Abstract
Growth of Bartonella henselae is strongly heme dependent, and B. henselae is unable to synthesize heme itself. At least five outer membrane-associated proteins from B. henselae bind hemin, including the 31-kDa protein designated Pap31. The gene of this protein was heterologously expressed in Escherichia coli M15(pREP4) and detected with monoclonal antibodies in the outer membrane fraction. Complementation of the hemA-deficient mutant E. coli K-12 EB53 (aroB tsx malT hemA) with pap31 demonstrated that this protein is involved in heme acquisition and may be an important virulence factor in the pathogenesis of B. henselae.
Bartonella henselae is a fastidious, pleomorphic, gram-negative rod; it is the major pathogen of cat scratch disease and is one of the agents of bacillary angiomatosis peliosis (23). Cats are the natural host of B. henselae (24). The bacterium shows an unusual erythrocyte parasitism (13, 21), and in vitro growth occurs only on media containing hemin or blood. In a recent study (25), it was demonstrated that hemin is essential for growth of B. henselae. The heme precursor protoporphyrin IX alone or in combination with FeCl2 or FeCl3 did not support growth, indicating that this bacterium cannot synthesize hemin itself. Neither transferrin nor lactoferrin can serve as a growth factor for B. henselae (25). It seems obvious that Bartonella spp. need a heme capture system to secure this essential factor for successful replication in their host.
Heme is a source of iron for a variety of gram-negative bacteria, including Yersinia spp., Escherichia coli, Neisseria spp., Haemophilus influenzae, Haemophilus ducreyi, Porphyromonas gingivalis, and Vibrio spp. (16, 28). The major mechanisms by which gram-negative bacteria acquire heme from their hosts involve either direct binding to specific outer membrane receptors or release of bacterial hemophores which interact in the extracellular environment with the heme source and present it to specific receptors (31).
For Bartonella quintana, as many as eight different hemin-binding proteins (HBPs) have been identified, including 30-, 42-, and 87-kDa proteins (6). The 30-kDa protein of B. quintana (HbpA) shows a close homology (49% identity) to a 31-kDa protein (Pap31) of B. henselae (6). The sequence of the pap31 gene was already described by Bowers et al. in 1998, but the protein was not further characterized (3). The Pap31 protein was originally suspected to be a phage-associated membrane protein in B. henselae and to play a role in the packaging of the 14-kb DNA within the phage. Bacteriophage-like particles are also known from Bartonella bacilliformis and seem to be present in B. quintana, Bartonella doshiae, and Bartonella grahamii (1).
Heme uptake mechanisms of B. henselae have not been characterized to date, and no HBPs are described from this pathogen. The goal of this study was to investigate the molecular mechanisms of heme acquisition by B. henselae. Using different methods, we detected at least five HBPs, including the Pap31 protein, in B. henselae. We present the characterization of Pap31 as an outer membrane HBP, the functional heterologous expression of Pap31 in E. coli, and the successful complementation of the hemA-deficient mutant E. coli K-12 EB53.
All bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Bartonella henselae | Type strain | ATCC 49882 |
| E. coli K-12 EB53 | aroB tsx malT hemA | Generous gift of Klaus Hantke, Tübingen, Germany |
| E. coli M15(pREP4) | Host for expression vectors pQE70 and pQE60 | Qiagen |
| E. coli M15 pap31 | As E. coli M15(pREP4) pRZn7-Pap31(ompT) | This study |
| E. coli EB53(pREP4) | As E. coli K-12 EB53 KmrlacI p15A ori | This study |
| E. coli EB53 pap31 | As E. coli EB53(pREP4) pRZn7-Pap31(ompT) Kmr | This study |
| Plasmids | ||
| pQE60 | Expression vector; ColE1 PT5 lacO Ampr | Qiagen |
| pQE70 | Expression vector; ColE1 PT5 lacO Ampr | Qiagen |
| pRZn7-Pap31 (ompT) | As pQE70 pap31(ompT) | This study |
| pREP4 | lacIq Kmr p14A ori | Qiagen |
Cell fractionation and detection of outer membrane proteins of B. henselae.
B. henselae (ATCC 49882) cultures were grown on chocolate agar plates as described previously (24). Cell fractionation was performed by the Sarkosyl method (32). In short, B. henselae organisms were harvested from 25 chocolate agar plates, washed in phosphate-buffered saline, resuspended in 10 ml of distilled water, and broken by sonication (three times for 60 s, with a 30-s cooling period between each burst) at 4°C. Sodium N-laurosyl sarcosinate (Serva, Heidelberg, Germany) was added to a final concentration of 2% (vol/vol), and the mixture was incubated at room temperature for 30 min. The outer membrane-peptidoglycan complex was recovered by centrifugation at 28,000 rpm in an SW28 rotor for 60 min, washed twice in distilled water containing 0.2 mM PMSF (phenylmethylsulfonyl fluoride) (Roche, Mannheim, Germany), resuspended in 0.5 ml of the same solution, and stored frozen at −20°C until use. The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (14) as described previously (26). The soluble and insoluble protein fractions of B. henselae are presented in Fig. 1.
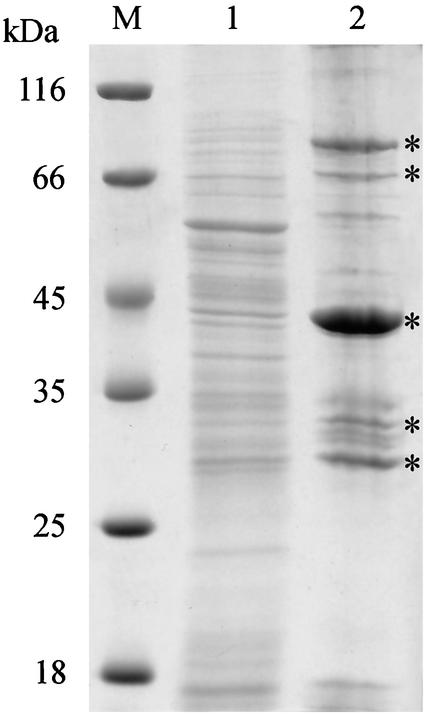
FIG. 1.
Coomassie blue-stained SDS-PAGE gel of the Sarkosyl-soluble and -insoluble fractions of B. henselae ATCC 49882. M, molecular mass standard (Peqlab); lane 1, Sarkosyl-soluble fraction; lane 2, Sarkosyl-insoluble outer membrane fraction. The five HBPs discussed in the text are marked by asterisks.
Identification of B. henselae HBPs and detection of Pap31 as an outer membrane protein.
HBPs of the outer membrane fraction of B. henselae were identified by hemin-binding blots as described by Carrol et al. (6) and by hemin-agarose binding assays (15). The proteins in the hemin-binding blot were detected with diaminobenzidine (DAB) (Roche Diagnostics).
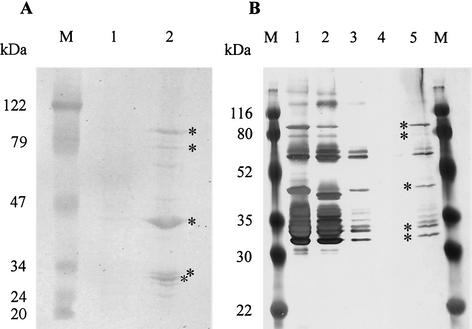
Multiple HBPs were detected with both methods in the outer membrane fraction of B. henselae (Fig. 2A, lane 2, and 2B, lane 5), but no strong reactions were seen in the cytoplasmic and inner membrane fractions (Fig. 2A, lane 1). We selected from the hemin-reactive proteins the five most dominant bands (the 31-, 34-, 43-, 80-, and 89-kDa proteins) for further studies. The 31- and 43-kDa proteins already showed a brown color prior to detection with DAB. The N-terminal sequences of the 31-, 34-, and 43-kDa proteins were determined by Edman degradation by using a model 477 A gas phase protein sequencer (Applied Biosystems) as described previously (26). BLASTp searches for the 31-, 34-, and 43-kDa proteins showed 100% matches to the Pap31 protein (for the 31- and 34-kDa proteins) (3) and to the Omp43 protein (for the 43-kDa protein) (4) of B. henselae. The amino acid sequence of Pap31 showed a 49% identity to the 30-kDa HBP HbpA from B. quintana (6). A very dominant HBP of 60 kDa was detectable especially in the hemin-agarose binding assay. N-terminal sequencing of this protein revealed that it was identical with the already known 60-kDa heat shock protein of B. henselae (11).
FIG. 2.
Identification of HBPs. (A) Hemin binding blot. M, prestained molecular mass standard (Peqlab, Germany); lane 1, cytoplasmic and inner membrane proteins; lane 2, outer membrane proteins. (B) Hemin-agarose binding assay (silver stained). M, prestained molecular mass standard (Bio-Rad); lane 1, outer membrane proteins; lanes 2 to 4, purification steps: lane 2, supernatant after incubation with hemin-agarose; lane 3, supernatant after washing in high-salt buffer; lane 4, supernatant after washing in low-salt buffer; lane 5, HBPs. The major HBPs of 89, 80, 43, 34, and 31 kDa are marked by asterisks.
mRNA expression of pap31 in B. henselae and of recombinant pap31 in E. coli. (i) Recombinant DNA techniques.
Plasmid extraction and plasmid transformation were described previously (2, 5, 29). Standard cloning, ligation, and restriction enzyme analysis were done according to the methods of Sambrook and Russell (22). PCR was performed by using the Pwo polymerase from Peqlab (Erlangen, Germany) in accordance with the manufacturer's specifications. Heterologous expression of the pap31 gene in E. coli M15(pREP4) was done in the expression vector pQE60 (Qiagen, Hilden, Germany), introducing an N-terminal NcoI restriction site and a C-terminal HindIII restriction site via PCR for the cloning procedure. Expression was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM. For the replacement of the Bartonella pap31 signal sequence with the E. coli ompT signal sequence, megaprimer PCR was done by amplifying the ompT signal sequence with the sense primer 5′-TGCAAGCCATTGCGAGGCCT-3′ and antisense primer 5′-AACGATAACATCAGCAGCAAAAGAGCTGATCGCAATAGGGGTTGTCAGGA-3′. The pap31 gene without the signal sequence was amplified with the sense primer 5′-ATCAGCTCTTTTGCTGCTGATGTTATCGTTCCTCATGAAGTAGCGCCAAC-3′ and the antisense primer 5′-ACCAAGCTTTAAATCTGTGTTAATTCA-3′. These two amplicons were combined and amplified with the sense primer 5′-GCATGCGGGCGAAACTTCTGGGA-3′ and the antisense primer 5′-ACCAAGCTTTAAATCTGTGTTAATTCA-3′. Ad-ditionally, SphI and HindIII restriction sites were included at the ends of the newly created pap31 gene [pap31(ompT)]. The pap31(ompT) gene was confirmed by sequence analysis by the dideoxynucleotide chain termination method (27). This construct was ligated into the expression vector pQE70 (Qiagen) to produce the vector pRZn7-Pap31(ompT), removing the His tag of the vector. Expression experiments were done in E. coli M15(pREP4) in accordance with the manufacturer's instructions. Oligonucleotide primers were purchased from MWG Biotech, Ebersberg, Germany.
(ii) RNA isolation and RT-PCR.
Total RNA was isolated from B. henselae ATCC 49882, E. coli M15(pREP4), E. coli M15(pREP4) harboring pQE70, and from induced and uninduced E. coli M15 pap31 by using a NucleoSpin RNAII kit (Clontech, Heidelberg, Germany) with an additional DNase I digestion (Ambion, Huntingdon, United Kingdom). Contaminating DNA was excluded by a PCR with the total RNA as the template and with the sense primer 5′-ATGCCTTATGTTGCTGGT-3′ and antisense primer 5′-GAATTTGTACGCTACACCAA-3′. Reverse transcription of 0.5 μg of total RNA was performed with the Omniscript reverse transcriptase (Qiagen) by using the primer 5′-GAATTTGTACGCTACACCAA-3′. Reverse transcriptase (RT)-PCR was done with 2 μl of RT reaction mixture and the Taq polymerase (Roche Diagnostics) by using the same primer as for the reverse transcription with the sense primer 5′-ATGCCTTATGTTGCTGGT-3′ for recombinant pap31 and the sense primer 5′-ATGTGAAACCGATGCGGT-3′ for wild-type pap31, which amplify 291- and 535-bp fragments, respectively.
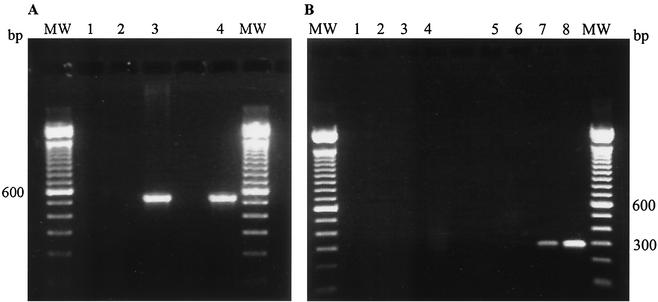
RT-PCR of pap31 in B. henselae is shown in Fig. 3A, demonstrating that this gene is transcribed in B. henselae. E. coli M15(pREP4) harboring the expression plasmid pQE60 with the pap31 gene showed no detectable synthesis of recombinant Pap31 and translocation of the protein to the outer membrane (data not shown). To ensure the surface exposure of Pap31, the signal sequence of B. henselae was omitted and replaced with the signal sequence of the outer membrane protease OmpT of E. coli (10), yielding pap31(ompT), which was ligated in the expression vector pQE70 to obtain pRZn7-Pap31(ompT). The expression in E. coli M15(pREP4) was not apparent in Coomassie blue-stained gels, but analysis of the cDNA by RT-PCR (Fig. 3B, lanes 7 and 8) showed a basal mRNA expression of pap31 before the induction with IPTG and a significantly increased expression after the induction.
FIG. 3.
mRNA expression of pap31 in B. henselae (A) and of recombinant pap31 in E. coli M15(pREP4) (B). The amplified PCR fragment from B. henselae is 535 bp long, and that from E. coli is 291 bp long. MW, 100-bp molecular weight standard. (A) pap31 expression in B. henselae. Lane 1, RNA-PCR; lane 2, negative control; lane 3, positive control; lane 4, RT-PCR. (B) pap31 expression in E. coli. Lanes 1 to 4, RNA-PCR; lanes 5 to 8, RT-PCR. Lanes 1 and 5, E. coli M15(pREP4); lanes 2 and 6, E. coli M15(pREP4) with pQE70; lanes 3 and 7, E. coli M15 pap31 uninduced; lanes 4 and 8, E. coli M15 pap31 induced.
Detection of Pap31 in B. henselae and of recombinant Pap31 in E. coli with monoclonal antibodies. (i) Generation of the monoclonal antibody VKS29.
Female, 6- to 8-week-old BALB/c mice were immunized with B. henselae (7) in accordance with standard protocols (33). In brief, B. henselae was grown for 5 days on Columbia agar plates. Bacteria were harvested in phosphate-buffered saline and heat killed for 2 h at 60°C. Mice were immunized with 100 μg of heat-killed B. henselae. The first immunization was performed with complete Freund's adjuvant. Immunizations on days 21, 42, and 63 were done with incomplete Freund's adjuvant. Booster injections on days 68, 69, and 70 were given without adjuvant. Hybridomas were produced by standard protocols (9). Identification of antibody-producing hybridomas was accomplished by immunoblotting by using B. henselae whole-cell lysates. One of the hybridomas (VKS29) reacted with a 31-kDa protein and was further characterized by Western blot analysis (30).
(ii) Two-dimensional electrophoretic protein pattern.
Two-dimensional gel electrophoresis was performed in a Mini-Protean II two-dimensional cell (Bio-Rad Laboratories, Munich, Germany). Isoelectric focusing was performed by using 60 μg of outer membrane protein and Servalyt ampholytes (Serva Electrophoresis Co., Heidelberg, Germany) with a pI (isoelectric point) range of 3 to 7. Gel preparation, buffers, and running conditions for the first dimension were as recommended by the manufacturer. The second-dimension 12% SDS-PAGEs were run by using a Mini-Protean II two-dimensional cell (Bio-Rad Laboratories), and the proteins were transferred to a polyvinylidene difluoride membrane. Protein spots were visualized by using the Pap31 monoclonal antibody.
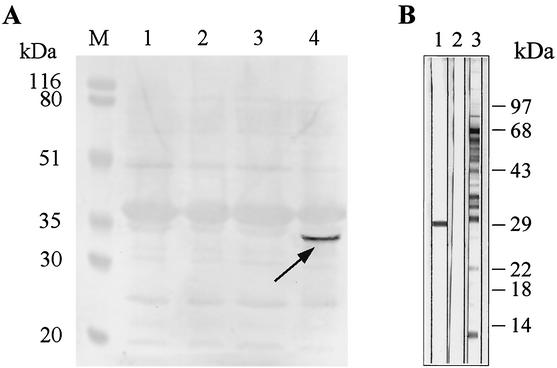
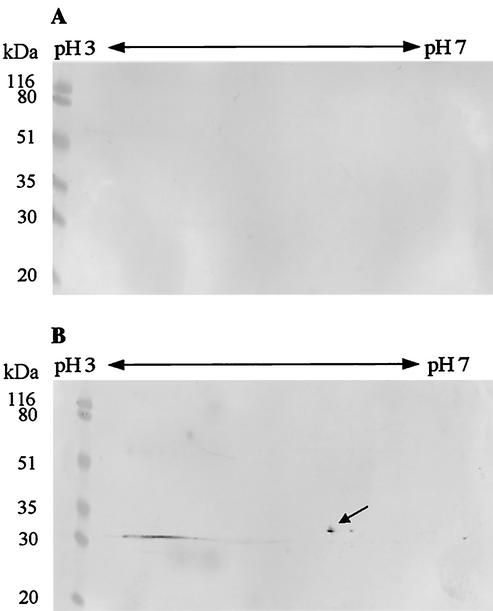
An outer membrane preparation of E. coli M15 pap31 (prepared by using the same protocol as for B. henselae) showed no visible synthesis of Pap31 in SDS-PAGE, but detection with the monoclonal antibody VKS29 against Pap31 clearly verified the synthesis of the recombinant Pap31 at 31 kDa in the induced state and not in the controls (Fig. 4A). The specificity of the monoclonal antibody VKS29 in a B. henselae whole-cell immunoblot for the 31-kDa protein is presented in Fig. 4B. To further prove the identity of recombinant Pap31 synthesized in E. coli, a two-dimensional SDS-PAGE of the outer membranes was performed with subsequent detection with the monoclonal antibody. Pap31 has a calculated pI of 5.2, which is in accordance with the protein spot on the two-dimensional PAGE gel (Fig. 5B, arrow). From these data, we conclude that Pap31 is synthesized in the outer membrane of E. coli M15 pap31.
FIG. 4.
Western blot analysis of B. henselae whole-cell protein and recombinant Pap31 in E. coli M15(pREP4) with monoclonal Pap31 antibody VKS29. (A) Outer membrane proteins. M, prestained molecular mass standard (Bio-Rad Laboratories); lane 1, E. coli M15(pREP4); lane 2, E. coli M15(pREP4) with pQE70; lane 3, E. coli M15 pap31 before induction; lane 4, E. coli M15 pap31 after induction. The arrow indicates the recombinant Pap31. (B) Bartonella henselae whole-cell protein sample immunoblots incubated with monoclonal antibody VKS29 (lane 1) and polyclonal immunserum (lane 3). Lane 2, negative control with secondary antibody and without primary antibody (horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G antibody).
FIG. 5.
Two-dimensional analysis of the recombinant Pap31 with the outer membrane proteins from E. coli M15 pap31 before and after induction. Pap31 has an calculated pI of 5.2 and was detected with the monoclonal Pap31 antibody VKS29. Shown are outer membrane proteins from E. coli M15 pap31 before (A) and after (B) induction. The arrow points to the recombinant Pap31 at the pI of 5. A molecular mass standard (Bio-Rad Laboratories) is shown at the left side of each panel.
Complementation of hemA-deficient mutant E. coli K-12 EB53.
The hemA-deficient mutant E. coli K-12 EB53 (aroB tsx malT hemA) (8) was grown on Luria-Bertani (LB) medium supplemented with 2 μg of δ-aminolevulinic acid (ALA) (Sigma Aldrich Chemicals Co., Deisenhofen, Germany)/ml at 37°C. E. coli K-12 EB53 (aroB tsx malT hemA) is unable to grow without supplementation of growth media with ALA, and due to the mutation of the aroB gene, this strain is unable to synthesize its own iron chelator enterochelin (8). The hemA gene encodes Glu-tRNA-reductase, the enzyme that initiates porphyrin synthesis by the alternative C5 pathway and is responsible for the synthesis of ALA from Glu-tRNA (20). The E. coli K-12 outer membrane is impermeable to extracellular hemin or its precursor protoporphyrin IX, and medium supplemented with hemin does not support growth of E. coli hemA mutants. The uptake of hemin through Pap31 should therefore complement the hemA mutation, and E. coli K-12 EB53 should grow on hemin-supplemented LB medium without ALA.
E. coli K-12 EB53 was first transformed with plasmid pREP4 (Qiagen) to repress the transcription of the pQE expression vectors, yielding E. coli K-12 EB53(pREP4). E. coli K-12 EB53(pREP4) was transformed with the vector pRZn7-Pap31(ompT) and designated E. coli EB53 pap31; the vector pQE70 was used as a negative control. To test for spontaneous revertants, all E. coli K-12 EB53 transformants were grown in LB medium without δ-aminolevulinate. The ability to use hemin was tested with EB53(pREP4) and EB53(pREP4) with pQE70 as negative controls and with E. coli EB53 pap31; each organism was tested with and without addition of IPTG. The complementation was done on solid media to minimize the reversion of the hemA mutation in the absence of ALA. The lack of reversion on solid agar was documented by the control plates by using LB without ALA and without hemin. Under these conditions, no growth occurred. Growth on hemin as the sole heme source was tested by spreading 2 μl of overnight culture in triplicate on LB agar plates without δ-aminolevulinate but supplemented with 5, 15, 30, 50, 100, and 150 μM hemin containing 1 mM IPTG and no IPTG. E. coli EB53 pap31 showed a clearly visible growth at hemin concentrations of 30 μM and higher in the induced conditions. Hemin concentrations of 5 μM and 15 μM could not complement the hemA mutation. No growth at all was seen in the controls, whereas minimal colonies appeared in the uninduced recombinant E. coli. We have shown in a recent study that at least 6 to 12 μM hemin was necessary for growth of B. henselae (25).
These experiments demonstrate that the pap31 gene was necessary and sufficient to restore growth of E. coli K-12 EB53 (aroB tsx malT hemA) cells on hemin-containing media. Since B. henselae is not able to synthesize heme itself and is not able to grow by using the direct precursor protoporphyrin IX, we conclude that Pap31 is one of the HBPs involved in heme acquisition and may be an important virulence factor in this human pathogen.
Interestingly, an alignment of the Pap31 amino acid sequence to related proteins in the sequence data bank gives no hint of hemin binding or iron acquisition proteins of other bacteria. Carroll et al. (6) found in the putative promoter region of hbpA in B. quintana a sequence that closely resembles the ferric uptake regulator (Fur) consensus sequence of E. coli (53% identity). A similar sequence is also found in B. henselae (ATAACTGTGGTATTTTT), showing an 82% identity to the putative Fur box of B. quintana (ATAACTGTGTCGTTTTT) (6) and 53% to the Fur box of E. coli.
Recently, the fur gene from B. henselae was characterized by Park et al. (19); the Fur box of this gene was not detected. However, an autoregulatory binding sequence is also absent in the regions upstream of fur in Bradyrhizobium japonicum (12) and Vibrio cholerae (17). Recently, Nienaber et al. (18) identified a heme uptake system in B. japonicum which is iron regulated but does not contain a Fur box homolog sequence in the promoter region. Considering the close genetic relation of the Bartonella species to Rhizobium species, it might be possible that the regulation of iron uptake or heme binding is similar to that of the Rhizobium species but different from that of other bacterial species. Unfortunately, the transcriptional start site is not yet identified in either B. henselae or in B. quintana and the −35/−10 type promoter is not obvious. The exact localization and regulation of the promoter region are currently under investigation.
Acknowledgments
The authors thank Wolfgang Bredt for continuous support of this work and Klaus Hantke, Lehrstuhl für Mikrobiologie, University of Tübingen, for providing the hemA-deficient E. coli K-12 EB53 strain.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) to A. Sander (SA 870/2-1).
REFERENCES
- 1.Barbian, K. D., and M. F. Minnick. 2000. A bacteriophage-like particle from Bartonella bacilliformis. Microbiology 146:599-609. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowers, T. J., D. Sweger, D. Jue, and B. Anderson. 1998. Isolation, sequencing and expression of the gene encoding a major protein from the bacteriophage associated with Bartonella henselae. Gene 206:49-52. [DOI] [PubMed] [Google Scholar]
- 4.Burgess, A. W. O., and B. E. Anderson. 2000. Isolation, sequencing and expression of Bartonella henselae omp43 and predicted membrane topology of the deduced protein. Microb. Pathog. 29:73-80. [DOI] [PubMed] [Google Scholar]
- 5.Calvin, N. M., and P. C. Hanawalt. 1988. High-efficiency transformation of bacterial cells by electroporation. J. Bacteriol. 170:2796-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, J. A., S. A. Coleman, L. S. Smitherman, and M. Minnick. 2000. Hemin-binding surface protein from Bartonella quintana. Infect. Immun. 68:6750-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drancourt, M., R. Birtles, G. Chaumentin, F. Vandenesch, J. Etienne, and D. Raoult. 1996. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet 347:441-443. [DOI] [PubMed] [Google Scholar]
- 8.Eberspächer, B., and V. Braun. 1980. The involvement of cytochromes in the uptake of ferrichrome by Escherichia coli K-12. FEMS Microbiol. Lett. 7:61-64. [Google Scholar]
- 9.Galfre, G., and C. Milstein. 1981. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 73(Pt. B):3-46. [DOI] [PubMed] [Google Scholar]
- 10.Grodberg, J., M. D. Lundrigan, D. L. Toledo, W. F. Mangel, and J. J. Dunn. 1988. Complete nucleotide sequence and deduced amino acid sequence of the ompT gene of Escherichia coli K-12. Nucleic Acids Res. 16:1209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haake, D. A., T. A. Summers, A. M. McCoy, and W. Schwartzman. 1997. Heat shock response and groEL sequence of Bartonella henselae and Bartonella quintana. Microbiology 143:2807-2815. [DOI] [PubMed] [Google Scholar]
- 12.Hamza, I., R. Hassett, and M. R. O'Brian. 1999. Identification of a functional fur gene in Bradyrhizobium japonicum. J. Bacteriol. 181:5843-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kordick, D. L., and E. B. Breitschwerdt. 1995. Intraerythrocytic presence of Bartonella henselae. J. Clin. Microbiol. 33:1655-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lee, B. C. 1992. Isolation of an outer membrane hemin-binding protein of Haemophilus influenzae type B. Infect. Immun. 60:810-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, B. C. 1995. Quelling the red menace: haem capture by bacteria. Mol. Microbiol. 18:383-390. [DOI] [PubMed] [Google Scholar]
- 17.Litwin, C. M., S. A. Boyko, and S. B. Calderwood. 1992. Cloning, sequencing and transcriptional regulation of the Vibrio cholerae fur gene. J. Bacteriol. 174:1897-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nienaber, A., H. Hennecke, and H.-M. Fischer. 2001. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41:787-800. [DOI] [PubMed] [Google Scholar]
- 19.Park, S. Y., K. L. Kelminson, A. K. Lee, P. Zhang, R. E. Warner, D. H. Rehkopf, S. B. Calderwood, and J. E. Koehler. 2001. Identification, characterization, and functional analysis of a gene encoding the ferric uptake regulation protein in Bartonella species. J. Bacteriol. 183:5751-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reidl, J., and J. J. Mekalanos. 1996. Lipoprotein e(P4) is essential for hemin uptake by Haemophilus influenzae. J. Exp. Med. 183:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolain, J. M., B. La Scola, Z. Liang, B. Davoust, and D. Raoult. 2001. Immunofluorescent detection of intraerythrocytic Bartonella henselae in naturally infected cats. J. Clin. Microbiol. 39:2978-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sander, A. 2001. Bartonellosis, p. 653-686. In N. Cimolai (ed.), Laboratory diagnosis of bacterial infections. Marcel Decker, New York, N.Y.
- 24.Sander, A., C. Bühler, K. Pelz, E. von Cramm, and W. Bredt. 1997. Detection and identification of two Bartonella henselae variants in domestic cats in Germany. J. Clin. Microbiol. 35:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander, A., S. Kretzer, W. Bredt, K. Oberle, and S. Bereswill. 2000. Hemin-dependent growth and hemin binding of Bartonella henselae. FEMS Microbiol. Lett. 189:55-59. [DOI] [PubMed] [Google Scholar]
- 26.Sander, A., A. Zagrosek, W. Bredt, E. Schiltz, Y. Piémont, C. Lanz, and C. Dehio. 2000. Characterization of Bartonella clarridgeiae flagellin (FlaA) and detection of anti-flagellin antibodies in patients with lymphadenopathy. J. Clin. Microbiol. 38:2943-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stojiljkovic, I., and K. Hantke. 1992. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 11:4359-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thung, W. L., and K.-C. Chow. 1995. A modified medium for efficient electrotransformation of E. coli. Trends Genet. 11:128-129. [DOI] [PubMed] [Google Scholar]
- 30.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 32.Williams, P., and L. Gledhill. 1991. Fractionation of bacterial cells and isolation of membranes and macromolecules, p. 87-108. In S. P. Denver and W. B. Hugo (ed.), Mechanisms of action of chemical biocides—their study and exploitation. Society for Applied Bacteriology Technicals Series, vol. 27. Blackwell Scientific Publications, Oxford, United Kingdom.
- 33.Wilske, B., S. Jauris-Heipke, R. Lobentanzer, I. Pradel, V. Preac-Mursic, D. Rossler, E. Soutschek, and R. C. Johnson. 1995. Phenotypic analysis of outer surface protein C (OspC) of Borrelia burgdorferi sensu lato by monoclonal antibodies: relationship to genospecies and OspA serotype. J. Clin. Microbiol. 33:103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]