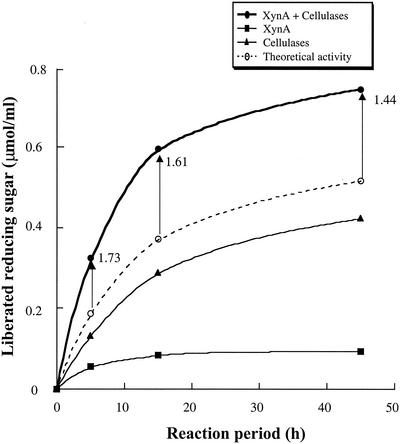
To determine the relationship between reaction period and synergy degree, corn cell walls were degraded by a mixture of XynA cellulosomes and cellulase cellulosomes for 5, 15, and 45 h. As shown in Fig. 3, the synergy degrees decreased from 1.73 (5 h) to 1.44 (45 h), according to the length of the reaction period. These results suggested that the synergistic effects between XynA cellulosomes and cellulase cellulosomes were more effective in the initial degradation steps than in the later steps.
FIG. 3.
Time course of corn cell wall degradation by a mixture of XynA cellulosomes and cellulase cellulosomes. The amount of reducing sugar liberated from corn cell walls was determined. Corn cell walls were degraded by 0.5 nmol/ml of XynA cellulosome (XynA), 1 nmol of cellulase cellulosomes (cellulases)/ml, or a mixture of 1 nmol of cellulase cellulosome and 0.5 nmol of XynA cellulosome (XynA plus cellulases)/ml at 37°C. The x axis indicates the reaction periods. The theoretical activity indicates the sum of individual activities of XynA cellulosomes and cellulase cellulosomes. The synergy degrees (the activity of the mixtures of XynA cellulosomes and cellulase cellulosomes divided by the corresponding theoretical activities) are indicated by arrow bars.
To further determine the synergistic effects between XynA cellulosomes and cellulase cellulosomes, sequential reactions were carried out. Corn cell walls were treated by either XynA or cellulase cellulosomes for 15 h in the first reaction. Then, the reaction mixtures were boiled for 20 min to inactivate the cellulosomes, which were used for the first reactions. By this heat treatment, the cellulosomes were inactivated completely (data not shown). After the reaction mixtures of the first reaction were boiled, the cellulosome that had not been used for the first reactions was added to the reaction mixtures, followed by incubation for an additional 15 h. The amount of liberated reducing sugars was then determined. The results are shown in Table 3. The amount of reducing sugars liberated by the same cellulosomes before or after the boiling treatment was almost the same, indicating that the boiling treatment had no effect on cellwallase activity. The synergistic degree of cellwallase activity in the “simultaneous reactions” was ca. 1.6. On the other hand, almost no synergistic effects were observed in the “sequential reactions.” These results indicated that XynA cellulosomes and cellulase cellulosomes degraded corn cell walls synergistically only in a simultaneous but not in a “sequential manner.”
TABLE 3.
Sequential and simultaneous reactions against plant cell wall by recombinant cellulosomes of the cellulase mixture and of XynAa
| Added cellulosome(s) | Mean amt (μmol/ml) of liberated reducing sugarb ± SD | Degree of synergyc | |
|---|---|---|---|
| First reaction | Second reaction | ||
| Simultaneous reaction | |||
| Cellulase + XynA | No enzyme | 0.622 ± 0.014 | 1.61 |
| No enzyme | Cellulase + XynA | 0.595 ± 0.043 | 1.59 |
| Sequential reaction | |||
| Cellulase | XynA | 0.407 ± 0.022 | 1.03 |
| XynA | Cellulase | 0.357 ± 0.048 | 0.98 |
| Control reaction | |||
| Cellulase | No enzyme | 0.300 ± 0.003 | |
| No enzymes | Cellulase | 0.278 ± 0.010 | |
| XynA | No enzyme | 0.087 ± 0.004 | |
| No enzyme | XynA | 0.096 ± 0.002 | |
| No enzyme | No enzyme | 0.007 ± 0.003 | |
Corn cell walls were degraded by 0.5 nmol of XynA cellulosome (XynA), 1 nmol of cellulase cellulosomes (cellulase), or 1 nmol of cellulase cellulosome plus 0.5 nmol of XynA cellulosome (XynA + cellulases)/ml at 37°C for 15 h. Between the first and the second reactions, the reaction mixtures were boiled for 20 min to inactivate the cellulosomes used for the first reaction.
The values are the average of three determinations.
The synergy degree values were calculated as the activity divided by the sum of the activities of the control reactions.
The contribution of mini-CbpA to cellwallase activity was also determined. The reducing sugar amount liberated by the mixture of XynA and cellulases without mini-CbpA was 0.378 μmol/ml (15-h reaction). Compared to the amount of reducing sugar liberated by the mixture of XynA and cellulase with mini-CbpA (0.622 μmol/ml), 40% of the cellwallase activity was lost by removal of mini-CbpA. These results suggested that mini-CbpA helped XynA and the cellulases to degrade corn cell walls.
DISCUSSION
To determine the mechanism of cell wall degradation by cellulosomes from C. cellulovorans, we recently determined the subunit composition and enzymatic activity of two cellulosome fractions that showed cellwallase activity (16). The xylanase XynA and the mannanase ManA, as well as three cellulases, were identified as major enzymatic subunits in the cellulosome fractions. Although both cellulosome fractions had the same level of cellulase activity, one of them possessed fourfold-higher cellwallase activity than the other. Moreover, the cellulosome fraction with higher cellwallase activity contained higher amounts of XynA and xylanase activity than the other. Xylan (ca. 20% of total weight), as well as cellulose (ca. 40%), is known to be a major component of the corn cell wall (2, 9). Therefore, we assumed that the difference of cellwallase activity between the two cellulosome fractions came from differences in xylanase activity. This assumption prompted the present study to determine the synergistic effects on cellwallase activity between XynA cellulosomes and cellulase cellulosomes.
As we expected, XynA cellulosomes and cellulase cellulosomes degraded corn cell walls synergistically. The cellwallase activity increased in relation to the amount of XynA cellulosomes present until the content of XynA cellulosomes reached 50% of the molar amount of cellulase cellulosomes (Fig. 1). This result implied that xylan degradation by XynA could be a rate-limiting factor for corn cell wall degradation by cellulosomes, when the amount of XynA was less than 50% of the amount of cellulase in cellulosomes. Actually, we showed previously that molar amounts of XynA in the higher and lower activity cellwallase cellulosome fractions were 14.5 and 3.5% of the molar amounts of cellulases, respectively (16). Therefore, the difference in XynA content appears to be one of the reasons for the difference in cellwallase activity between the two cellulosome fractions that were tested previously (16).
The synergistic degradation of cellulose by several cellulases is a well-known phenomenon (5, 6, 8, 18, 26, 28), although there have been no reports to our knowledge about synergism between cellulosomal xylanases and cellulosomal cellulases on plant cell wall degradation. Recently, we demonstrated that three cellulosomal cellulases from C. cellulovorans (EngE, EngH, and ExgS) degraded crystalline cellulose synergistically (17). In cellulase synergism, the synergistic reaction between endoglucanases and exoglucanases was considered to be a mechanism in which endoglucanases initially nicked the crystalline cellulose surface, and this was followed by exoglucanases liberating cellobiose from these nicks (27). As expected from this mechanism, synergistic effects are observed not only in “simultaneous reactions” but also in “sequential reactions,” in which endoglucanases are used for the first reaction followed by the exoglucanase reaction (17-19). This feature was considered to be one of the major reasons for synergism of cellulases.
Compared to the synergism of cellulases, the synergistic effect on cellwallase activity between XynA and cellulases has unique features. These features are as follows: (i) XynA and cellulases degraded different substrates in the cell walls (xylan by XynA and cellulose by cellulases); (ii) synergistic effects were found only against complex substrates (corn cell walls) but not against pure substrates (xylan or crystalline cellulose); and (iii) synergistic effects were observed only in the “simultaneous” but not in the “sequential” reactions. These features suggested that the synergistic effect on cellwallase activity between XynA and cellulases had a different mechanism from that of synergy between cellulases.
The structure of plant cell walls gives insight into the mechanism of synergistic effects between XynA and cellulases. In cell walls, xylan chains (arabinoxylans) are considered to hydrogen bind to cellulose microfibril surfaces and connect several cellulose microfibrils to generate cross-linked structures (2). This cell wall structure model suggests that degradation of xylan networks between cellulose microfibrils by xylanases might allow cellulases to access and degrade cellulose microfibrils embedded in the deeper structure. Also, degradation of cellulose microfibrils in the deeper structure might help xylanase to access and degrade xylan chains further in the deeper structure. This degradation model could explain the unique features of synergism between XynA and cellulases. Therefore, the mechanism proposed above could be a possible mechanism for the synergistic effect between XynA cellulosomes and cellulase cellulosomes.
One of our goals is to prepare “designer cellulosomes,” which could efficiently degrade plant cell walls to fermentable sugars for the production of energy-yielding compounds such as ethanol. Although we found a synergistic effect between XynA cellulosomes and cellulase cellulosomes, the specific activity for cellwallase of the recombinant cellulosome mixture of XynA and cellulases was still 10-fold lower than that of purified cellulosomes from C. cellulovorans culture broths (data not shown). Other hemicellulases might increase the specific activity of the cellwallase of recombinant cellulosomes. In addition to the XynA gene, we have also isolated cellulosomal mannanase ManA gene (21) and pectate lyase PelA (22) genes. Moreover, we recently have found noncellulosomal arabinofuranosidase activity in the cultural broth of C. cellulovorans (11). These hemicellulases might also act synergistically against corn cell walls, since mannan, pectin, and arabinose in arabinoxylan are also components of corn cell walls. We are proceeding to determine the effects of these hemicellulases on plant cell wall degradation.
Acknowledgments
We thank T. Nojiri at Meiji Seika Kaisha, Ltd., for supplying the powder from corn stem. We are grateful to Helen Chan for skillful technical assistance.
This study was supported in part by Department of Energy grant DE-DDF03-92ER20069.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Carpita, N. 1996. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:445-476. [DOI] [PubMed] [Google Scholar]
- 3.Doi, R. H., J. S. Park, C. C. Liu, L. M. Malburg, Y. Tamaru, A. Ichiishi, and A. Ibrahim. 1998. Cellulosome and noncellulosomal cellulases of Clostridium cellulovorans. Extremophiles 2:53-60. [DOI] [PubMed] [Google Scholar]
- 4.Foong, F. C., T. Hamamoto, O. Shoseyov, and R. H. Doi. 1991. Nucleotide sequence and characteristics of endoglucanase gene engB from Clostridium cellulovorans. J. Gen. Microbiol. 137:1729-1736. [DOI] [PubMed] [Google Scholar]
- 5.Gaudin, C., A. Belaich, S. Champ, and J. P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrissat, B., H. Driguez, C. Viet, and M. Schulein. 1985. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Bio/Technology 3:722-726. [Google Scholar]
- 7.Himmel, M. E., M. F. Ruth, and C. E. Wyman. 1999. Cellulase for commodity products from cellulosic biomass. Curr. Opin. Biotechnol. 10:358-364. [DOI] [PubMed] [Google Scholar]
- 8.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression, and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 9.Kaar, W. E., and M. T. Holtzapple. 1998. Benefits from tween during enzymic hydrolysis of corn stover. Biotechnol. Bioeng. 59:419-427. [DOI] [PubMed] [Google Scholar]
- 10.Kosugi, A., K. Murashima, Y. Tamaru, and R. H. Doi. 2002. Cell-surface-anchoring role of N-terminal surface layer homology domains of Clostridium cellulovorans EngE. J. Bacteriol. 184:884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosugi, A., K. Murashima, Y. Tamaru, and R. H. Doi. 2002. Xylanase and acetyl xylan esterase activities of XynA, a key subunit of the Clostridium cellulovorans cellulosome for xylan degradation. Appl. Environ. Microbiol. 68:6399-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Liu, C. C., and R. H. Doi. 1998. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosome. Gene 211:39-47. [DOI] [PubMed] [Google Scholar]
- 14.Lynd, L. R., J. H. Cushman, R. J. Nichols, and C. E. Wyman. 1991. Fuel ethanol from cellulosic biomass. Science 251:1318-1323. [DOI] [PubMed] [Google Scholar]
- 15.Murashima, K., C. L. Chen, A. Kosugi, Y. Tamaru, R. H. Doi, and S. L. Wong. 2002. Heterologous production of Clostridium cellulovorans engB, using protease-deficient Bacillus subtilis, and preparation of active recombinant cellulosomes. J. Bacteriol. 184:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Determination of subunit composition of Clostridium cellulovorans cellulosomes that degrade plant cell walls. Appl. Environ. Microbiol. 68:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Synergistic effects on crystalline cellulose degradation between cellulosomal cellulases from Clostridium cellulovorans. J. Bacteriol. 184:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nidetzky, B., W. Steiner, M. Hayn, and M. Claeyssens. 1994. Cellulose hydrolysis by the cellulases from Trichoderma reesei: a new model for synergistic interaction. Biochem. J. 298:705-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Streamer, M., K. E. Eriksson, and B. Pettersson. 1975. Extracellular enzyme system utilized by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cellulose: functional characterization of five endo-1,4-β-glucanases and one exo-1,4-β-glucanase. Eur. J. Biochem. 59:607-613. [DOI] [PubMed] [Google Scholar]
- 20.Tamaru, Y., and R. H. Doi. 1999. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J. Bacteriol. 181:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamaru, Y., and R. H. Doi. 2000. The engL gene cluster of Clostridium cellulovorans contains a gene for cellulosomal manA. J. Bacteriol. 182:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamaru, Y., and R. H. Doi. 2001. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc. Natl. Acad. Sci. USA 98:4125-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamaru, Y., S. Ui, K. Murashima, A. Kosugi, H. Chan, R. H. Doi, and B. Liu. 2002. Formation of protoplasts from cultured tobacco cells and. Arabidopsis thaliana by the action of cellulosomes and pectate lyase from Clostridium cellulovorans. Appl. Environ. Microbiol. 68:2614-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood, T. M., and K. M. Bhat. 1988. Methods for measuring cellulase activities. Methods Enzymol. 160:87-112. [Google Scholar]
- 26.Wood, T. M., and S. I. McCrae. 1972. The purification and properties of the C1 component of Trichoderma koningii cellulase. Biochem. J. 128:1183-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood, T. M., and S. I. McCrae. 1979. Synergism between enzymes involved in the solubilization of native cellulose. Adv. Chem. Ser. 181:181-209. [Google Scholar]
- 28.Wood, T. M., S. I. McCrae, and K. M. Bhat. 1989. The mechanism of fungal cellulase action: synergism between enzyme components of Penicillium pinophilum cellulase in solubilizing hydrogen bond-ordered cellulose. Biochem. J. 260:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]