Abstract
The swimming behavior of Escherichia coli at any moment is dictated by the intracellular concentration of the phosphorylated form of the chemotaxis response regulator CheY, which binds to the base of the flagellar motor. CheY is phosphorylated on Asp57 by the sensor kinase CheA and dephosphorylated by CheZ. Previous work (Silversmith et al., J. Biol. Chem. 276:18478, 2001) demonstrated that replacement of CheY Asn59 with arginine resulted in extreme resistance to dephosphorylation by CheZ despite proficient binding to CheZ. Here we present the X-ray crystal structure of CheYN59R in a complex with Mn2+ and the stable phosphoryl analogue BeF3−. The overall folding and active site architecture are nearly identical to those of the analogous complex containing wild-type CheY. The notable exception is the introduction of a salt bridge between Arg59 (on the β3α3 loop) and Glu89 (on the β4α4 loop). Modeling this structure into the (CheY-BeF3−-Mg2+)2CheZ2 structure demonstrated that the conformation of Arg59 should not obstruct entry of the CheZ catalytic residue Gln147 into the active site of CheY, eliminating steric interference as a mechanism for CheZ resistance. However, both CheYE89A and CheYE89Q, like CheYN59R, conferred clockwise flagellar rotation phenotypes in strains which lacked wild-type CheY and displayed considerable (≈40-fold) resistance to dephosphorylation by CheZ. CheYE89A and CheYE89Q had autophosphorylation and autodephosphorylation properties similar to those of wild-type CheY and could bind to CheZ with wild-type affinity. Therefore, removal of Glu89 resulted specifically in CheZ resistance, suggesting that CheY Glu89 plays a role in CheZ-mediated dephosphorylation. The CheZ resistance of CheYN59R can thus be largely explained by the formation of the salt bridge between Arg59 and Glu89, which prevents Glu89 from executing its role in catalysis.
In Escherichia coli chemotaxis, cells swim towards attractants and away from repellants by regulating the frequency of switching between smooth swims (counterclockwise flagellar rotation) and reorienting tumbles (clockwise flagellar rotation), so that the duration of smooth swims is lengthened when swimming in a favorable direction. The series of molecular events that mediate this flow of information have been characterized in considerable detail (6, 8), and bacterial chemotaxis has long served as a prototype for the study of two-component regulatory systems, a large family of signal transduction systems found in bacteria, fungi, and plants (26).
In chemotaxis, the sensor kinase CheA associates with the cytoplasmic portion of transmembrane chemoreceptors and autophosphorylates a histidyl residue (His48) at a rate which is regulated by the occupancy of the extracellular binding sites of the receptors with attractant or repellant molecules. CheA then transfers the phosphoryl group to an aspartyl residue (Asp57) of the freely diffusible response regulator protein CheY. Phosphorylated CheY (CheY-P) binds to the flagellar switch, inducing clockwise flagellar rotation. Removal of the phosphoryl group from CheY occurs via catalysis by the accessory protein CheZ.
Extensive structural and biochemical investigation has led to considerable insight into the phosphorylation and dephosphorylation chemistry of the response regulator CheY. CheY is a small (128-residue), single-domain, nearly spherical protein with a (βα)5 “Rossman” fold (27). The active site for phosphorylation comprises an Mg2+ ion and five residues which are conserved among response regulators: Asp12, Asp13, the site of phosphorylation Asp57, Thr87, and Lys109. The active-site residues reside either on the C terminus of a β strand or on a loop connecting the C terminus of a β strand to the next α-helix.
Both CheY phosphorylation (by CheA or a variety of small molecules) and autodephosphorylation are believed to occur by analogous in-line nucleophilic substitution reactions which go through a pentavalent phosphorous transition state (20, 25). The carboxylate groups of Asp12 and Asp13 are involved in coordination of the Mg2+ ion (25), which is required for phosphorylation, autodephosphorylation, and dephosphorylation by CheZ (15). Lys109, Thr87, and Mg2+ interact directly with fluorine atoms in the CheY-BeF3−-Mn2+ structure (12) and are believed to assist in catalysis of phosphorylation and dephosphorylation through neutralization of negative charge and/or transition state stabilization (14, 25).
The fundamental mechanism of CheZ-mediated dephosphorylation of CheY has recently been elucidated with the solution of the cocrystal structure of CheZ with CheY-BeF3−-Mg2+, a stable structural and functional analogue of CheY-P (29). The four-helix-bundle domain of the CheZ dimer interacts directly with the active-site region of CheY so that the CheZ Gln147 side chain inserts into the CheY active site. CheZ Gln147 may aid the CheY autodephosphorylation mechanism by positioning the attacking water molecule in the appropriate geometry for in-line attack of the phosphorus.
A previous investigation from this laboratory (24) included characterization of a CheY mutant with an arginine residue substituted for asparagine at position 59 (CheYN59R). Asn59 is not conserved among CheY proteins from different bacterial species but is located on the β3α3 loop at the active site, with its side chain pointing into the channel where the phosphodonor enters. The arginine substitution at this position resulted in extreme resistance to CheZ-mediated dephosphorylation despite binding CheZ with the same affinity as activated wild-type CheY (24). Substitution with an alanine residue at position 59 did not affect CheZ sensitivity, indicating that the CheZ resistance was due to introduction of the arginine and not the removal of the asparagine.
The present study was directed at elucidating the structural and mechanistic basis for the strong CheZ resistance of CheYN59R. The solution of the X-ray crystal structure of this mutant in a complex with BeF3− and Mn2+ reported here, coupled with the recent structure of the (CheY-BeF3−-Mg2+)2CheZ2 complex (29), ruled out the possibility of either steric hindrance of CheZ Gln147 or introduction of an interaction between Arg59 and the phosphoryl group as explanations for the CheZ resistance of CheYN59R. However, the CheYN59R-BeF3−-Mn2+ structure implicated the possible importance of the β4α4 loop of CheY in the CheZ-mediated dephosphorylation mechanism. Together with additional biochemical analysis, the data suggest that CheY Glu89, located on the β4α4 loop, plays a role in CheZ-mediated dephosphorylation.
MATERIALS AND METHODS
Site-directed mutagenesis and protein purification.
Plasmids encoding cheYE89A, cheYE89Q, cheYE89H, cheYE89K, cheZR151A, and cheZR151K were prepared by PCR mutagenesis (11) of the pRBB40 plasmid (5). Wild-type and mutant forms of CheY (10) and CheZ (24) were purified as described from E. coli strain K0641recA (5) carrying the appropriate pRBB40 plasmid.
Crystallization of CheYN59R-BeF3−-Mn2+.
Crystals were prepared at room temperature by the hanging-drop vapor diffusion method. A solution containing the assembled complex [CheYN59R (10 mg/ml), MnCl2 (1 mM), NaF (10 mM), and BeCl2 (1 mM)] was mixed with an equal volume of well solution [100 mM Tris (pH 8.4), 1.8 M (NH4)2SO4, 5% (vol/vol) glycerol] and placed over the same well solution. Crystals appeared within 24 h. The crystals were gradually equilibrated with a cryoprotectant solution consisting of well solution supplemented with 25% (vol/vol) glycerol, as described before (12). Crystals were flash frozen in liquid nitrogen.
Structure determination.
Data were collected at 100 K with CuKα X-rays on a Rigaku RU-H3R/R-Axis IV++ equipped with Osmic Confocal “Blue” optics. One-degree oscillation frames were processed and scaled with DENZO and SCALEPACK (19). The coordinates of the wild-type CheY-BeF3−-Mn2+ structure (12) (PDB code 1FQW) were used as a starting model for molecular replacement with the Amore program (18). Two major peaks in the rotation function map were used to locate the positions of two independent molecules in the asymmetric unit, with data between 8 and 4 Å. Rigid body refinement gave an R-factor of 27% and an Amore correlation coefficient of 75%. Refinement was carried out with the CNS package (9). Twofold noncrystallographic symmetry restraints were used in earlier cycles of the refinement but were released in the last cycles of torsion angle-simulated annealing refinement. Data processing and structural refinement statistics are shown in Table 1.
TABLE 1.
Summary of crystallographic analysis
| Parameter | Value |
|---|---|
| Statistics | |
| Space group | P212121 |
| Unit cell dimensions (Å) | a = 53.04, b = 53.615, c = 160.89 |
| Resolution limits (Å) | 40-2.7 |
| Total no. of measurements | 87,457 |
| No. of unique reflections | 13,060 |
| Rsymma | 4.4% (21.3%)b |
| Average redundancy (completeness) | 3 (98.3%) |
| Average I/(σ I) | 27 (6.4)b |
| Refinement | |
| No. of atoms in asymmetric unit | 2,032 (includes 44 water and 3 sulfate) |
| No. of reflections in working set (test set) | 12,392 (668) |
| Resolution used | All with bulk solvent correction |
| R-factorc | 22.6% |
| Rfreec | 27.7% |
| RMS deviation from ideality | |
| Bonds | 0.006Å |
| Angles | 1.23° |
Rsymm = 100 × Σ|I − <I>|/ΣI, where I is the integrated intensity of a reflection.
Numbers in parentheses refer to the highest-resolution shell (2.8 to 2.7 Å).
R = Σ|Fo − Fc|/ΣFo, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is calculated by using 5% of reflections set aside and never used during refinement.
Cell behavioral assays.
Bacterial swarming was assayed on semisolid agar (1), and the direction of flagellar rotation was assessed by tethered-cell analysis (7).
CheZ sensitivity assay.
The sensitivity of wild-type and mutant CheY to dephosphorylation by CheZ was assessed spectroscopically by measurement of the rate of Pi release with the Enzchek Pi assay kit (Molecular Probes). Application of this assay to assess the flow of phosphoryl groups on and off CheY has been described previously in detail (24). Briefly, CheY (final concentration, 4.5 μM) was added to a cuvette containing phosphoimidazole (3 mM), CheZ (if appropriate), and assay kit components, and the absorbance at 360 nm was monitored continuously. A previously determined extinction coefficient (0.0091 μM Pi−1 cm−1) was used to convert the observed slope to the Pi release rate.
Kinetics of CheY autophosphorylation and autodephosphorylation.
Time courses of CheY phosphorylation with phosphoramidate or acetyl phosphate were monitored by tryptophan fluorescence (24). Reactions were initiated with a stopped-flow apparatus (Applied Photophysics RX2000). Apparent first-order rate constants (kobs) were determined with an exponential curve fit, and reported kobs values represent the averages of three independent measurements. CheY autodephosphorylation rates were determined by measurement of levels of [32P]CheY-P as a function of time by Phosphorimager analysis of electrophoretic gels, as previously described (23). The buffer was 100 mM Tris (pH 7.5)-10 mM MgCl2. Two independent time courses were measured, and the average kdephosph value is reported.
Fluorescence anisotropy.
Binding of activated wild-type or mutant CheY to CheZ was assessed by measurement of the change in anisotropy of fluoresceinated CheZ from Salmonella enterica serovar Typhimurium as previously described (3, 24). Acetyl phosphate (17 mM) or BeF3− (0.1 mM BeCl2, 10 mM NaF) was initially added to the fluoresceinated CheZ, followed by successive additions of CheY until the signal became saturated.
RESULTS
Crystal structure of CheYN59R complexed with BeF3− and Mn2+.
In order to elucidate the structural basis for the extreme CheZ resistance of CheYN59R, the X-ray crystal structure of CheYN59R complexed with the stable phosphoryl analogue BeF3− and the divalent cation Mn2+ was solved (Table 1, Fig. 1). The complex was crystallized under conditions similar to those used for the wild-type CheY-BeF3−-Mn2+ complex (12), and the crystals had the same overall shape (square bipyramidal), the same P212121 space group, and nearly identical unit cell dimensions (Table 1) as the wild-type complex (12). The intermolecular crystal contacts for the CheYN59R-BeF3−-Mn2+ complex were the same as those described for wild-type CheY-BeF3−-Mn2+ (12) and did not involve residues in the vicinity of the active-site region. Therefore, structural differences observed between the complexes likely reflect true differences in protein structure and not changes due to differential crystal packing.
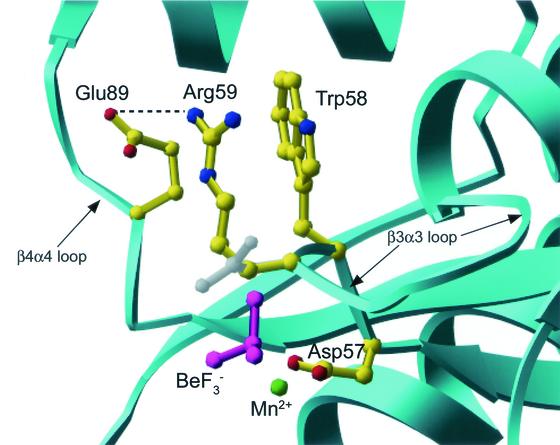
FIG. 1.
Structural features of active-site region of CheYN59R-BeF3−-Mn2+. The secondary structure of CheYN59R is shown in ribbon representation (cyan), and select residues are shown in ball-and-stick representation. A salt bridge (dashed line) is present between Glu89 (yellow) and Arg59 (yellow), the site of the substitution. The conformation of Asn59 in the superimposed wild-type CheY-BeF3−-Mn2+ structure (PDB code 1FQW) is shown in gray. BeF3− is magenta, and Mn2+ is green.
The entire backbone and all side chains of CheYN59R were visible. In addition, 25 water molecules and four sulfate ions were modeled into positive peaks (greater than 3σ) in the electron density maps. None of the sulfate ions and one of the water molecules (see below) were located near the active site. For both of the CheY molecules which constituted the asymmetric unit, the conformation of the alpha carbon backbone was very similar (root mean square deviation = 0.2 Å) to that of the analogous complex with wild-type CheY (PDB code 1FQW). Many features of the active-site architecture in CheYN59R-BeF3−-Mn2+ were indistinguishable from those of wild-type CheY-BeF3−-Mn2+. Difference Fo − Fc electron density maps calculated with only protein atoms clearly showed the BeF3− attached to Asp57 and the Mn2+ coordinated with the same six ligands as observed in the wild-type CheY-BeF3−-Mn2+ structure (one fluorine atom, carboxylate groups of Asp57 and Asp13, the backbone carbonyl of residue 59, and two water molecules). The characteristic conformations of the Thr87 and Tyr106 side chains observed in wild-type CheY-BeF3−-Mn2+, believed to be a hallmark of CheY activation, were clearly present in the CheYN59R-BeF3−-Mn2+ complex. These observations are consistent with the conclusion that CheYN59R-P behaves like activated wild-type CheY in its interaction with the flagellar switch (24).
The only significant differences between the mutant and wild-type structures were localized at the site of the substitution (Fig. 1). Whereas the side chain nitrogen of Asn59 forms a hydrogen bond with one of the fluorine atoms in the wild-type structure, the backbone carbonyl of Arg59 in the mutant structure is within hydrogen-bonding distance (2.8 Å) of the same fluorine atom. The side chain of Arg59 extends towards the surface of the CheY molecule essentially parallel to the Trp58 side chain. The nitrogen atom of the guanidinium group is approximately 3.0 Å from a carboxylate oxygen of Glu89, thus forming a new salt bridge. In molecule A of the asymmetric unit, this distance is 2.9 Å, and in molecule B it is 3.2 Å.
There were several additional subtle differences between the two molecules (A and B) which constitute the asymmetric unit in the region of the Arg59-Glu89 salt bridge. First, in molecule A, the side chain of Glu89 was rotated slightly towards Arg59 and the active-site region relative to its position in the wild-type structure. Second, while the electron density for Arg59 in molecule B is very well defined, in molecule A there was a small negative peak (about −3σ) in the Fo − Fc electron density map centered on the NΕ atom of Arg59 but no nearby positive density to suggest an alternative conformation. In addition, the refined atomic B-factors for the more distal side chain atoms of Arg59 are slightly higher than the average for neighboring residues (48 to 50 versus 30's and lower 40's), suggesting that the Arg59 side chain may spend a small portion of time in disordered conformations and the rest in the discrete conformation in association with Glu89. Finally, a water molecule (3.2 σ) is located within 3.0 Å of both Glu89 OΕ1 and Arg59 NΕ in molecule B but not A. The OΕ1 of Glu89 in molecule A is shifted closer to the position of this water, possibly replacing it. An analogous difference of one water molecule was present in the wild-type CheY-BeF3−-Mg2+ structure (PDB code 1FQW).
Modeling CheYN59R-BeF3−-Mn2+ structure into the (CheY-BeF3−-Mg2+)2CheZ2 crystal structure.
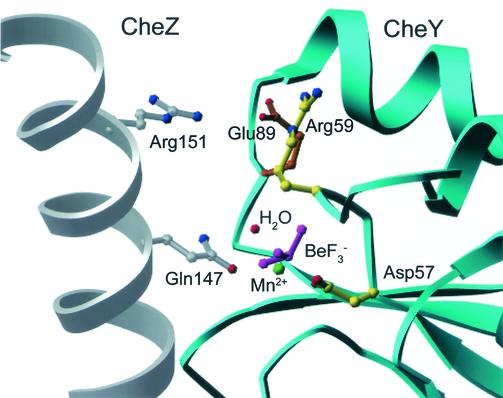
As a first step in the assessment of a possible mechanism for the strong CheZ resistance of CheYN59R (24), the CheYN59R-BeF3−-Mn2+ structure was modeled into the CheY site of the (CheY-BeF3−-Mg2+)2CheZ2 complex structure (29) (Fig. 2). In the current model of CheZ-dependent dephosphorylation, CheZ Gln147, which is inserted into the active site of CheY, acts as an essential catalytic residue, possibly for orientation of the water molecule which attacks the aspartyl phosphate group during dephosphorylation. One possible explanation for the CheZ resistance of CheYN59R is that Arg59 physically blocks the Gln147 side chain from insertion into the position required for catalysis (24). However, modeling (Fig. 2) predicts that the arginine at position 59 is pointing away from the active site to form the salt bridge with Glu89. This would not be expected to physically block CheZ Gln147, and thus, this explanation for the CheZ resistance of CheYN59R can be ruled out. Similarly, the structure also precludes the possibility that interactions between the Arg59 side chain and the phosphoryl group cause the CheZ resistance of CheYN59R. The Arg59 side chain is pointed away from the BeF3−, with 6.7 Å separating the closest guanidinium nitrogen and fluorine atoms.
FIG. 2.
Active-site region of CheYN59R-BeF3−-Mn2+ structure modeled into the (CheY-BeF3−-Mg2+)2CheZ2 crystal structure (PDB code 1KMI). The secondary structures of CheY (cyan) and CheZ (gray) are in ribbon representation. Select residues in ball-and-stick representation are CheZ Gln147 (gray) and Arg151 (gray), CheY Asp57 and Arg59 (yellow), Glu89 (orange), BeF3− (magenta), and Mn2+ (green). A water molecule (red) has been modeled into the active site in the appropriate geometry for in-line attack of the BeF3− (analogue of PO32−) and within hydrogen-bonding distance of CheZ Gln 147 (29). Only one chain of the CheZ dimer is shown for clarity.
Characterization of CheY Glu89 mutants.
The most notable difference between the CheY59NR-BeF3−-Mn2+ structure and the analogous wild-type CheY structure is the introduction of a salt bridge between Arg59 and Glu89 in the mutant CheY structure. Glu89 is located on the β4α4 loop, a mobile loop which undergoes the most significant change in position upon CheY activation (12, 13). Therefore, it is plausible that introduction of an attractive interaction between Arg59 (on the β3α3 loop) and Glu89 (on the β4α4 loop) could affect the mobility and/or accessibility of the Glu89 side chain and somehow inhibit CheZ-mediated dephosphorylation.
To investigate a possible causal relationship between the Arg59-Glu89 salt bridge observed in CheYN59R-BeF3−-Mn2+ and the CheZ resistance of CheYN59R, mutants of CheY with substitutions at Glu89 were characterized genetically and biochemically. Strains containing cheYE89Q, cheYE89A, cheYE89H, and cheYE89K were unable to mediate chemotaxis, as measured in a cell swarming assay (data not shown). Failure to swarm reflects the inability to switch rapidly between clockwise and counterclockwise flagellar rotation. Cell-tethering assays demonstrated that cells containing CheYE89Q, CheYE89A, CheYE89H, or CheYE89K had strong clockwise biases, indistinguishable from the behavior of cells containing CheYN59R (24).
The cause of the clockwise flagellar rotation of cells containing CheY with substitutions at position 89 was explored by biochemical analysis of CheYE89A and CheYE89Q. Clockwise swimming behavior would result if the intracellular concentration of CheY-P was increased relative to that of wild-type CheY as a result of an increased rate of phosphorylation and/or a decreased rate of dephosphorylation. Measurements of the kinetics of autophosphorylation and autodephosphorylation revealed modest differences between wild-type CheY, CheYE89A, and CheYE89Q (Table 2). For autophosphorylation with acetyl phosphate or phosphoramidate, CheYE89A had rates very similar to those of wild-type CheY, whereas CheYE89Q autophosphorylated about threefold faster than wild-type CheY for both phosphodonors. Autodephosphorylation rate constants were similar for CheYE89Q and wild-type CheY, but CheYE89A had an autodephosphorylation rate constant about two- to threefold slower (Table 2).
TABLE 2.
Rate constants for phosphorylation and autodephosphorylation of CheY mutants at position 89
| CheY |
kobs for phosphorylation (s−1)a
|
kdephosph (s−1)b | |
|---|---|---|---|
| Acetyl phosphate (50 mM) | Phosphoramidate (50 mM) | ||
| Wild-type | 0.46 | 0.61 | 0.065 |
| E89Q | 1.7 | 1.6 | 0.065 |
| E89A | 0.67 | 0.66 | 0.025 |
kobs = kphosp [phosphodonor]/Ks + kdephosph (22). kobs values were measured at 25°C.
kdephosph values were measured at room temperature.
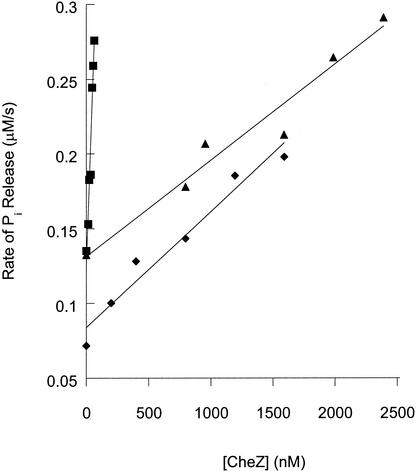
In contrast to the subtle differences in phosphorylation and autodephosphorylation rates, CheYE89A and CheYE89Q displayed substantial CheZ resistance (Fig. 3). Titration of the Pi release rate of reactions containing CheY (wild-type, E89Q, or E89A) and monophosphoimidazole with CheZ gave an approximately linear relationship for subsaturating CheZ concentrations (24). However, the slopes of the lines were different, indicating that both CheYE89Q and CheYE89A required more CheZ than wild-type CheY to obtain the same degree of rate enhancement. Taking the ratio of the slopes as an indicator of CheZ sensitivity indicated that CheYE89A and CheYE89Q were 35- and 43-fold more resistant to CheZ, respectively, than wild-type CheY. Therefore, like CheYN59R, both CheYE89A and CheYE89Q were resistant to the activity of CheZ. However, the magnitude of CheZ resistance was greater for CheYN59R, which required >250-fold more CheZ than the wild type for a similar response (24). The clockwise behavior of cells containing CheYE89A or CheYE89Q can thus be explained by the CheZ resistance, which would result in higher CheY-P levels. A similar magnitude of CheZ resistance for CheYN23D results in exclusively clockwise flagellar rotation (21, 24).
FIG. 3.
Sensitivity of wild-type and mutant CheY proteins to dephosphorylation by CheZ. The rates of release of Pi in mixtures containing CheY (4.5 μM), phosphoimidazole (3 mM), and various amounts of CheZ were measured spectroscopically with the Enzchek Pi assay kit (Molecular Probes). Data are shown for wild-type CheY (squares), CheYE89Q (triangles), and CheYE89A (diamonds). The lines were generated by least-squares fitting, and the slopes were used as an indicator of CheZ sensitivity.
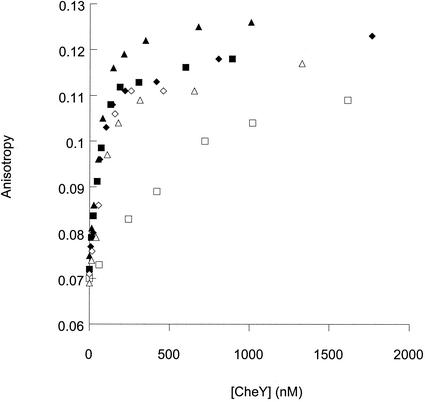
The resistance to the dephosphorylation activity of CheZ displayed by CheYE89Q and CheYE89A (Fig. 3) could be due to decreased affinity for CheZ, as with CheYN23D (21), or to resistance to the catalytic activity of CheZ, as with CheYN59R (24). Fluorescence anisotropy measurements of fluoresceinated CheZ in the presence of increasing amounts of wild-type and mutant CheY were used to assess relative binding abilities (Fig. 4). It has been shown previously that the anisotropy curves for wild-type CheY-BeF3− and CheYN59R-P are shifted to lower CheY concentrations relative to wild-type CheY-P. This is because, under the conditions of this assay, CheZ is present in sufficient concentration to significantly decrease the steady-state concentration of wild-type CheY-P but does not affect the concentrations of CheY-BeF3− or CheYN59R-P, which bind to CheZ with high affinity (Kd < 250 nM). Curves for both CheYE89Q-P and CheYE89A-P were similarly shifted to lower CheY concentrations relative to wild-type CheY and were superimposable, within experimental error, on the curves obtained for wild-type CheY-BeF3−, CheYE89A-BeF3−, and CheYE89Q-BeF3−. This behavior is consistent with the CheZ resistance of these mutants (Fig. 3) and demonstrates tight binding of these mutant proteins to CheZ.
FIG. 4.
Binding of wild-type and mutant CheY to fluoresceinated CheZ. The steady-state fluorescence anisotropy of fluoresceinated CheZ (0.13 μM) was measured in the presence of increasing concentrations of activated CheY. Activation was accomplished by the addition of acetyl phosphate (12 mM), which phosphorylates CheY, or BeF3− (0.1 mM BeCl2, 10 mM NaF). Measurements are shown for wild-type CheY (squares), CheYE89A (diamonds), and CheYE89Q (triangles). Open symbols show activation by phosphorylation, and solid symbols show activation by BeF3−.
Investigation of tentative CheY Glu89-CheZ Arg151 salt bridge.
In the (CheY-BeF3−-Mg2+)2CheZ2 cocrystal structure, the side chain of Glu89 was not visible in the electron density map and was modeled from its position in the CheY-BeF3−-Mg2+ structure (29). In this position, the Glu89 carboxylate oxygen was 3.6 Å from CheZ Arg151, suggesting the possibility of a salt bridge between these two residues. Position 151 is conserved as either an arginine or lysine in all 14 of the CheZ sequences currently available. In order to assess the importance of a potential salt bridge between CheY Glu89 and CheZ Arg151, we made the mutations cheZR151A and cheZR151K. Both of these mutations conferred wild-type chemotaxis, as judged by both the swarming and tethering assays (data not shown), consistent with wild-type-like CheZ sensitivity. This implies either that the CheZ Arg151-CheY Glu89 salt bridge does not form in the CheY/CheZ complex or that it forms but is not essential for CheZ activity. Therefore, it is not likely that the role of CheY Glu89 in CheZ-mediated dephosphorylation of CheY involves a critical interaction with CheZ Arg151.
DISCUSSION
CheY Glu89 has a role in CheZ-mediated dephosphorylation.
It has previously been shown that a residue on CheZ, Gln147, plays a direct role in the catalysis of the CheZ-mediated dephosphorylation of CheY (29). The primary finding of the current study was identification of a residue on CheY, Glu89 on the β4α4 loop, as also playing a role in dephosphorylation by CheZ. Mutation of Glu89 to either glutamine or alanine resulted in ≈40-fold resistance to the dephosphorylation activity of CheZ. The resistance to CheZ was not due to diminished binding ability, as the mutants in their activated form (CheY-P or CheY-BeF3−) bound to CheZ with an affinity similar to that of activated wild-type CheY (CheY-BeF3−).
CheYE89A and CheYE89Q behaved similarly to wild-type CheY in all other activities that were examined. The proteins displayed autophosphorylation and autodephosphorylation rates which were within a factor of 3 of those of wild-type CheY. Cells containing CheYE89A or CheYE89Q exhibited exclusively clockwise flagellar rotation, which demonstrated that the phosphorylated form of the proteins interacted effectively with the flagellar switch. Whereas the CheZ resistance of CheYN59R was due to the presence of the arginine (CheYN59A was CheZ sensitive), the behavior of the substitutions at position 89 appears to be due to the removal of Glu89, based on the similar behavior of CheYE89A and CheYE89Q. Furthermore, the strong clockwise bias of cells containing CheYE89H or CheYE89K is a strong indication that these substitutions resulted in the same resistance to CheZ. The similarity between CheYN59R and CheYE89A/E89Q suggests that the behavior of CheYN59R is a result of the salt bridge formed between Arg59 and Glu89. The salt bridge could potentially immobilize Glu89, keeping it from executing a role necessary in CheZ-mediated catalysis.
Currently, 14 bacterial species whose genomic sequences encode CheZ have been identified. The CheY proteins in all of these species contain a glutamate at the position corresponding to position 89 of E. coli CheY, whereas this CheY residue varies considerably among species which do not encode CheZ. This trend is consistent with a role for CheY Glu89 in CheZ-mediated dephosphorylation. The entire β4α4 loop of CheY (residues 87 to 91) is conserved as Thr-Ala-Glu-Ala-Lys in all but three of these CheY proteins.
Possible roles of Glu89 in CheZ-mediated dephosphorylation.
The (CheY-BeF3−-Mg2+)2CheZ2 structure shows two surfaces of interaction between CheY and CheZ (29). The interaction between the carboxy-terminal α-helix of CheZ and the α4β5α5 surface of CheY is critical for binding of CheZ and CheY-P (2, 16, 29). The other interaction surface involves a region on the four-helix-bundle domain of CheZ and the active-site region of CheY. This interface includes insertion of the CheZ Gln147 side chain into the active site of CheY. Gln147 is an essential catalytic residue and may position the attacking water molecule in the correct orientation for in-line attack of the phosphoryl group (29). The uncertainty of the precise position of Glu89 in the (CheY-BeF3-Mg2+)2CheZ2 structure (29) (see Results) limits the use of the structural data to indicate a specific role for CheY Glu89 in catalysis. We were able to eliminate a possible salt bridge between CheY Glu89 and CheZ Arg151 as being necessary for CheZ activity based on the observation that CheZ mutants with substitutions at Arg151 displayed wild-type chemotactic swarming.
The general location of Glu89 in the (CheY-BeF3−-Mg2+)2CheZ2 structure, coupled with the biochemical data presented here, is consistent with several possible roles for Glu89 in CheZ-mediated dephosphorylation. One possibility is that Glu89 also interacts with the nucleophilic water molecule, as proposed for CheZ Gln147. Modest rotation of the Glu89 side chain (23° for χ1 and 19° for χ2) from its currently modeled position in the (CheY-BeF3−-Mg2+)2CheZ2 structure (PDB code 1KMI) would put a carboxylate oxygen atom within hydrogen-bonding distance (3.0 Å) of a water molecule modeled into the appropriate position for in-line attack (data not shown). Because Glu89 is anionic, the Glu89 carboxylate could act as a general base, activating water to hydroxide, a more potent nucleophile. This possibility is supported by the observation that CheYE89Q displayed the same level of CheZ resistance as CheYE89A (Fig. 3), which suggests that the role of Glu89 is dependent on the negative charge (necessary for action as a base) and not simply hydrogen-bonding potential (necessary for positioning a water molecule). Glu89 could also interact indirectly with the attacking water molecule, such as through an intermediary second water molecule, as has been proposed for a glutamate residue in the phosphoserine phosphatase from Methanococcus jannaschii (28).
Another role for Glu89 could be participation in interactions which contribute to transition state stabilization in the CheZ-mediated dephosphorylation reaction. This possibility, however, is not supported by the observation that substitution of Glu89 did not significantly impact the rates of the autophosphorylation or autodephosphorylation reactions, whose transition states are likely similar to the transition state for dephosphorylation by CheZ (29). However, due to uncertainty about the position of Glu89, we cannot rule out such an interaction purely on structural terms, although this would require a large CheZ-dependent shift in conformation of the β4α4 loop. Finally, rather than direct participation in catalysis, the CheY Glu89 may contribute to a hydrogen-bonding network which is essential for positioning of CheZ Gln147 in the active site. Whatever the exact role of CheY Glu89 in CheZ-mediated dephosphorylation, the observation that CheZQ147A has undetectable dephosphorylation activity (29) indicates that Glu89 is not capable of carrying out its catalytic role in the absence of CheZ Gln147.
Mobility of the CheY β4α4 loop.
The magnitude of the CheZ resistance of CheYN59R (>250-fold) was significantly greater than that of CheYE89A and CheYE89Q (≈40-fold). This difference suggests that the Arg59 substitution may result in more than preventing Glu89 from carrying out a role in catalysis. The salt bridge between Arg59 and Glu89 in CheYN59R would be expected to decrease the mobility of the β4α4 loop, which could play an additional role in the CheZ reaction. The β4α4 loop has a high degree of conformational flexibility in unactivated CheY (17), and activation of CheY by BeF3− results in a repositioning of the loop, with the Glu89 Cα atom moving 4.5 Å relative to its position in unactivated CheY (12). A hydrogen bond between the Tyr106 hydroxyl and the backbone carbonyl of Glu89 stabilizes this conformation (12).
One explanation for the higher degree of CheZ resistance for CheYN59R compared to CheYE89A and CheYE89Q is that mobility of the β4α4 loop is necessary for CheZ activity. This would be the case if CheY needed to relax back to its inactive conformation before dissociation from CheZ. This notion is supported by the observation that full-length CheZ (4) as well as the peptide corresponding to the carboxy terminus of CheZ (16) bind more tightly to activated than to unactivated CheY. Mobility of the β4α4 loop may be necessary not only to bring Glu89 into position for catalysis (which neither CheY59NR nor the Glu89 mutants can do) but may also be necessary for a subsequent step in CheZ-mediated dephosphorylation (which is additionally lacking in CheY59NR).
A more general role of the mobility of the CheY β4α4 loop in the CheZ mechanism is supported by the observation that other mutations on the β4α4 loop, like those at Glu89, also lead to CheZ resistance. Xhu et al. (30) reported a 50- and 5-fold decrease in CheZ sensitivity for CheYA88T and CheYA90T, respectively, despite wild-type binding to CheZ. The CheZ resistance of mutants which contain substitutions at Ala88 and Ala90 may indicate that these residues need to be small to allow the full loop mobility required for CheZ sensitivity. The dynamics of the β4α4 loop may also explain why Glu89 does not appear to play a role in autodephosphorylation of CheY. Binding of CheY to CheZ may be required to immobilize the loop sufficiently so that Glu89 is in the proper position for catalysis.
Acknowledgments
We thank Veronica Morris for construction and characterization of CheZ Arg151 mutants.
This work was supported by grant GM50860 from the National Institute of General Medical Science to R.B.B.
Footnotes
For a commentary on this article, see page 1492 in this issue.
REFERENCES
- 1.Appleby, J. L., and R. B. Bourret. 1998. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J. Bacteriol. 180:3563-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blat, Y., and M. Eisenbach. 1996. Conserved C-terminus of the phosphatase CheZ is a binding domain for the chemotactic response regulator CheY. Biochemistry 35:5679-5683. [DOI] [PubMed] [Google Scholar]
- 3.Blat, Y., and M. Eisenbach. 1996. Oligomerization of the phosphatase CheZ upon interaction with the phosphorylated form of CheY, the signal protein of bacterial chemotaxis. J. Biol. Chem. 271:1226-1231. [DOI] [PubMed] [Google Scholar]
- 4.Blat, Y., and M. Eisenbach. 1994. Phosphorylation-dependent binding of the chemotaxis signal molecule CheY to its phosphatase, CheZ. Biochemistry 33:902-906. [DOI] [PubMed]
- 5.Bourret, R. B., J. F. Hess, and M. I. Simon. 1990. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc. Natl. Acad. Sci. USA 87:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 7.Bray, D., R. B. Bourret, and M. I. Simon. 1993. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol. Biol. Cell 41:469-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunger, A., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, Kuszewski, J., M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 10.Hess, J. F., R. B. Bourret, and M. I. Simon. 1991. Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in E. coli. Methods Enzymol. 200:188-204. [DOI] [PubMed] [Google Scholar]
- 11.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension with the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Lee, S. Y., H. S. Cho, J. G. Pelton, D. Yan, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of activated CheY. Comparison with other activated receiver domains. J. Biol. Chem. 276:16425-16431. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. Y., H. S. Cho, J. G. Pelton, D. Yan, R. K. Henderson, D. S. King, L. Huang, S. Kustu, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 14.Lukat, G. S., B. H. Lee, J. M. Mottonen, A. M. Stock, and J. B. Stock. 1991. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J. Biol. Chem. 266:8348-8354. [PubMed] [Google Scholar]
- 15.Lukat, G. S., A. M. Stock, and J. B. Stock. 1990. Divalent metal binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry 29:5436-5442. [DOI] [PubMed] [Google Scholar]
- 16.McEvoy, M. M., A. Bren, M. Eisenbach, and F. W. Dahlquist. 1999. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J. Mol. Biol. 289:1423-1433. [DOI] [PubMed] [Google Scholar]
- 17.Moy, F. J., D. F. Lowry, P. Matsumura, F. W. Dahlquist, J. E. Krywko, and P. J. Domaille. 1994. Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein with three- and four-dimensional heteronuclear (13C, 15N) NMR spectroscopy. Biochemistry 33:10731-10742. [DOI] [PubMed] [Google Scholar]
- 18.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 19.Otwinowski, Z., and W. Minor. 1996. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276A:307-326. [DOI] [PubMed] [Google Scholar]
- 20.Robinson, V. L., D. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7:626-633. [DOI] [PubMed] [Google Scholar]
- 21.Sanna, M. G., R. V. Swanson, R. B. Bourret, and M. I. Simon. 1995. Mutations on the chemotactic response regulator, CheY, that confer resistance to the phosphatase activity of CheZ. Mol. Microbiol. 15:1069-1079. [DOI] [PubMed] [Google Scholar]
- 22.Schuster, M., R. Zhao, R. B. Bourret, and E. J. Collins. 2000. Correlated switch binding and signaling in bacterial chemotaxis. J. Biol. Chem. 275:19752-19758. [DOI] [PubMed] [Google Scholar]
- 23.Silversmith, R. E., J. L. Appleby, and R. B. Bourret. 1997. The catalytic mechanism of phosphorylation and dephosphorylation of CheY: Kinetic characterization of imidazole-phosphates as phosphodonors and the role of acid catalysis. Biochemistry 36:14965-14974. [DOI] [PubMed] [Google Scholar]
- 24.Silversmith, R. E., J. G. Smith, G. P. Guanga, J. T. Les, and R. B. Bourret. 2001. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. J. Biol. Chem. 276:18478-18484. [DOI] [PubMed] [Google Scholar]
- 25.Stock, A. M., E. Martinez-Hackert, B. F. Rasmussen, A. H. West, J. B. Stock, D. Ringe, and G. A. Petsko. 1993. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry 32:13375-13380. [DOI] [PubMed] [Google Scholar]
- 26.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 27.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 28.Wang, W., H. S. Cho, R. Kim, J. Jancarik, H. Yokota, H. T. Nguyen, I. V. Grigoriev, D. E. Wemmer, and S.-H. Kim. 2002. Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic “snapshots” of intermediate states. J. Mol. Biol. 319:421-431. [DOI] [PubMed] [Google Scholar]
- 29.Zhao, R., E. J. Collins, R. B. Bourret, and R. E. Silversmith. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol. 9:570-575. [DOI] [PubMed] [Google Scholar]
- 30.Zhu, X., K. Volz, and P. Matsumura. 1997. The CheZ-binding surface of CheY overlaps the CheA- and FliM-binding surfaces. J. Biol. Chem. 272:23758-23764. [DOI] [PubMed] [Google Scholar]