Abstract
The waa gene cluster is responsible for the biosynthesis of the lipopolysaccharide (LPS) core region in Escherichia coli and Salmonella. Homologs of the waaZ gene product are encoded by the waa gene clusters of Salmonella enterica and E. coli strains with the K-12 and R2 core types. Overexpression of WaaZ in E. coli and S. enterica led to a modified LPS structure showing core truncations and (where relevant) to a reduction in the amount of O-polysaccharide side chains. Mass spectrometry and nuclear magnetic resonance spectroscopy were used to determine the predominant LPS structures in an E. coli isolate with an R1 core (waaZ is lacking from the type R1 waa gene cluster) with a copy of the waaZ gene added on a plasmid. Novel truncated LPS structures, lacking up to 3 hexoses from the outer core, resulted from WaaZ overexpression. The truncated molecules also contained a KdoIII residue not normally found in the R1 core.
Lipopolysaccharide (LPS) is the major constituent in the outer membrane of gram-negative bacteria (32). LPS is essential for the integrity of the outer membrane and for cell viability, making it a potential target for the development of novel therapeutics. In pathogens, LPS molecules are important virulence determinants; they are responsible for endotoxicity and their surface exposure makes them significant cell-surface antigens. Studies of LPS chemistry have been driven by the importance of the molecule in host interactions and by practical considerations in the development of effective vaccines. The general features of many LPS structures have been characterized. In the Enterobacteriaceae, LPS consists of three regions: (i) lipid A, the hydrophobic membrane anchor; (ii) a short core oligosaccharide (core OS); and (iii) a polymer of glycosyl (repeat) units known as O polysaccharide (O-PS).
The core OS region is conceptually divided into two regions, the inner and outer core, on the basis of sugar composition. The inner core region contains 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) and l-glycero-d-manno-heptopyranose (Hep), and the outer region is mainly composed of hexose and acetamidohexose sugars (22). The inner core contains structural elements that are conserved among gram-negative enteric bacteria, possibly due to constraints imposed by its function in outer membrane stability (32). In the Enterobacteriaceae, the basic inner core OS structure can be modified by replacement of the heptose-Kdo backbone with residues including rhamnose, galactose, glucosamine, N-acetylglucosamine, Kdo, phosphate, and phosphorylethanolamine (PEtN) depending on the core type. Modification of heptose residues with phosphate has been shown to play a fundamental role in maintaining membrane stability (46, 47), but the biological role(s) of the remaining substitutions is unknown. In contrast to the inner core, the outer core shows a relatively higher degree of diversity. Five different core types have been identified in Escherichia coli (designated K-12, R1, R2, R3, and R4) and two core types have been identified in Salmonella, based predominantly on differences in outer core structure (20, 22). Nevertheless, the overall outer core OS structure shows conserved structural themes.
The genes responsible for core OS biosynthesis in E. coli and Salmonella are located in the waa locus. Many of the genes carried by the operons in this cluster encode glycosyltransferases responsible for the sequential elongation of the core OS on a lipid A acceptor (20). Genes required for outer core biosynthesis, as well as those involved in the modification or decoration of the inner core, are carried by the long central operon defined by its initial gene, waaQ. In some cases, functional assignments of gene products have been based primarily on sequence similarity shared with enzymes of known function or by complementation studies which used mutant strains with known core defects (20). However, the roles of some waa gene products have not been predicted due to a lack of obvious or uninformative similarities in database searches. One example is the waaZ gene product found in the waa locus of the E. coli K-12 and R2 core types, as well as in Salmonella enterica serovar Typhimurium and all other serovars of S. enterica examined to date (19, 28). The waaZ gene is not found in E. coli strains with the R1, R3, and R4 core types. The fact that the waaZ sequence is highly conserved in isolates with a particular core structure suggests a shared function in core OS assembly. Significantly, a still unassigned function shared in the core biosynthesis of Salmonella and the E. coli K-12 and R2 core types is the nonstoichiometric addition of KdoIII to KdoII (Fig. 1). The objective of this study was to establish the contribution of WaaZ to the assembly of the inner core OS, and we show that the waaZ gene product is required for the addition of KdoIII.
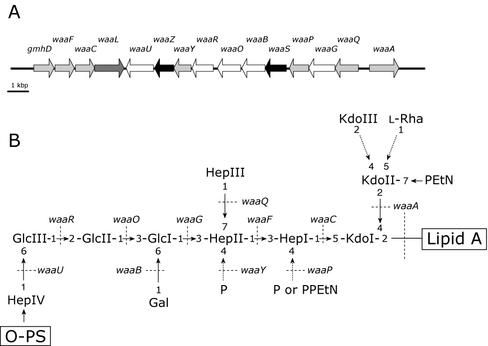
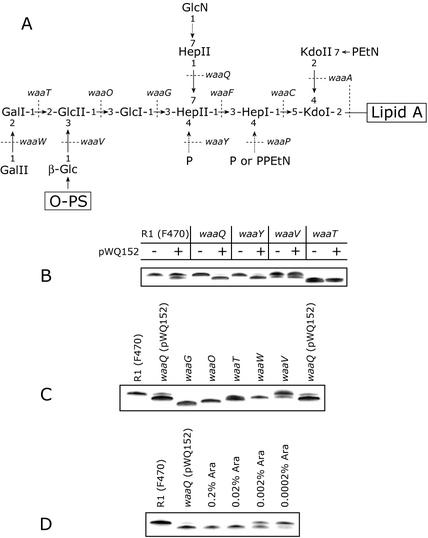
FIG. 1.
Genetic organization of the core OS biosynthetic cluster and the structure of the core OS for E. coli K-12. (A) The waa cluster from E. coli K-12. Genes involved in inner core backbone synthesis and decorations of the inner core are shown in light grey, and genes responsible for outer core synthesis are shown in white. Genes of unknown function are highlighted in black, and waaL, involved in O-PS ligation to the core OS, is in dark gray. (B) Structure of the E. coli K-12 core OS. Dashed arrows indicate nonstoichiometric substitutions. Dotted lines identify the known or predicted genetic determinants involved in the indicated linkages.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are summarized in Table 1. Bacteria were grown in Luria-Bertani (LB) broth at 37°C. Growth media were supplemented with ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), gentamicin (30 μg/ml), or kanamycin (50 μg/ml) as necessary.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype, serotype, or description | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | K-12 φ80d deoR lacZΔM15 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169 F− | 34 |
| BL21(λDE3) | F−ompT hsdSB(rB− mB−) gal dcm (λDE3) | Novagen |
| W3100 | E. coli K-12 core OS prototype R-LPS, IN(rrnD-rrnE)1 | CGSC4466a |
| F470 | E. coli R1 core OS prototype, R-LPS derivative of O8:K27 | 19, 37, 47 |
| F632 | E. coli R2 core OS prototype, R-LPS derivative of O100:K? (B):H2 | 17, 18 |
| F653 | E. coli R3 core OS prototype, R-LPS derivative of O111:K58− | 35 |
| F2513 | E. coli R4 core OS prototype, R-LPS derivative of O14:K7 | 19, 36 |
| CWG28 | trp his lac rpsL cpsK30-1 derivative of strain E69 (O9:K−) | 45 |
| CWG296 | waaP::aacC1 derivative of F470, Gmr | 47 |
| CWG297 | waaQ::aacC1 derivative of F470, Gmr | 47 |
| CWG303 | waaG::aacC1 derivative of F470, Gmr | 19 |
| CWG308 | waaO::aacC1 derivative of F470, Gmr | 19 |
| CWG309 | waaT::aacC1 derivative of F470, Gmr | 19 |
| CWG310 | waaW::aacC1 derivative of F470, Gmr | 19 |
| CWG311 | waaV::aacC1 derivative of F470, Gmr | 19 |
| CWG312 | waaY::aacC1 derivative of F470, Gmr | 47 |
| CWG345 | waaZ::aphA-3 derivative of W3100, Kmr | This study |
| S. enterica serovar Typhimurium LT2 SL3770 | waa+, S-LPS | SGSC225b |
| Plasmids | ||
| pBAD18-Ap | Arabinose-inducible expression vector, Apr | 15 |
| pBAD24 | Arabinose-inducible expression vector, Apr | 15 |
| pET28a(+) | IPTG-inducible expression vector, Kmr | Novagen |
| pYA3265 | Source of the nonpolar aphA-3 cassette conferring kanamycin resistance, Kmr | A. Honeyman via E. Vimr |
| pMAK705 | pUC19-pMAK700 hybrid suicide vector containing the temperature-sensitive pSC101 replicon, Cmr | 16 |
| pWQ152 | pBAD24-derivative carrying the waaZ coding region from W3100 as an NcoI-HindIII fragment, Apr | This study |
| pWQ153 | pET28a(+) derivative carrying the EcoRI-HindIII fragment containing the waaZ coding region from pWQ152 | This study |
| pWQ154 | pBAD18-Ap derivative containing an XbaI-HindIII fragment from pWQ153 and expressing the His6-tagged WaaZ gene product | This study |
| pWQ160 | pBAD24 derivative carrying the waaF coding region from W3100 as an NcoI-HindIII fragment, Apr | This study |
E. coli Genetic Stock Center.
Salmonella Genetic Stock Center.
DNA methods.
PCR amplification was carried out in a Perkin-Elmer GeneAmp PCR System 2400 thermocycler. Amplification reactions were done in 50-μl volumes with PwoI DNA polymerase (Roche) and conditions optimized for the primer pair. Oligonucleotide primer synthesis and automated DNA sequencing were performed at the Guelph Molecular Supercentre (University of Guelph, Guelph, Ontario, Canada). All PCR products were sequenced to verify that they were error free. The QIAquick PCR purification kit (Qiagen, Inc., Mississauga, Ontario, Canada) or the Geneclean III kit (Bio101) was used to purify PCR products and plasmid DNA fragments. Restriction endonuclease digestions and ligation reactions were performed by using standard methods, as directed by the manufacturer. Transformation was carried out by electroporation with a Gene Pulser from Bio-Rad Laboratories (Hercules, Calif.) and methods described elsewhere (6). Plasmid DNA was isolated by using QIAprep spin columns from Qiagen, and chromosomal DNA was prepared according to the method of Hull et al. (27) by using the InstaGene kit (Bio-Rad Laboratories) according to the manufacturer's directions.
Computer analysis.
Routine sequence analysis was performed with the MacVector and AssemblyLIGN software packages (International Biotechnologies, Inc., New Haven, Conn.). Homology searches of nucleotide and amino acid sequences in the National Center for Biotechnology Information databases were detected with BLAST (1) and PSI-BLAST (2). Multiple-sequence alignments and percent identity and similarity scores were calculated by using CLUSTAL_W (http://www.ebi.ac.uk/clustalw/).
In vitro mutagenesis and allelic exchange.
Individual genes were independently mutated by insertion of a nonpolar antibiotic resistance cassette into the target open reading frame (ORF). The nonpolar cassette used was the aphA-3 (kanamycin) resistance cassette. The mutated gene was transferred to the chromosome by homologous recombination, as described previously (4), by using the temperature-sensitive suicide delivery vector pMAK705 (16).
To construct the waaZ::aphA-3 mutant, the waaZ coding region was first PCR amplified from E. coli W3100 chromosomal DNA with the primers K-12 waaZ5 (5′-AGTTTCAGGAAGCTTAATTAGCATAAG-3′) and K-12 waaZ6 (5′-CACAGGTTTACCATGGAGAATATTAGA-3′). The 875-bp PCR product was digested with HindIII and NcoI at sites introduced by the primers (underlined) and ligated to the similarly digested pBAD24 vector, generating plasmid pWQ152. The waaZ gene was removed as a 931-bp BamHI-HindIII fragment and ligated to the suicide-delivery vector pMAK705 at the same sites. The HincII fragment containing the aphA-3 cassette from pYA3265 was then ligated into a HincII site within the waaZ ORF. A derivative with the aphA-3 cassette cloned in the same orientation as waaZ was selected. This pMAK705 derivative was transferred into E. coli W3100 by electroporation. Allelic exchange was performed as described previously (4, 16), and mutants were selected by kanamycin resistance and chloramphenicol sensitivity. The presence of the mutated gene on the chromosome was confirmed by PCR and sequencing of the insertion junctions in the amplified PCR product. One representative mutant E. coli CWG345 was selected for further analysis.
Construction of the waaF expression vector.
The waaF gene was amplified from E. coli W3100 chromosomal DNA with the primers K-12 waaF1 (5′-CGACGCATAAGAATTCTGCATG-3′) and K-12 waaF2 (5′-TCCACCACCCAGTCAAGCTTAATC-3′). The 1,182-bp PCR product was digested with the EcoRI and HindIII sites (underlined) and ligated into the same sites in pBAD24 to make plasmid pWQ160.
Complementation studies and overexpression of WaaZ in various E. coli and S. enterica strains.
The pBAD derivatives were used for expression of WaaZ and WaaF in different core backgrounds. The plasmid pBAD24 belongs to a family of expression vectors that uses the arabinose-inducible and glucose-repressing arabinose promoter (15). Repression from the araC promoter was achieved by growth in 0.4% (wt/vol) glucose and induction was obtained by adding l-arabinose to a final concentration of 0.02% (wt/vol) (unless otherwise indicated). Briefly, a culture was grown at 37°C in LB supplemented with ampicillin and 0.4% glucose for 18 h. This culture was diluted 1:100 in fresh medium and grown until the culture reached an optical density at 600 nm of 0.2. This culture was then induced and grown for another 2 h. Repressed controls were continuously grown in glucose-containing media.
Protein expression and analysis.
The waaZ gene was isolated from pWQ152 as an 859-bp EcoRI-HindIII fragment and cloned into pET28a(+), generating plasmid pWQ153. This plasmid expresses WaaZ with an in-frame His6 tag fused to the N terminus (plasmid pWQ153). The protein was overexpressed in E. coli BL21(λDE3)(pWQ153). This strain was grown at 37°C in LB supplemented with kanamycin for 18 h and was then diluted 1:50 in fresh medium and grown until the culture reached an optical density at 600 nm of 0.6. Expression of His6-WaaZ was induced by adding isopropyl-1-thio-β-d-galactopyranoside (IPTG) to the culture at a final concentration of 1 mM and continuing incubation for another 1.5 h. The cells were harvested, washed once in phosphate-buffered saline, resuspended in a lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole), and sonicated. Unbroken cells and large cellular debris were removed from the lysate by low-speed centrifugation (10 min at 4,000 × g). Cell membranes were then separated from the soluble fraction by ultracentrifugation (90 min at 100,000 × g) of the cell-free lysate. The soluble and membrane fractions were solubilized in sample buffer by boiling at 100°C for 15 min and then separated on 12% polyacrylamide gels by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Proteins were visualized by Coomassie brilliant blue staining or by Western immunoblotting. Western immunoblotting was carried out with Qiagen anti-pentahistamine mouse monoclonal antibodies according to the manufacturer's instructions.
To verify whether the His6-tagged WaaZ was still functional, the His6-tagged waaZ coding region was removed from pWQ153 as an XbaI-HindIII fragment and ligated to a similarly cut pBAD18-Ap expression vector to give pWQ154. The plasmid pWQ154 was then transformed into CWG345 (waaZ::aphA-3), and the LPS of the transformant was analyzed by silver-stained Tricine-SDS-PAGE.
SDS-PAGE analysis of LPSs.
For SDS-PAGE, LPS was isolated on a small scale from proteinase K-digested whole-cell lysates as described by Hitchcock and Brown (21). The LPS was then separated on a 10 to 20% gradient Tricine SDS-PAGE gel from Novex (San Diego, Calif.) and visualized by silver staining (41).
Core OS isolation.
E. coli strain F470(pWQ152) was grown in a fermentor (2 × 10 liters) in LB for 21 h, harvested, and then lyophilized (13.53 g, dry mass). The LPS was isolated from the dried cells by the phenol-chloroform-light petroleum method (13), giving 585.2 mg of LPS, a yield of 4.3% of the bacterial dry mass. The LPS (326 mg) was de-O-acylated with hydrazine at 37°C (yield, 163 mg, 50% of the LPS) and then de-N-acylated by KOH treatment (yield, 67.5 mg, 41.5% of the LPS) (23). The oligosaccharide sample was desalted by using gel permeation chromatography on a Sephadex G10 column (3 by 60 cm) eluted in pyridine:acetic acid:water (4:10:1,000, by volume). The eluant was monitored with a differential refractometer (Knauer). Individual oligosaccharides were then separated by preparative high-performance anion-exchange chromatography (HPAEC) with a CarboPac PA1 column (9 mm by 25 cm; DIONEX Corp.) and eluted by a linear gradient of 0 to 100% 1 M sodium acetate in water (pH 6.0) over 70 min (23). Four major phosphorylated oligosaccharides were isolated and desalted (twice, as described above). The 1H nuclear magnetic resonance (NMR) spectra of the four fractions showed that two oligosaccharides (oligosaccharides 2 and 4) (1.2 and 1.4 mg and 0.43 and 0.37% of the LPS, respectively) possessed three Kdo residues. All four oligosaccharides were used for mass spectrometry (MS) analysis, but only oligosaccharides 2 and 4 were used for further structural investigation by NMR.
MS.
Matrix-assisted laser desorption-ionization (MALDI-MS) analyses of de-O-acylated LPS were performed with a Bruker-ReflexII spectrometer (Bruker-Franzen Analytik, Bremen, Germany) in linear and/or reflection time of flight (TOF) configuration and delayed ion extraction at an acceleration voltage of 20 kV. Routinely, the compounds (<10 nmol) were dissolved in 15 μl of distilled water and treated with small amounts of cation exchanger [Amberlite IR-120 (H+); Merck, Darmstadt, Germany] to remove interfering cations. An aliquot (1 μl) of the sample was then mixed with 1 μl of 0.5 M matrix solution of 2,5-dihydroxybenzoic acid (gentisic acid, DHB; Aldrich, Steinheim, Germany) in methanol, and aliquots of 0.5 μl were deposited on a metallic sample holder and dried in a stream of air. The samples were analyzed in the negative-ion mode. The mass spectra shown are the average of at least 50 single analyses. Mass-scale calibration was performed externally with similar LPS-derived oligosaccharides of known chemical structure. Details of the applied methods are given elsewhere (30).
Electrospray ionization (ESI) MS was performed by using a Fourier transform ion cyclotron (FT-ICR) mass analyzer (ApexII; Bruker Daltonics, Billerica, Mass.) equipped with a 7-T actively shielded magnet and an Apollo electrospray ion source. Samples were dissolved in a mixture of 2-propanol, water, and triethylamine (30:30:0.01, by volume) at a concentration of ∼20 ng/μl and sprayed with a flow rate of 2 μl/min. Capillary skimmer dissociation (CSD) was induced by increasing the capillary exit voltage from −100 to −350 V.
NMR spectroscopy.
For structural assignments of oligosaccharides 2 and 4, one-dimensional and two-dimensional 1H NMR spectra were recorded for solutions of 1.2 and 1.4 mg, respectively, in 0.5 ml of 2H2O with a Bruker DRX 600 spectrometer (operating frequencies: 1H, 600.13 MHz; 13C, 150.90 MHz) at 27°C. Measurements were made relative to internal acetone (1H, 2.225 ppm; 13C, 34.5 ppm). 31P NMR spectra of the samples were recorded at an operating frequency of 242.97 MHz at 27°C with 85% phosphoric acid as an external standard. The correlation (COSY) and total correlation spectroscopy (TOCSY), the 1H, 13C, and 1H,31P heteronuclear multiple-quantum coherence (HMQC), and the rotating frame nuclear Overhauser effect spectroscopy (ROESY) spectra were all measured with standard Bruker software. In the latter, a mixing time of 300 ms was employed.
RESULTS
Characterization of the waaZ gene product.
The waaZ gene is located in the waa cluster of E. coli isolates with core types K-12 and R2 and in the waa clusters of S. enterica (20, 28). The WaaZ gene products of E. coli K-12 (accession number AAC76648.1) and E. coli R2 (accession number AAC69650.1) share 86.2% amino acid identity and 91.4% total similarity. The E. coli K-12 protein shares 68.4 and 66.4% identity and 82.2 and 80.6% similarity with homologs from S. enterica serovar Typhimurium (accession number AAL22574.1) and S. enterica serovar Typhi (accession number AL627280), respectively. The E. coli K-12 protein also shares weak similarity to putative glycosyltransferases from Klebsiella pneumoniae (ORF 4) (accession number AAD37764.1) and Sinorhizobium meliloti (accession number AL603644.1) (20 and 21% identity and 44 and 39% similarity, respectively). The K. pneumoniae ORF 4 protein is encoded within the waa gene cluster (33) and is involved in the addition of an outer core Kdo residue (E. Frirdich, E. Vinogradov, and C. Whitfield, unpublished data). The S. meliloti ORF is carried on the pSymB megaplasmid and belongs to a cluster of proteins that are thought to be involved in LPS core or lipid A assembly (12). However, neither of these proteins have definitively assigned glycosyltransferase functions. Sequences corresponding to waaZ are missing from the waa gene clusters of E. coli prototype strains with R1, R3, and R4 core OS types. To test the possibility that these strains possessed a waaZ gene at an unlinked locus, chromosomal DNA from the prototype strain of each core type was digested with restriction enzymes and examined by Southern hybridization with a probe derived from the E. coli K-12 waaZ gene. No hybridization was detected at either high or low stringency (data not shown).
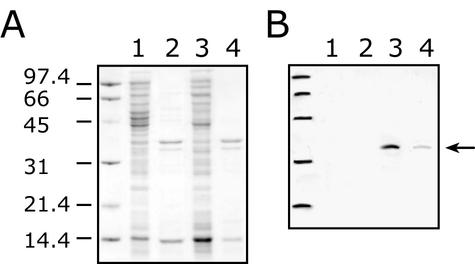
The DNA sequence predicts that waaZ encodes a protein with a molecular mass of 32,917 Da. The waaZ ORF is preceded by a potential ribosome binding site (AGGT) located 5 bp upstream of an ATG codon. The WaaZ protein has no signal sequence and is predicted to be a soluble protein with a pI of 9.41. To examine the cellular location of the waaZ gene product, the gene was cloned into pBAD24 as plasmid pWQ152. Whole-cell lysates of strains containing pWQ152 were examined by SDS-PAGE, but no polypeptide corresponding to WaaZ was evident after Coomassie blue staining (data not shown). Therefore, in order to trace WaaZ expression, the waaZ ORF was cloned into pET28a(+) to generate pWQ153, which encodes a WaaZ derivative with a hexahistidine tag fused to the N terminus (His6-WaaZ). Although His6-WaaZ was also not identified in a Coomassie-stained SDS-PAGE gel, it was detected with a corresponding Western blot probed with monoclonal antibodies specific for the His6 tag (Fig. 2). From the blot, His6-WaaZ shows an apparent molecular mass consistent with its molecular mass predicted from sequence data of 35,081 Da. A few minor and faster migrating bands appear in some preparations and likely represent degradation products derived from His6-WaaZ. There was no apparent requirement for degradation products in the phenomena that follow. From a cell-free lysate, the soluble and membrane fractions were separated. Based on the profiles on Western blots, the majority of His6-WaaZ is present in the soluble fraction, but a significant amount is also located in the membrane fraction.
FIG. 2.
Overexpression of His6-tagged WaaZ and cellular location of the protein. (A) Coomassie-stained SDS-PAGE. (B) Corresponding Western immunoblot probed with anti-His5 monoclonal antibody. The analysis was done with BL21(λDE3) containing pWQ153 with and without 1 mM IPTG induction. Cell lysates were prepared by sonication, and the membrane fraction was separated from the soluble fraction by ultracentrifugation. Lanes 1 and 2 represent the soluble and membrane fractions of the uninduced controls, respectively. Lanes 3 and 4 represent the soluble and membrane fractions of the IPTG-induced samples, respectively. The membrane and soluble fractions were prepared such that they were directly comparable in terms of the number of original cells providing the lysate. For loading on SDS-PAGE, the ratio of the protein content in the soluble fraction to that in the membrane fraction was 1:2.5. The numbers on the left represent molecular mass in kilodaltons. The arrow indicates the position of His6-WaaZ.
Analysis of the phenotype resulting from a chromosomal waaZ mutation.
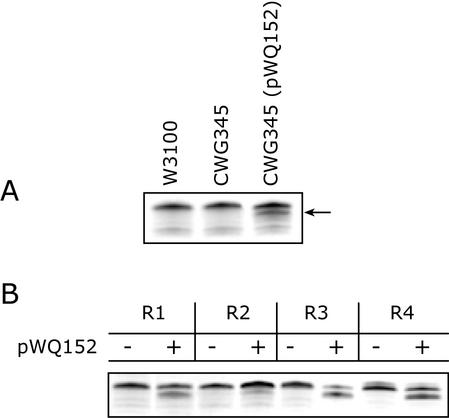
In an attempt to establish the function of WaaZ in core OS biosynthesis, a chromosomal waaZ mutation was constructed in E. coli K-12 strain W3100. Strain W3100 was chosen because the structure of its core region had been fully characterized (25, 26). The waaZ gene is located in the central waaQ operon (Fig. 1) and was inactivated by insertion of the nonpolar aphA-3 cassette in order to maintain expression of downstream genes. The LPS of the waaZ mutant (CWG345) was examined by electrophoresis on Tricine-PAGE and showed a migration indistinguishable from that of the wild-type strain W3100 (Fig. 3A).
FIG. 3.
Tricine-SDS-PAGE gel showing the migration of LPS from E. coli CWG345 (waaZ::aphA-3) and the effects of WaaZ overexpression on LPS mobility in strains representing different core types. LPS samples were separated on a 10 to 20% Tricine gel by SDS-PAGE and visualized by silver staining. All strains produce R-LPS, therefore only the region of the gel containing the lipid A core species is shown. (A) LPS mobility of CWG345 (waaZ) with (+) and without (−) plasmid-borne waaZ (pWQ152) compared to the wild-type strain W3100. The arrow indicates a faster-migrating species resulting from overexpression of WaaZ. (B) The effect on LPS migration of adding multicopy WaaZ to the representative prototype strains for the E. coli core types. Expression from pWQ152 was induced by the addition of 0.02% l-arabinose. − and +, uninduced and induced cultures, respectively.
Influence of waaZ overexpression on LPS profiles in Tricine-PAGE.
There are several potential interpretations for the phenotype of CWG345: (i) WaaZ may play no role in LPS biosynthesis or (ii) WaaZ mediates the addition of a nonstoichiometric substitution in amounts too small to generate a unique and detectable band in Tricine-PAGE gels. Reasoning that multicopy WaaZ might generate a higher amount of modified LPS leading to an alteration detectable on Tricine-PAGE gels, plasmid pWQ152 was introduced into the waaZ mutant (CWG345) and expression was induced with l-arabinose. Surprisingly, the LPS of the complemented strain displayed two lipid A core species. One comigrated with the major band found in the parental strain, and the other was of faster mobility (Fig. 3A). An identical phenotype was obtained when the parent strain W3100 was transformed with pWQ153 expressing His6-WaaZ, indicating that the N-terminal His6-tag did not influence function (data not shown).
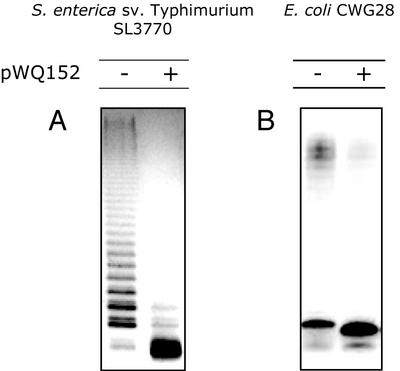
To determine whether the influence of WaaZ overexpression on LPS species was confined to E. coli K-12, plasmid pWQ152 was introduced into the prototype strains for E. coli core types R1 through R4, and S. enterica serovar Typhimurium SL3770. Shifts in LPS mobility indicative of novel lower-molecular-mass lipid A core species were seen in all of the E. coli core type representatives, although the alteration in the R2 core type representative was not as evident as for the others (Fig. 3B). S. enterica serovar Typhimurium SL3770 and E. coli CWG28 (O9:K−, R1 core OS) produce smooth (O-PS substituted) LPS. After transformation with pWQ152, the amount of O-PS ligated to the cell surface decreased significantly upon induction of waaZ expression (Fig. 4). The reduction in core OS size was therefore concluded to reflect a core truncation affecting the O-PS ligation site, rather than only representing a reduction or elimination of inner core OS modifications in molecules with a full-length core backbone. Some O-PS was still ligated to the core OS in the smooth strains overexpressing WaaZ, indicating that there was still some full-length core OS species present, but truncated molecules clearly predominate.
FIG. 4.
Silver-stained SDS-PAGE analysis showing the decrease in O-PS ligation in strains overexpressing WaaZ. S. enterica serovar Typhimurium SL3770 and E. coli CWG28 (O9:K−, R1 core) produce S-LPS. (A) S. enterica. (B) E. coli CWG28 Expression from pWQ152 was induced by the addition of 0.02% l-arabinose. − and +, uninduced and induced cultures, respectively.
Effect of waaZ overexpression in R1-type core OS mutants.
WaaZ was expressed in a series of defined core mutants available in the R1 core OS background. This provided the opportunity to determine the point in core elongation at which truncations occur. Changes in LPS migration were seen only in derivatives containing a full-length R1 core backbone, F470, CWG297 (waaQ), and CWG312 (waaY) (Fig. 5A and B). The core mutants CWG297 (waaQ) and CWG312 (waaY) have altered modifications in the inner core. Both lack both a phosphate residue on HepII, but CWG297 (waaQ) is also missing HepIII (47) (Fig. 5A). By comparison to core mutant standards, the truncation caused by overexpressed WaaZ in F470, CWG297, and CWG311 appears to be equivalent to approximately three sugars, i.e., similar to the waaT mutant (Fig. 5C). The waaT mutant lacks the terminal two hexoses in the core as well as the β-linked glucose side branch (19).
FIG. 5.
The structure of the R1 core OS and the effect of waaZ overexpression in R1 core OS mutants on LPS migration in SDS-PAGE. (A) Structure of the E. coli R1 core OS. Dashed arrows indicate nonstoichiometric substitutions. Dotted lines identify the genetic determinants involved in the indicated linkages (19). In panels A, B, and C, LPS samples were separated on a 10 to 20% Tricine gel by SDS-PAGE and visualized by silver staining. All strains produce R-LPS, therefore only the region of the gels containing the lipid A core is shown. Expression from pWQ152 in panels B and C was induced by the addition of 0.02% l-arabinose. Panel B shows the effect of addition of multicopy WaaZ on LPS profiles in a series of R1 core OS mutants. − and +, uninduced and induced cultures, respectively. (C) Comparison of waaQ (pWQ152) LPS to core OS LPS mutant standards. The truncated LPS resulting from WaaZ overexpression comigrates with waaT LPS, a form lacking 3 sugars (note that the order synthesis also results in loss of the WaaV-added β-glucosyl residue in the waaT mutant) (19). (D) Induction of waaZ expression in waaQ (pWQ152) with various concentrations of arabinose.
The effects of WaaZ can be titrated by inducing CWG297(pWQ152) with various concentrations of arabinose, with an increase in truncated core molecules resulting from higher amounts of arabinose inducer. This confirmed that the effect is due specifically to WaaZ levels (Fig. 5D). To further confirm that the truncations caused by WaaZ were WaaZ-specific, the plasmid pWQ160 (overexpressing waaF) was introduced in multicopy into the E. coli K-12 strain W3100 and into the R1 strain F470. The waaF gene encodes the heptosyltransferase II (7, 14). This produced no change in LPS mobility (data not shown). Furthermore, in previous work with a variety of different outer core OS glycosyltransferase genes, no similar truncation was observed (data not shown).
Structural analysis of LPS core modification in F470 (R1 core OS) overexpressing waaZ. (i) MALDI-LIN-TOF MS of de-O-acylated LPS from F470 and F470(pWQ152).
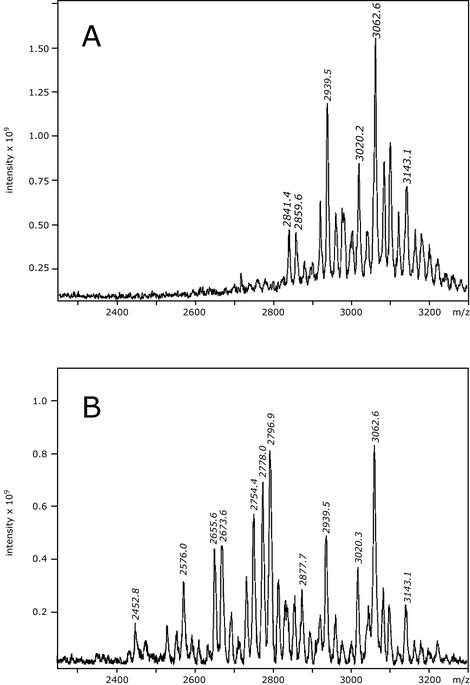
To determine the changes in LPS structure resulting from WaaZ overexpression, LPS was extracted from strains F470 and F470(pWQ152) and MS and NMR methods were used to determine the carbohydrate backbone structure. Despite the apparent simplicity of Tricine-PAGE profiles of rough LPSs, it is well established that isolated LPSs contain a heterogeneous mixture of molecules. MALDI-TOF MS was used to identify the spectrum of core OS species found in de-O-acylated LPS from F470 and F470(pWQ152). This approach is important, because nonstoichiometric substitutions can result in unique species representing a small percentage of isolated core OS molecules that can easily be missed in NMR studies. The de-O-acylated LPSs were analyzed by using MALDI-TOF in the negative-ion mode. The spectra are shown in Fig. 6, and the corresponding deduced carbohydrate backbone structures for the major peaks are summarized in Table 2.
FIG. 6.
Negative-ion mode MALDI-TOF (linear configuration) mass spectra of de-O-acylated LPS. (A) E. coli F470. (B) E. coli F470(pWQ152).
TABLE 2.
Structural assignments for the major molecular ions from negative-ion MALDI-LIN-TOF mass spectra of de-O-acylated LPS from E. coli F470 and F470(pWQ152)b
| Observed mass (m/z) | Structural assignmenta | Theoretical mass (m/z) | Present in:
|
|
|---|---|---|---|---|
| F470 | F470 (pWQ152) | |||
| 3,143.1 | (lipidA-Hy37)-Kdo2-Hep3-Hex5-HexN-P-PEtN | 3,143.8 | + | + |
| 3,062.6 | (lipidA-Hy37)-Kdo2-Hep3-Hex5-P2-PEtN | 3,062.6 | + | + |
| 3,020.3 | (lipidA-Hy37)-Kdo2-Hep3-Hex5-P3 | 3,019.5 | + | + |
| (lipidA-Hy37)-Kdo2-Hep3-Hex5-HexN-P | 3,020.7 | |||
| 2,939.5 | (lipidA-Hy37)-Kdo2-Hep3-Hex5-P2 | 2,939.5 | + | + |
| 2,859.6 | (lipidA-Hy37)-Kdo2-Hep3-Hex5-P | 2,859.6 | + | + |
| 2,796.9 | (lipidA-Hy37)-Kdo3-Hep3-Hex2-P2-PEtN | 2,796.5 | − | + |
| (lipidA-Hy37)-Kdo3-Hep3-Hex3-PEtN | 2,798.7 | |||
| 2,778.0 | (lipidA-Hy37)-Kdo2-Hep3-Hex4-P2 | 2,777.5 | − | + |
| 2,754.4 | (lipidA-Hy37)-Kdo3-Hep3-Hex2-P3 | 2,753.4 | − | + |
| (lipidA-Hy37)-Kdo3-Hep3-Hex3-P1 | 2,755.6 | |||
| 2,673.6 | (lipidA-Hy37)-Kdo3-Hep3-Hex2-P2-PEtN2 | 2,673.4 | − | + |
| 2,655.6 | (lipidA-Hy37)-Kdo2-Hep3-Hex2-P3-PEtN | 2,656.3 | − | + |
| 2,576.0 | (lipidA-Hy37)-Kdo2-Hep3-Hex2-P2-PEtN | 2,576.3 | − | + |
| 2,452.8 | (lipidA-Hy37)-Kdo2-Hep3-Hex2-P2 | 2,453.2 | − | + |
lipidA-Hy37 refers to the de-O-acylated lipid A backbone with the structure (GlcN-C14:0-OH)2.
Data are shown in Fig. 6.
The mass spectrum of F470 (Fig. 6A) contains a series of intense [M − H]− pseudomolecular ions in the region with an m/z of 2,800 to 3,200. The most intense ion at an m/z of 3,062.6 corresponds to a species with the composition (lipid A-Hy37)-Kdo2-Hep3-Hex5-P2-PEtN, where lipidA-Hy37 refers to the de-O-acylated lipid A backbone. Previous compositional analysis of F470 LPS indicated that the majority of molecules contained a complete core OS (44), and the molecular ion at an m/z of 3,062.6 reflects this structure. The R1 LPS contains a minor component in which a GlcN residue is added to HepIII (Fig. 6A) (44), and this nonstoichiometric substitution is missing in the ion at an m/z of 3,062.6. The species represented by the ion at an m/z of 3,143.1 corresponds to the R1 core bearing the GlcN substitution but lacking a phosphate residue, in comparison to the peak at an m/z of 3,062.6. All remaining ions are variations of these two species, and as can be seen in Table 2, they differ only in the number of phosphate and PEtN groups.
The spectrum of F470(pWQ152) (Fig. 6B) contains additional novel ions in the m/z range of 2,400 to 3,200, consistent with the truncations resulting from WaaZ overexpression and visualized in Tricine-PAGE gels. The relatively small amount of full-length core is predicted by the limited ligation of O-PS to complete core OS seen in S. enterica serovar Typhimurium and E. coli CWG28 (Fig. 4). This spectrum shows ions in the region of an m/z of 2,800 to 3,200 at an m/z of 2,939.5, 3,020.3, 3,062.6, and 3,143.1, corresponding to the full-length R1 core species with or without the GlcN substitution on HepIII (see Table 2 for structures). Ions in the region of an m/z of 2,400 to 2,800 correspond to truncated core molecules. There are four major core species in this region of the F470(pWQ152) spectrum. The smallest molecular ions at an m/z of 2,452.8, 2,576.0, and 2,655.6 correspond to a species with the carbohydrate backbone (lipid A-Hy37)-Kdo2-Hep3-Hex2, lacking three of the outer core OS glycoses. The three peaks differ in the number and type of phosphoryl substitutions. Peaks at an m/z of 2,673.6, 2,754.4, and 2,796.9 all represent species containing three Kdo residues with either two or three hexose residues. The presence of three Kdo residues reflects an activity unique to WaaZ, since there is no evidence for KdoIII in the MS profile of the wild-type F470 LPS or in previous NMR determinations of the complete R1 core OS structure (44). The ion at an m/z of 2,778.0 represents the only peak with the composition (lipidA-Hy37)-Kdo2-Hep3-Hex4-P2, and it lacks only one hexose sugar in comparison to the five hexoses of the wild type. Interestingly, no significant molecular ions were identified in F470(pWQ152) containing only one hexose sugar.
(ii) ESI FT-ICR MS of deacylated LPS and HPAEC isolated fractions from F470(pWQ152).
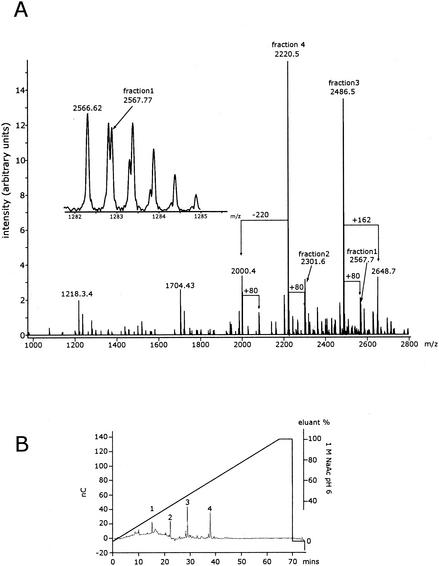
ESI FT-ICR MS was used to further investigate the LPS of F470(pWQ152), since it showed appreciable differences to that of the wild-type F470. The negative-ion ESI FT-ICR broad-band mass spectrum of the deacylated LPS of F470(pWQ152) yielded a complex pattern of mainly doubly and triply charged molecular ion peaks. To better visualize the biological heterogeneity in the sample, the charge-deconvoluted spectrum is given in Fig. 7A. Four major phosphorylated oligosaccharides were separated from deacylated F470(pWQ152) by HPAEC (Fig. 7B). Peaks derived from these oligosaccharides appear in the ESI FT-ICR spectrum and are labeled as fractions 1 to 4 (Fig. 7A). The peaks of minor intensity could not be isolated by HPAEC, as they were present in small amounts. However, they could easily be identified as species differing from the isolated fractions because they possessed an additional phosphate group or hexose or were missing a Kdo residue, resulting in mass differences with a Δm/z of +80, +162, and −220, respectively. Additional unlabeled minor peaks in the ESI FT-ICR spectrum originate from sodium and potassium cationization. The insert in Fig. 7A shows the expanded molecular ion region of fraction 1, indicating the presence of two overlapping molecular species. The isotopic peak at an m/z of 2,566.62 can be assigned to a molecule identical to that of fraction 3 but with an additional phosphate group, whereas the isotopic peak at an m/z of 2,567.77 consists of a separate species that was isolated by HPAEC as fraction 1.
FIG. 7.
Charge-deconvoluted negative-ion ESI FT-ICR mass spectrum of deacylated LPS from E. coli F470(pWQ152) and the major fractions separated by HPAEC. Panel A shows the mass spectrum of the E. coli F470(pWQ152) LPS and the contributions of the 4 HPAEC fractions from panel B. The insert shows the expanded molecular ion region of fraction 1. Panel B shows the HPAEC of the deacylated E. coli F470(pWQ152) LPS. The four oligosaccharide fractions that were isolated by preparative HPAEC are indicated. Peaks 2 and 4 contained 3 Kdo residues and were therefore used for subsequent detailed structural analysis by NMR.
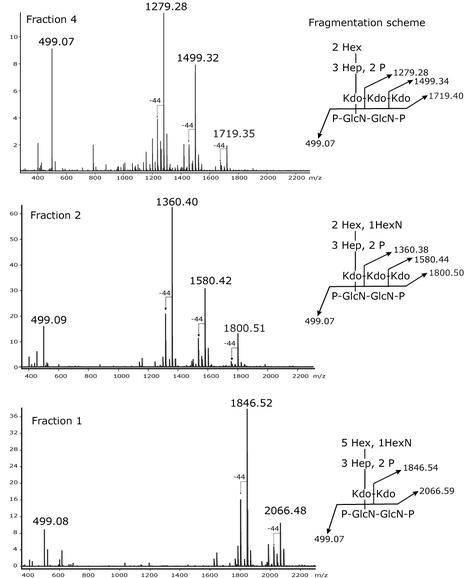
The measured molecular masses of the mono-isotopic peaks from the HPAEC-purified fractions are in excellent agreement with the calculated molecular masses for molecules consisting of the residues listed in Table 3. According to these assignments, fractions 2 and 4 should contain three Kdo residues. Additional confirmation for the presence of the three Kdo residues, as well as information regarding the arrangement and branching points of the Kdos, is provided by negative-ion ESI spectra after CSD (Fig. 8). The spectra provide characteristic fragment ions of diagnostic importance, as depicted in their respective fragmentation schemes. It is known from the analysis of core OS molecules from other bacterial species that under the applied conditions, fragmentation between Kdo-GlcN is generated, which is accompanied by decarboxylation (Δm/z of −44) and the removal of the side chain Kdo (29). This can be clearly observed in the fragmentation pattern for fractions 2 and 4. However, in fraction 1, cleavage of only one Kdo residue (Δm/z of 220) can be observed, as predicted for a core OS structure containing only two Kdo residues. The fragment ion at an m/z of 499.08 is present in all spectra, indicating that the lipid A backbone, including the number of phosphate groups, is identical in all fractions.
TABLE 3.
Chemical composition of each of the four fractions of deacylated F470(pWQ152) LPS separated by HPAECa
| Fraction | Observed mass (m/z) | Chemical composition | Theoretical mass (m/z) |
|---|---|---|---|
| 1 | 2,567.68 | 3 HexN, 2 Kdo, 3 Hep, 5 Hex, 3 P | 2,567.69 |
| 2 | 2,301.56 | 3 HexN, 3 Kdo, 3 Hep, 2 Hex, 3 P | 2,301.58 |
| 3 | 2,486.57 | 2 HexN, 2 Kdo, 3 Hep, 5 Hex, 4 P | 2,486.58 |
| 4 | 2,220.47 | 2 HexN, 3 Kdo, 3 Hep, 2 Hex, 4 P | 2,220.48 |
The molecular mass was determined by charge-deconvoluted negative-ion ESI FT-ICR MS (spectra not shown).
FIG. 8.
Negative-ion CSD ESI FT-ICR mass spectra of HPAEC fractions 1, 2, and 4 of deacylated LPS from E. coli F470(pWQ152). Inserts show the fragmentation schemes for each fraction along with the calculated ion masses.
NMR spectroscopy of oligosaccharides 2 and 4.
The precise structures of oligosaccharides 2 and 4 were established by 1H, 13C, and 31P NMR spectroscopy. Chemical shifts were assigned by using COSY, TOCSY, ROESY, and HMQC experiments and by comparison with published data (24, 31, 44). The NMR data are presented in Tables 4 and 5 and, together with the MS data, yielded the core OS structure given in Fig. 9. The anomeric region of the 1H NMR spectrum of oligosaccharide 4 contained 7 signals, representing three heptose, two hexose, and two hexosamine residues. Their identification was possible by the assignment of most signals. One hexosamine residue (residue B) (Fig. 9) possessed the β-gluco configuration, which was supported by a ROESY experiment that yielded an intraresidual nuclear Overhauser enhancement spectroscopy (NOE) connectivity from H-1 to H-3. The other hexosamine (residue A) and the hexoses (residues I and K) possessed the α-gluco configuration, and the heptoses (residues F, G, and H) possessed the α-manno configuration, which, in the latter case, was also established by the small J1,2 coupling constant of ∼1 Hz. The characteristic signals of H-3 of three Kdo residues were present at 1.984 ppm (H-3ax) and 2.196 ppm (H-3eq) (residue C), 1.820 ppm (H-3ax) and 2.187 ppm (H-3eq) (residue D), and 1.820 ppm (H-3ax) and 2.295 ppm (H-3eq) (residue E). Their α configurations were established on the basis of the chemical shift of their 3eq protons. All of these sugars were pyranoses. Thus, oligosaccharide 4 represents a decasaccharide.
TABLE 4.
1H NMR chemical shifts of sugar residues of oligosaccharides 2 and 4 of LPS from E. coli F470(pWQ152)d
| Residue and oligosaccharide |
1H NMR chemical shift (ppm) of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H-1 | H-2 | H-3ax | H-3eq | H-4 | H-5 | H-6(a) | H-6b | H-7(a) | H-7b | H-8a | H-8b | |
| A | ||||||||||||
| 2 | 5.728 | 3.471 | 3.953 | 3.670 | 4.170 | 4.324 | 3.836 | |||||
| 4 | 5.754 | 3.499 | 3.960 | 3.680 | 4.156 | 4.344 | 3.822 | |||||
| B | ||||||||||||
| 2 | 4.887 | 3.135 | 3.906 | 3.863 | 3.798 | NDa | ND | |||||
| 4 | 4.881 | 3.162 | 3.925 | 3.868 | 3.712 | 3.499 | 3.715 | |||||
| C | ||||||||||||
| 2 | 1.964 | 2.168 | 4.163 | 4.300 | 3.670 | 3.896 | ND | ND | ||||
| 4 | 1.984 | 2.196 | 4.166 | 4.304 | ND | 3.892 | ND | ND | ||||
| D | ||||||||||||
| 2 | 1.828 | 2.189 | 4.169 | 4.086 | 3.668 | ND | 3.957 | 3.828 | ||||
| 4 | 1.820 | 2.187 | 4.153 | 4.099 | 3.677 | 3.950 | 3.964 | 3.830 | ||||
| E | ||||||||||||
| 2 | 1.828 | 2.266 | 4.138 | 4.057 | 3.675 | ND | ND | 3.752 | ||||
| 4 | 1.820 | 2.295 | 4.140 | 4.054 | ND | ND | ND | ND | ||||
| F | ||||||||||||
| 2 | 5.335 | 4.113 | 4.205 | 4.492 | 4.274 | 4.196 | ND | ND | ||||
| 4 | 5.333 | 4.113 | 4.179 | 4.472 | 4.284 | 4.149 | 3.897 | 3.897 | ||||
| G | ||||||||||||
| 2 | 5.212 | 4.428 | 4.055 | 4.055 | 3.745 | 4.150 | 3.798b | 3.705b | ||||
| 4 | 5.135 | 4.438 | 4.163 | 4.472 | 3.901 | 4.296 | 3.745 | 3.745 | ||||
| H | ||||||||||||
| 2 | 4.957 | 4.030 | 3.899 | 3.825 | 3.676 | 4.148 | 3.751 | 3.751 | ||||
| 4 | 4.984 | 4.017 | 3.912 | 3.904 | 3.678 | 4.052 | 3.793 | 3.793 | ||||
| I | ||||||||||||
| 2 | 5.275 | 3.693 | 3.984 | 3.735 | 3.907 | ND | ND | |||||
| 4 | 5.228 | 3.702 | 4.022 | 3.754 | 3.914 | 3.938c | 3.761c | |||||
| K | ||||||||||||
| 2 | 5.419 | 3.735 | 3.822 | 3.484 | 4.038 | 3.851 | 3.812 | |||||
| 4 | 5.483 | 3.730 | 3.817 | 3.464 | 4.085 | 3.938c | 3.859c | |||||
| L | ||||||||||||
| 2 | 5.242 | 3.384 | 3.965 | 3.542 | 3.803 | ND | ND | |||||
ND, not detected.
Determined in the ROESY spectrum.
Signals may be interchanged.
Spectra were recorded from a solution in 2H2O at 600.13 MHz at 27 °C. Chemical shifts are expressed relative to that of acetone (1H, 2.225 ppm). Monosaccharide units are as shown in Fig. 9.
TABLE 5.
13C NMR chemical shifts of sugar residues of oligosaccharides 2 and 4 of LPS from E. coli F470(pWQ152)d
| Residue and oligosaccharide |
13C NMR chemicals shift (ppm) of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | C-7 | C-8 | |
| A | ||||||||
| 2 | 92.47 | 55.91 | 70.86 | 70.96 | 73.87 | 71.14 | ||
| 4 | 92.63 | 55.59 | 70.56 | 71.06 | 73.88 | 70.87 | ||
| B | ||||||||
| 2 | 100.85 | 56.92 | 73.33 | 73.60 | 75.11 | NDa | ||
| 4 | 100.39 | 56.89 | 73.19 | 75.56 | ND | 63.55 | ||
| C | ||||||||
| 2 | ND | ND | 35.86 | 71.96 | 70.50 | 73.42 | 71.23 | ND |
| 4 | ND | ND | 35.73 | 71.89 | 70.60 | ND | 67.61 | 65.21b |
| D | ||||||||
| 2 | ND | ND | 36.04 | 73.87 | 68.04 | ND | ND | 64.52 |
| 4 | ND | ND | 35.86 | 73.91 | 67.86 | 72.81 | 70.56 | 64.47 |
| E | ||||||||
| 2 | ND | ND | 36.04 | 67.40 | 67.49 | ND | ND | 64.29 |
| 4 | ND | ND | 35.86 | 67.37 | 67.61 | ND | ND | 64.11b |
| F | ||||||||
| 2 | 100.22 | 72.18 | 77.64 | 71.63 | 73.78 | 69.44 | ND | |
| 4 | 100.23 | 72.20 | 79.02 | 71.05 | ND | 70.34 | 63.90 | |
| G | ||||||||
| 2 | 103.41 | 70.69 | 80.42 | 67.06 | 73.49 | 70.29 | 71.05 | |
| 4 | 103.89 | 71.05 | 80.58 | 70.89 | 73.22 | 68.72 | 69.73 | |
| H | ||||||||
| 2 | 102.40 | 71.26 | 71.34 | ND | 73.32 | 67.26 | 71.26 | |
| 4 | 101.35 | 71.26 | 71.79 | 67.54 | 72.85 | 70.56 | 64.31 | |
| I | ||||||||
| 2 | 102.13 | 71.23 | 81.11 | 71.17 | 73.21 | ND | ||
| 4 | 102.55 | 71.05 | 79.11 | 71.38 | 73.14 | 61.75c | ||
| K | ||||||||
| 2 | 97.94 | 71.22 | ND | 70.97 | 72.90 | 61.71 | ||
| 4 | 96.98 | 73.61 | 72.89 | 71.30 | 72.40 | 61.56c | ||
| L | ||||||||
| 2 | 97.39 | 55.46 | 70.86 | 70.89 | 73.41 | ND | ||
ND, not detected.
Signals may be interchanged.
Signals may be interchanged.
Spectra were recorded from a solution in 2H2O at 150.90 MHz at 27°C. Chemical shifts are expressed relative to that of acetone (13C, 34.5 ppm). Monosaccharide units are as shown in Fig. 9.
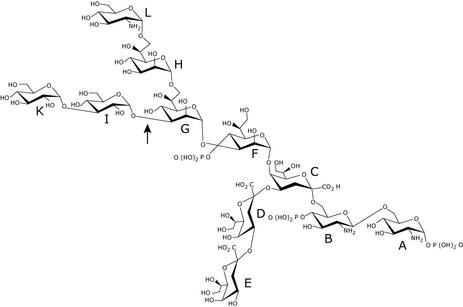
FIG. 9.
Structures of oligosaccharides 2 and 4. The structure of oligosaccharide 2 is shown in detail. In oligosaccharide 4, residue L is lacking and O-4 of heptose residue G (indicated by the arrow) contains an additional phosphate group.
The 13C NMR chemical shifts were assigned by an HMQC experiment by using the interpreted 1H NMR spectrum. Seven anomeric signals were identified (Table 5). Anomeric signals for the Kdo residues C, D, and E were not detected. Low field-shifted signals indicated substitutions at O-6 (residues A and B), O-4 and O-5 (residue C), O-3 and O-4 (residue F), O-3, O-4, and O-7 (residue G), O-4 (residue D), and O-3 (residue I). Residues E, H, and K were terminal sugars.
The sequences of the monosaccharide residues were determined by using data obtained from a ROESY experiment. NOE contacts between anomeric and trans-glycosidic protons were observed for all hexose and heptose residues, and for GlcN residue B. An interresidual NOE contact was observed between H-1 of GlcN residue B to H-6a of GlcN residue A, thus establishing the β-(1→6) linkage of the lipid A backbone. Since Kdo possesses no anomeric proton, it was not possible to deduce the linkage of Kdo residue C to GlcN residue B by an NOE contact. However, since all other linkages of the hexoses and heptoses could be identified by NOE connectivities, the (2→6) linkage of residue C to residue B and the residue sequence E-D-C could be established by the downfield 13C chemical shifts of C-6 of residue B (63.55 ppm) (Table 4), C-4 of residue C (71.89 ppm), and C-4 of residue D (73.91 ppm). No NOE contacts between deoxy-protons H-3ax and H-3eq of Kdo residues C and D and H-6 of Kdo residues D and E, respectively (24), were identified. Further NOE contacts were identified between H-1 of residue F and H-5 (strong) and H-7 of residue C (strong), H-1 of residue G and H-2 (weak) and H-3 (medium) of residue F, H-1 of residue H and H-7a,b of residue G (medium), H-1 of residue I and H-3 of residue G (strong), and H-1 of residue K and H-3 of residue I (strong).
In the 31P NMR spectrum of oligosaccharide 2, four signals of monophosphate groups were identified at −0.60, 0.37, 1.17, and 1.40 ppm. A 31P-1H HMQC experiment gave cross-peaks between the signal at −0.60 ppm and H-1 of GlcN residue A (5.754 ppm) and the signal at 0.37 ppm with H-4 of GlcN residue B (3.868 ppm), establishing the 1,4′-bisphosphorylated and β-(1-6)-linked lipid A backbone. The other two signals at 1.17 and 1.40 ppm connected with the H-4 protons of heptose residues F and G (unresolved at 4.472 ppm), proving that both heptoses are replaced at O-4 by phosphate.
The anomeric region of the 1H NMR spectrum of oligosaccharide 2 contained 8 signals, representing three heptose, two hexose, and three hexosamine residues. The additional hexosamine was an α-linked GlcN residue (residue L). Chemical shift data (Tables 4 and 5) and results of a ROESY experiment were similar to those obtained for oligosaccharide 4, with two exceptions. First, the signal of C-7 of heptose residue H was significantly shifted to the lower field (71.26 ppm in comparison to 64.31 ppm of residue H in oligosaccharide 4). This together with an NOE connectivity (medium) between H1 of residue L and H-7a,b of residue H indicated that oligosaccharide 2 is an undecasaccharide that possesses the carbohydrate backbone shown in Fig. 9. Second, the signal of C-4 of heptose residue G was not shifted downfield (67.06 ppm in comparison to 70.89 ppm in oligosaccharide 1), indicating that residue G is not substituted with a phosphate residue.
The quality of the 31P NMR spectrum of oligosaccharide 2 was rather low, and only three poorly resolved signals of monophosphate groups could identified at ∼0.45, 0.80, and 1.40 ppm, consistent with the presence of the bisphosphorylated lipid A backbone (the first two signals) and of only one phosphorylated heptose residue.
Structural data obtained by MS and NMR spectroscopy was combined to give the structures of oligosaccharides 2 and 4 that are shown in Fig. 9. With the exception of the KdoIII residue resulting from WaaZ overexpression, the structures of the core OS fractions 2 and 4 of F470(pWQ152) LPS were both found in the complete structure of the core OS from F470 (44).
DISCUSSION
The objective of this study was to examine the role of waaZ in the assembly of the LPS inner core in E. coli and S. enterica. The waaZ gene is located in the waaQ operon of the waa gene cluster, which encodes glycosyltransferases responsible for outer core assembly and enzymes involved in modifications of the inner core. The outer core transferases responsible for core elongation were previously assigned, as were the gene products involved in the substitutions of the heptose region (WaaP, WaaQ, and WaaY) (20, 47). However, the genes involved in modifications of the inner core Kdo residues with l-rhamnose or KdoIII were not established. The waaZ gene is found in the E. coli K-12 and R2 core types and in the core of S. enterica serovars. A substitution common to these core types is the nonstoichiometric addition of a KdoIII to the KdoII residue of the inner core (22). The E. coli R3 core type was originally reported to contain KdoIII, but recent structural analyses have shown that not to be the case (O. Holst, personal communication) and isolates with R3 cores lack the waaZ gene (20). Based on the correlation between the distribution of the waaZ gene and the KdoIII residue, it was hypothesized that waaZ may be involved in the addition of KdoIII and structural analyses confirmed this to be the case.
A putative glycosyltransferase (ORF 4) in K. pneumoniae shows limited similarity to the E. coli WaaZ protein and is encoded by the waa cluster of K. pneumoniae (33). In a reinvestigation of the core OS structure of K. pneumoniae, it was found that an inner core KdoIII residue was not present (42, 43), contrary to earlier suggestions (40). However, recent data (Frirdich et al., unpublished) implicate ORF 4 in the addition of a Kdo residue to the outer core in K. pneumoniae, where it provides the attachment site for O antigens (42, 43). While it is tempting to speculate that WaaZ and ORF 4 are Kdo transferases, there is currently no direct evidence to support such assignments. Attempts to verify Kdo transferase activity in vitro have been unsuccessful. Such experiments are complicated by the low yields of soluble WaaZ, the instability of the CMP-Kdo substrate (39) that must be enzymatically synthesized in situ (14), and a lack of information regarding the structure of the appropriate LPS acceptor. At this stage it remains a formal possibility that WaaZ is a regulator of KdoIII transferase activity, rather than it being a transferase per se. For example, it is conceivable that WaaZ modifies the action of the WaaA Kdo transferase that is normally bifunctional in E. coli (Fig. 1) (reviewed in reference 32). However, there is no precedent for such a function in core OS biosynthesis in E. coli and Salmonella. Glycosyltransferases which use nucleotide diphospho-sugar, nucleotide monophospho-sugars, and sugar phosphates are classified into families based on sequence similarity (8). WaaZ and its homologs in E. coli K-12 and R2 and in Salmonella were submitted to the CAZy (carbohydrate-active enzymes) server that classifies structurally similar enzymes involved in the synthesis, degradation, or modification of glycosidic bonds (9, 10). These enzymes were not classified with WaaA (family 30), the bifunctional WaaA Kdo transferase (Fig. 1), or any other known families. The lack of relationship between WaaZ and WaaA might reflect constraints imposed by bifunctional versus putative monofunctional action. If WaaZ homologues are indeed monofunctional Kdo transferases, the relatively weak similarity observed between the E. coli WaaZ and the K. pneumoniae ORF 4 product may be explained by their different acceptor requirements. Definitive assignment of WaaZ as a glycosyltransferase awaits further biochemical investigation.
In order to assess the role waaZ plays in inner core assembly, an E. coli K-12 strain was constructed in which waaZ was insertionally inactivated (CWG345). The phenotype of the CWG345 LPS was indistinguishable from that of the wild type by Tricine-PAGE analysis. In retrospect, this was not entirely surprising, as the WaaZ substituents were found to be present in very small amounts in the parental strain; KdoIII is found in only 10 to 15% of the LPS molecules on the cell surface of E. coli K-12 (O. Holst, personal communication). Rather than attempt to isolate and structurally characterize fractions from the waaZ mutant strain (CWG345) and its parent and look for the loss of a residue that is normally present in such small amounts in a heterogeneous population of parental LPS species, an alternative approach was adopted in which WaaZ was overexpressed. This was based on the rationale that overexpression of WaaZ should cause an increase in KdoIII-substituted LPS molecules. Overexpression of WaaZ in a strain with an R1 core type was chosen because: (i) the strain lacks waaZ; (ii) the structure of the F470 LPS, the prototype R1 core OS, was previously determined (44); and (iii) the R1 core has no other modifications at KdoII, potentially simplifying the analysis. That WaaZ overexpression would result in a truncation of the core backbone was quite unexpected, but this effect did provide material for structural characterization and showed that WaaZ overexpression was correlated with the appearance of KdoIII in the core OS structure. Detailed structural analysis carried out on the LPS core of E. coli F470(pWQ152) highlighted the range of core OS species found in F470 and showed the significant shift in species in the truncated core OS of F470(pWQ152). Molecules corresponding to full-length core species were limited in F470(pWQ152), and several of the truncated core molecules were predicted to contain a KdoIII residue. For the 2 major species (fractions 2 and 4) isolated as single molecular species from HPAEC, the presence of KdoIII was unequivocally confirmed by ESI FT-ICR MS, ESI spectra after CSD (which provides a fragmentation pattern for the fraction), and NMR. Both oligosaccharides only contain two of the five outer core hexoses, further establishing the extent of the core truncations that result from WaaZ overexpression.
The truncation of LPS resulting from elevated activity of a core transferase has not been previously observed. In control experiments, we overexpressed the waaF gene product, encoding heptosyltransferase II, and there were no truncations evident in SDS-PAGE gels (E. Frirdich and C. Whitfield, unpublished data). The WaaZ-mediated core truncation appears to occur at the step catalyzed by WaaT, resulting in a structure lacking the terminal two Gal residues in the core as well as the β-linked glucose side branch (19). Most surprisingly, the effect of WaaZ is confined to strains whose LPS molecules contain a full-length backbone; overexpression had no effect on the already truncated core OSs from waaV and waaW mutants (lacking 1 and 2 sugars, respectively). There are two possible explanations for the truncations caused by WaaZ. Transferases involved in core elongation are thought to form a complex at the inner leaflet of the cytoplasmic membrane, although there is no direct experimental data to support this model. A significant amount of WaaZ localizes to the membrane, despite the fact that it is predicted to be a soluble protein. It is conceivable that overexpressed WaaZ interferes with necessary protein-protein interactions between the transferases forming this complex, preventing the addition of core residues by late-acting transferases, thereby providing indirect support for the existence of a coordinated complex. In this possible scenario, chromosomal copy-level WaaZ would not have an obvious effect in E. coli and Salmonella isolates due to the low amounts of protein made. Notably, the amount of WaaZ produced from constructs in high-level expression vectors is lower than that of many other core OS assembly enzymes studied in this laboratory (data not shown). An alternative explanation for the WaaZ phenotype is that it alters the core OS acceptor for a later transferase by the addition of KdoIII. This seems unlikely, given that KdoIII addition causes truncations in the outer core (spatially removed from the Kdo region), but it must still be considered. In either model, the KdoIII residues in the LPS from the wild-type strain should also be confined to shorter molecules. The low levels of naturally occurring KdoIII preclude detailed structural studies to unequivocally resolve this, but MS results (data not shown) are consistent with this assumption. These data suggest that the increase in KdoIII arising from WaaZ overexpression is not simply due to the truncated core being a better acceptor for KdoIII transfer. If this were the case, KdoIII should be increased in other waa mutants of E. coli K-12 affected in core backbone assembly. However, analysis of an E. coli K-12 waaFC mutant with a highly truncated core did not show significant amounts of KdoIII (7). KdoIII status has not been investigated in other mutants with truncated core backbones, primarily because of the difficulty in obtaining structural data for this region of the inner core.
The results presented here provide the first evidence for an alteration in inner core structure (albeit resulting from WaaZ overexpression) being correlated with a change in overall LPS architecture. It has been suggested previously that, in E. coli, WaaS, WaaZ, and WaaQ may have an integral role in the production of a lipooligosaccharide-like form of LPS that does not serve as an acceptor for O-PS (38). The use of the lipooligosaccharide term in the context of E. coli and Salmonella is potentially confusing since it is generally reserved to describe the LPS of mucosal pathogens, such as Haemophilus influenzae and Neisseria species that naturally lack O-PS (32). The lipooligosaccharide proposal was based on SDS-PAGE profiles of LPS from various mutants, and independent supporting data (e.g., precise structures and/or enzymatic activities) was not available. It is evident that these genes are not essential for production of smooth LPS (S-LPS), because LPS molecules in waaQ mutants can serve as acceptors for O-PS (47) and the E. coli K-12 waaZ mutant can express S-LPS when a plasmid carrying O-antigen biosynthesis genes is introduced (Frirdich and Whitfield, unpublished). However, the effect could be more subtle, as all bacteria with S-LPS produce a certain amount of rough LPS (R-LPS); small differences in inner core OS modifications could potentially dictate the balance between R-LPS and S-LPS on the cell surface. In this scenario, the balance would be lost if a critical enzyme (perhaps WaaZ) were overexpressed. While modulations in the ratio of S-LPS to R-LPS would influence virulence (through altered complement resistance), it should be noted that WaaZ itself is not present in all E. coli isolates. Notably, it is absent in isolates expressing the R1 and R3 core OS types. The R1 core OS is found in the more-virulent extraintestinal pathogens, and R3 is associated with verotoxigenic E. coli (3, 5, 11). The R2 and K-12 core types are, in contrast, confined to commensal E. coli. Therefore, while there is conclusive structural evidence implicating WaaZ at some level in the reaction that transfers KdoIII to KdoII, it is more difficult to determine what part this substitution may play in the biology of E. coli and Salmonella.
Acknowledgments
We thank Sylvia Düpow, Regina Engel, Helga Lüthje, and Antje Müller for technical assistance and Hans-Peter Cordes for recording the NMR spectra.
This work was financially supported by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Bacterial Diseases Network (to C.W.) and by the Deutsche Forschungsgemeinschaft (grant LI-448/1-1 [to B.L.]). C.W. is a holder of a Canada Research Chair, and E.F. received a postgraduate scholarship from NSERC and CIHR.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amor, K., D. E. Heinrichs, E. Frirdich, K. Ziebell, R. P. Johnson, and C. Whitfield. 2000. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 68:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amor, P. A., and C. Whitfield. 1997. Molecular and functional analysis of genes required for expression of group IB K antigens in Escherichia coli: characterization of the his-region containing gene clusters for multiple cell-surface polysaccharides. Mol. Microbiol. 26:145-161. [DOI] [PubMed] [Google Scholar]
- 5.Appelmelk, B. J., Y.-Q. An, T. A. M. Hekker, L. G. Thijs, D. M. MacLaren, and J. de Graaf. 1994. Frequencies of lipopolysaccharide core types in Escherichia coli strains from bacteraemic patients. Microbiology 140:1119-1124. [DOI] [PubMed] [Google Scholar]
- 6.Binotto, J., P. R. MacLachlan, and P. R. Sanderson. 1991. Electrotransformation of Salmonella typhimurium LT2. Can. J. Microbiol. 37:474-477. [DOI] [PubMed] [Google Scholar]
- 7.Brabetz, W., S. Müller-Loennies, O. Holst, and H. Brade. 1997. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur. J. Biochem. 247:716-724. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326:929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In G. D. H. J. Gilbert, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 10.Coutinho, P. M., and B. Henrissat. 1999. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach, p. 15-23. In K. H. K. Ohmiya, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co., Tokyo, Japan.
- 11.Currie, C. G., and I. R. Poxton. 1999. The lipopolysaccharide core type of Escherichia coli O157:H7 and other non-O157 verotoxin-producing E. coli. FEMS Immunol. Med. Microbiol. 24:57-62. [DOI] [PubMed] [Google Scholar]
- 12.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galanos, C., O. Luderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245-249. [DOI] [PubMed] [Google Scholar]
- 14.Gronow, S., W. Brabetz, and H. Brade. 2000. Comparative functional characterization in vitro of heptosyltransferase I (WaaC) and II (WaaF) from Escherichia coli. Eur. J. Biochem. 267:6602-6611. [DOI] [PubMed] [Google Scholar]
- 15.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton, C. A., M. Aldea, B. K. Washburn, P. Babtizke, and S. R. Kushner. 1989. New method of generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hämmerling, G., O. Lüderitz, O. Westphal, and P. H. Mäkelä. 1971. Structural investigations on the core polysaccharide of Escherichia coli O100. Eur. J. Biochem. 22:331-344. [DOI] [PubMed] [Google Scholar]
- 18.Heinrichs, D. E., M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. The assembly system for the lipopolysaccharide R2 core-type of Escherichia coli is a hybrid of those found in Escherichia coli K-12 and Salmonella enterica. J. Biol. Chem. 273:8849-8859. [DOI] [PubMed] [Google Scholar]
- 19.Heinrichs, D. E., J. A. Yethon, P. A. Amor, and C. Whitfield. 1998. The assembly system for the outer core portion of R1- and R4-type lipopolysaccharides of Escherichia coli. J. Biol. Chem. 273:29497-29505. [DOI] [PubMed] [Google Scholar]
- 20.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 21.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holst, O. 1999. Chemical structure of the core region of lipopolysaccharides, p. 115-154. In H. Brade, S. M. Opal, S. N. Vogel, and D. C. Morrison (ed.), Endotoxin in health and disease. Marcel Dekker, Inc., New York, N.Y.
- 23.Holst, O. 2000. Deacylation of lipopolysaccharides and isolation of oligosacharide phosphates, p. 345-353. In O. Holst (ed.), Methods in molecular biology, bacterial toxins: methods and protocols, vol. 145. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 24.Holst, O., K. Bock, L. Brade, and H. Brade. 1995. The structures of oligosaccharide bisphosphates isolated from the lipopolysaccharide of a recombinant Escherichia coli strain expressing the gene gseA [3-deoxy-d-manno-octulopyranosonic acid (Kdo) transferase] of Chlamydia psittaci 6BC. Eur. J. Biochem. 229:194-200. [PubMed] [Google Scholar]
- 25.Holst, O., and H. Brade. 1992. Chemical structure of the core region of lipopolysaccharides, p. 134-170. In D. C. Morrison and J. L. Ryan (ed.), Bacterial endotoxic lipopolysaccharides, vol. I. CRC Press, Boca Raton, Fla.
- 26.Holst, O., U. Zähringer, H. Brade, and A. Zamojski. 1991. Structural analysis of the heptose/hexose region of the lipopolysaccharide from Escherichia coli K-12 strain W3100. Carbohydr. Res. 215:323-335. [DOI] [PubMed] [Google Scholar]
- 27.Hull, R. A., R. E. Gill, P. Hsu, B. H. Minshew, and S. Falkow. 1981. Construction and expression of recombinant plasmids encoding type I or mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 33:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaniuk, N. A., M. A. Monteiro, C. T. Parker, and C. Whitfield. 2002. Molecular diversity of the genetic loci responsible for lipopolysaccharide core oligosaccharide assembly within the genus Salmonella. Mol. Microbiol. 46:1305-1318. [DOI] [PubMed] [Google Scholar]
- 29.Knirel, Y. A., O. V. Bystrova, A. S. Shashkov, B. Lindner, N. A. Kocharova, S. N. Senchenkova, H. Moll, U. Zahringer, K. Hatano, and G. B. Pier. 2001. Structural analysis of the lipopolysaccharide core of a rough, cystic fibrosis isolate of Pseudomonas aeruginosa. Eur. J. Biochem. 268:4708-4719. [DOI] [PubMed] [Google Scholar]
- 30.Lindner, B. 2000. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of lipopolysaccharides, p. 311-325. In O. Holst (ed.), Methods in molecular biology, bacterial toxins: methods and protocols, vol. 145. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 31.Müller-Loennies, S., O. Holst, B. Lindner, and H. Brade. 1999. Isolation and structural analysis of phosphorylated oligosaccharides obtained from Escherichia coli J-5 lipopolysaccharide. Eur. J. Biochem. 260:235-249. [DOI] [PubMed] [Google Scholar]
- 32.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regué, M., N. Climent, N. Abitiu, N. Coderch, S. Merino, L. Izquierdo, M. Altarriba, and J. M. Tomás. 2001. Genetic characterization of the Klebsiella pneumoniae waa gene cluster, involved in core lipopolysaccharide biosynthesis. J. Bacteriol. 183:3564-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Schmidt, G., I. Fromme, and H. Mayer. 1970. Immunochemical studies on core lipopolysaccharides of Enterobacteriaceae of different genera. Eur. J. Biochem. 14:357-366. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt, G., B. Jann, and K. Jann. 1974. Genetic and immunochemical studies on Escherichia coli O14:K7:H−. Eur. J. Biochem. 42:303-309. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt, G., B. Jann, and K. Jann. 1969. Immunochemistry of R lipopolysaccharides of Escherichia coli. Different core regions in the lipopolysaccharides of O group 8. Eur. J. Biochem. 10:501-510. [DOI] [PubMed] [Google Scholar]
- 38.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugai, T., C. H. Lin, G. J. Shen, and C. H. Wong. 1995. CMP-KDO synthetase: overproduction and application to the synthesis of CMP-KDO and analogs. Bioorg. Med. Chem. 3:313-320. [DOI] [PubMed] [Google Scholar]
- 40.Süsskind, M., L. Brade, H. Brade, and O. Holst. 1998. Identification of a novel heptoglycan of α1→2-linked d-glycero-d-manno-heptopyranose. Chemical and antigenic structure of lipopolysaccharides from Klebsiella pneumoniae ssp. pneumoniae rough strain R20 (O1−:K20−). J. Biol. Chem. 273:7006-7017. [DOI] [PubMed] [Google Scholar]
- 41.Tsai, G. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 42.Vinogradov, E., M. Cedzynski, A. Ziolkowski, and A. Swierzko. 2001. The structure of the core region of the lipopolysaccharide from Klebsiella pneumoniae O3. Eur. J. Biochem. 268:1722-1729. [DOI] [PubMed] [Google Scholar]
- 43.Vinogradov, E., and M. B. Perry. 2001. Structural analysis of the core region of the lipopolysaccharides from eight serotypes of Klebsiella pneumoniae. Carbohydr. Res. 335:291-296. [DOI] [PubMed] [Google Scholar]
- 44.Vinogradov, E. V., K. Van Der Drift, J. E. Thomas-Oates, S. Meshkov, H. Brade, and O. Holst. 1999. The structures of the carbohydrate backbones of the lipopolysaccharides from Escherichia coli rough mutants F470 (R1 core type) and F576 (R2 core type). Eur. J. Biochem. 261:629-639. [DOI] [PubMed] [Google Scholar]
- 45.Whitfield, C., G. Schoenhals, and L. Graham. 1989. Mutants of Escherichia coli O9:K30 with altered synthesis and expression of the capsular K antigen. J. Gen. Microbiol. 135:2589-2599. [DOI] [PubMed] [Google Scholar]
- 46.Yethon, J. A., J. S. Gunn, R. K. Ernst, S. I. Miller, L. Laroche, D. Malo, and C. Whitfield. 2000. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect. Immun. 68:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yethon, J. A., D. E. Heinrichs, M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273:26310-26316. [DOI] [PubMed] [Google Scholar]