Abstract
Supernatants of rhamnose-induced Erwinia chrysanthemi strain 3937 cultures contain a principal secreted protein named RhiE. A rhiE mutant has been found among a set of rhamnose-induced MudI1681 lacZ fusions. RhiE is a 62-kDa protein that has rhamnogalacturonate lyase activity on rhamnogalacturonan I (RG-I). It does not require a divalent cation for its activity and has an optimal pH of 6.0. rhiE expression is strongly induced in the presence of rhamnose but is also regulated by PecT and Crp, two regulators of the transcription of pectinolytic enzyme genes. RhiE is secreted through the type II Out secretion pathway. RhiE has no disulfide bond. The absence of RhiE secretion in a dsb mutant indicated that disulfide bond formation is required for the biogenesis of the secretion apparatus. RhiE was searched for in several E. chrysanthemi strains by using antibodies, and it was found to be present in one-third of the strains tested. However, the reduced virulence of the rhiE mutant indicates that degradation of the RG-I region of pectin is important for full virulence of E. chrysanthemi.
Pectin is one of the main components present in plant cell walls. The pectin molecule can have three types of backbone: homogalacturonan, rhamnogalacturonan I (RG-I), and RG-II. Homogalacturonan (polygalacturonate [PGA]) is a polymer of galacturonic acid residues linked by α 1-4 bonds which are partially methylated and acetylated. Homogalacturonan forms the smooth regions of pectin which are interspersed with hairy regions of RG-I and RG-II. RG-I has a backbone composed of as many as 100 repeats of the disaccharide α-l-rhamnose-α-1,4-galacturonic acid. Side chains of arabinan or galactan are attached to O-4 of the rhamnosyl residues. RG-II is a complex polysaccharide with a short homogalacturonan backbone to which are linked side chains containing more than 10 different types of sugars (1).
Plant-pathogenic microorganisms often secrete enzymes that are able to degrade the polysaccharides of cell walls in order to disorganize the plant tissue and obtain carbon sources for their growth. The enzymes that degrade the smooth regions of pectin have been extensively studied (26). The action of pectin methylesterases and of pectin acetylesterases liberates PGA. PGA is then degraded by pectate lyases or polygalacturonases that cleave either in an endo or in an exo mode. These enzymes have been characterized in bacteria as well in fungi. Much less is known about the enzymes degrading RG-I and RG-II. It seems that the presence of side chains prevents the degradation of the homogalacturonan backbone of RG-II by polygalacturonase and pectate lyases, and the debranching enzymes have not yet been characterized. Only a few enzymes that are able to cleave the RG-I backbone have so far been characterized, mostly in Aspergillus aculeatus. A rhamnogalacturonate lyase (18) cleaves inside the chain between a rhamnose and a galacturonate, creating an unsaturation on the galacturonate at the nonreducing end. A rhamnogalacturonate rhamnohydrolase (17) and a rhamnogalacturonate galacturonohydrolase (16) cleave the rhamnose and galacturonate residues, respectively, present at the nonreducing end of the rhamnogalacturonate chain. Other enzymes with putative rhamnogalacturonase activity have been described for Trametes sanguinea (28) and Neurospora crassa (20). Up to now, only one bacterial enzyme that is able to cleave RG-I has been described: the rhamnogalacturonate lyase of Pseudomonas cellulosa (14).
Erwinia chrysanthemi is an enterobacterium that provokes the soft rot disease of many plant species. It secretes, in the outer medium, enzymes that degrade the components of the cell walls: pectinases, cellulases, and proteases. At least eight pectinases and the cellulase Cel5 are secreted by the type II Out secretion system, while four proteases are secreted by a type I secretion machinery (26). A type III secretion system allows for secretion of the harpin HrpN (2). E. chrysanthemi possesses a battery of enzymes that allow it to degrade the PGA region of pectin and to then use it as the sole carbon source for its growth. A pectin acetylesterase (PaeY), two pectin methylesterases (PemA and PemB), eight endo-pectate lyases (PelA, PelB, PelC, PelD, PelE, PelI, PelL, and PelZ), two exo-pectate lyases (PelW and PelX), and three polygalacturonases (PehV, PehW, and PehX) have been characterized (26). The oligogalacturonates produced are then metabolized and enter the bacterial general metabolism. Synthesis of all of these enzymes is finely controlled by a set of transcriptional regulators (KdgR, PecS, PecT, cyclic AMP receptor protein [CRP], and ExpR) that respond to environmental signals (9, 32). Thus, the E. chrysanthemi PGA pathway is very elaborate and allows complete degradation of the smooth region of pectin. It would seem unlikely that E. chrysanthemi could degrade the smooth regions of pectin to completion but leave the RG-I regions intact. The synthesis of enzymes involved in PGA degradation is induced in the presence of galacturonate. To identify the enzymes involved in RG-I degradation, we looked for rhamnose-inducible secreted proteins. We characterized the gene rhiE, encoding a secreted rhamnogalacturonate lyase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. E. chrysanthemi and Escherichia coli cells were grown at 30 and 37°C, respectively, in Luria broth (LB) medium, M63 minimal medium, or MacConkey medium (15) supplemented with a carbon source (0.2%) and, when required, with antibiotics at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; and chloramphenicol, 20 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Relevant characteristics | Reference or source |
|---|---|---|
| Bacterial strains | ||
| E. chrysanthemi 3937 derivatives | ||
| A350 | lmrTclacZ | 11 |
| A837 | lmrTclacZ kdgR | 8 |
| A1524 | lmrTclacZ pecS::Mu-Cmr | 25 |
| A2174 | lmrTclacZ pecT::Cmr | 7 |
| A2507 | lmrTclacZ crp::Cmr | 24 |
| A2757 | lmrTclacZ expI::Cmr | 19 |
| A2107 | lmrTclacZ dsbA::uidA-kan dsbC-Cmr | Laboratory collection |
| A3657 | lmrTclacZ pecS::Mu-Cmr ΔoutD | V. E. Shevchik |
| A3749 | lmrTclacZ rhiE::MudI1681 | This work |
| A3884 | lmrTclacZ rhiE::Cmr | This work |
| A3946 | lmrTclacZ rhiE::uidA-Km | This work |
| A3981 | lmrTclacZ kdgR::Tn5-B21 ΔoutC | V. E. Shevchik |
| A3982 | lmrTclacZ kdgR::Tn5-B21 ΔPDZoutC | V. E. Shevchik |
| Other E. chrysanthemi strains | ||
| EC16 | Laboratory collection | |
| ENA49 | Laboratory collection | |
| ET3 | E. Lojkowska | |
| 4073 | E. Lojkowska | |
| 1240 (ICPB EC174) | E. Lojkowska | |
| SF109-1 | E. Lojkowska | |
| 4072 | E. Lojkowska | |
| 1271 (NCPPB 1065) | E. Lojkowska | |
| 4040 | E. Lojkowska | |
| 722 | E. Lojkowska | |
| 1270 (NCPPB) | E. Lojkowska | |
| 4062 | E. Lojkowska | |
| SH230-C143 | E. Lojkowska | |
| 1342 (ICPB EC239) | E. Lojkowska | |
| 1891 | E. Lojkowska | |
| 4060 | E. Lojkowska | |
| 4061 | E. Lojkowska | |
| 4065 | E. Lojkowska | |
| 3367 | E. Lojkowska | |
| 3716 | E. Lojkowska | |
| CIP366 | E. Lojkowska | |
| E. carotovora subsp. odorifera 56-487 | Laboratory collection | |
| E. carotovora subsp. betavasculorum 52-479 | Laboratory collection | |
| E. carotovora subsp. carotovora SCRI193 | Laboratory collection | |
| E. carotovora subsp. atroseptica SCRI31 | Laboratory collection | |
| E. rhapontici 49-421 | Laboratory collection | |
| E. cypripedi 51-440 | Laboratory collection | |
| E. carnegiana 55-483 | Laboratory collection | |
| E. coli | ||
| NM522 | Δ(lac-proAB) thi hsd-5 supE (F′ proAB+lacIqΔM15) | Stratagene |
| MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 ΔlacX74 rpsL hsdR2 (rK− mK−) mcrA mcrB1 | Laboratory collection |
| BL21(DE3) | E. coli B; F−dcm ompT hsdS gal λ(DE3); T7 polymerase gene under the control of the lacUV5 promoter | Laboratory collection |
| Plasmids | ||
| pBluescript | Apr | Stratagene |
| pT7-5 | T7Φ10; Apr | 33 |
| pT7-RhiE | pT7-5 bearing rhiE | This work |
| Phages | ||
| ΦEC2 | E. chrysanthemi generalized transducing phage | 23 |
| MudI1681 | Mu cts62 derivative (′lacZYA Kmr) | 6 |
Genetic techniques.
Transduction with phage φEC2 was carried out as described by Résibois et al. (23). Marker exchange recombinations were obtained after growth in low-phosphate medium, as described by Roeder and Collmer (27). Mutagenesis was performed with miniMu MudI1681 (6). A 0.2-ml portion of the lysate of phage MudI1681 was added to 0.2 ml of an overnight culture of strain A350. After 20 min, the bacteria were spread onto LB-kanamycin agar plates. Colonies formed after 24 h were replica plated onto MacConkey-lactose and MacConkey-lactose-rhamnose agar plates.
Recombinant DNA techniques.
Preparations of chromosome and plasmid DNAs, restriction digestions, ligations, DNA electrophoresis, transformations, and electroporations were carried out as described by Sambrook et al. (30). Sequencing was performed by Genome Express SA (Grenoble, France). Site-directed mutagenesis was performed with the QuickChange kit (Stratagene).
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (13). After electrophoresis, the proteins were electroblotted onto nitrocellulose in a semidry apparatus at 2mA/cm2 for 20 min in transfer buffer containing 40 mM glycine, 50 mM Tris, 0.4% SDS, and 10% methanol. The nitrocellulose was then saturated with gelatin, incubated with antibodies, and developed with the ECL enhanced chemiluminescence detection kit (Pharmacia-Amersham Biotech). Anti-RhiE antibodies were diluted 1:2,000.
Preparation of RhiE antibodies.
Strain A350 was grown overnight in LB medium containing rhamnose. After centrifugation, the proteins contained in the supernatants were precipitated in 50% ethanol and loaded onto preparative SDS-10% polyacrylamide gels. The band containing RhiE was cut out, and the protein was electroeluted. The protein was injected into a rabbit for antibody production by Valbex (Villeurbanne, France).
Protein labeling.
Overexpression and exclusive labeling of plasmid-encoded proteins were carried out with the T7 promoter-T7 polymerase system of Tabor and Richardson (33).
Pathogenicity test.
Pathogenicity tests were performed on chicory leaves. Ten microliters of M63 medium-grown bacteria diluted to an optical density at 600 nm of 0.05 were inoculated into a leaf wounded with a scalpel. Leaves were maintained for 40 h in a dew chamber, and the length of rotted tissue was measured.
Enzyme assays.
β-Glucuronidase assays were performed with toluenized cells grown to exponential phase with p-nitrophenyl-β-d-glucuronate as the substrate (21). β-Galactosidase assays were performed with toluenized cells grown to exponential phase with o-nitrophenyl-β-d-galactose as the substrate (15). Rhamnogalacturonate lyase activity was monitored by measuring the increase of absorbance at 230 nm in a reaction medium containing 0.05% RG-I and enzymatic extract in 100 mM Tris-HCl or citrate-phosphate buffer at 37°C. RG-I was prepared as described by Bonnin et al. (3). Briefly, sugar beet pectin was dispersed in 0.1 M NaOH at 90°C. After 2 h, the pH was adjusted to 6 with HCl and the extracted polymers were precipitated with 3 volumes of ethanol. The precipitate was hydrolyzed with 0.5 M HCl at 80°C. After dialysis and freeze-drying, rhamnogalacturonan (rhamnose/galactose/galacturonic acid molar ratio of 1:1.2:2.8) was recovered.
Nucleotide sequence accession number.
The complete DNA sequence of rhiE will appear in nucleotide databases under accession number AJ438339.
RESULTS
E. chrysanthemi secretes a rhamnose-inducible protein.
While galacturonate is metabolized by E. chrysanthemi, rhamnose, the other main component of pectin, is not. None of the 20 E. chrysanthemi strains tested was able to use rhamnose as the sole carbon source for its growth (Table 2). In addition, no mutant able to grow on agar plates containing rhamnose as the sole carbon source was observed even after several days, in contrast to observations with lactose, 2-keto-3-deoxygluconate, or glucuronate, for which the degradation pathway exists but is not expressed in the wild-type strain (8, 11, 12). This suggests that E. chrysanthemi does not possess a complete rhamnose degradation pathway. However, when the proteins present in the supernatant of a culture of strain A350 grown in the absence or presence of rhamnose were analyzed by SDS-PAGE, a 62-kDa rhamnose-induced protein could be observed, indicating that rhamnose can enter the bacteria and turn on the expression of some genes.
TABLE 2.
Presence of RhiE in several Erwinia strains
| Strain | Host | Geographic origin | RFLP groupa | Growth on rhamnose | Presence of RhiEb |
|---|---|---|---|---|---|
| E. chrysanthemi | |||||
| 3937 | Chrysanthemum | France | NAc | − | + |
| ET3 | Tomato | Martinique | 33 | − | + |
| 4073 | Carnation | Great Britain | NA | − | − |
| 1240 | Carnation | Denmark | 33 | − | − |
| SF109-1 | Sunflower | France | 43 | − | + |
| 4072 | Corn | India | NA | − | − |
| 1271 | Corn | Egypt | 42 | − | − |
| 4040 | Potato | Peru | NA | − | − |
| 722 | Tomato | France | NA | − | − |
| 1270 | Parthenium | Denmark | NA | − | − |
| 4062 | Agalonema | St. Lucia | 46 | − | + |
| SH230 | Tomato | Cuba | 44 | − | + |
| 1342 | Chrysanthemum | Italy | NA | − | + |
| 1891 | Tobacco | United States | 39 | − | − |
| 4060 | Begonia | The Netherlands | NA | − | − |
| 4061 | Colocasia | Solomon Islands | 34 | − | + |
| 4065 | Banana | Colombia | NA | − | − |
| 3367 | Dahlia | France | NA | − | − |
| 3716 | Kalanchoe | France | NA | − | + |
| CIP366 | Potato | Peru | NA | − | − |
| ENA49 | Diffenbachia | Brazil | NA | − | + |
| EC16 | ? | ? | NA | − | + |
| E. carotovora subsp. odorifera 56-487 | + | − | |||
| E. carotovora subsp. betavasculorum 56-479 | + | − | |||
| E. carotovora subsp. carotovora SCRI193 | − | − | |||
| E. carotovora subsp. atroseptica SCRI31 | − | − | |||
| E. carnegiani 55-483 | + | − | |||
| E. cypripedi 51-440 | − | − | |||
| E. rhapontici 49-421 | − | − |
The RFLP groups were defined by Waleron et al. (37).
The presence of RhiE was determined by immunodetection in supernatants of culture grown in the presence of rhamnose.
NA, not applicable.
To find the gene of this secreted protein, we performed mutagenesis of strain A350 with phage MudI1681, which allows for the formation of a transcriptional fusion with the lacZ reporter gene. We looked for colonies which were white on MacConkey-lactose plates and pink or red on MacConkey-lactose-rhamnose plates. Ten mutants containing fusions in rhi (rhamnose-inducible) genes were found. The supernatants of cultures of these 10 mutants grown in LB-rhamnose medium were analyzed by SDS-PAGE. In one of them, A3749, the 62-kDa rhamnose inducible protein was absent. The mutated gene was named rhiE.
Cloning and sequence of rhiE.
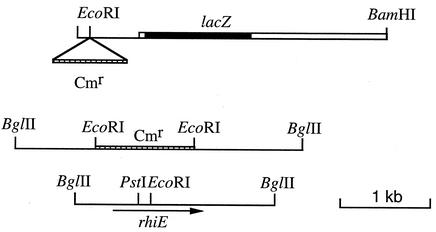
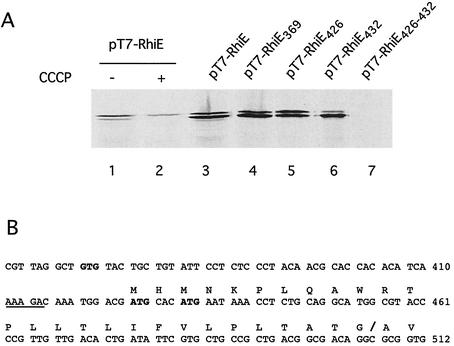
To clone the wild-type allele of rhiE, we first cloned the region upstream of the point of insertion of MudI1681. The rhiE::MudI1681 allele was cloned in vivo on an R110 plasmid. The R′rhiE plasmid was digested with BamHI, and the fragments were ligated in pBluescript. Plasmids containing the lacZ gene and the adjacent E. chrysanthemi DNA were identified as blue colonies after the transformation of E. coli MC1061 and selection on LB-ampicillin-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates. Analysis of the plasmid content of Lac+ colonies obtained by BamHI digestion showed that their insert contained, besides the lacZ gene, 1.7 kb of E. chrysanthemi DNA (Fig. 1). Establishment of a restriction map of this DNA fragment showed that it contained a unique EcoRI site. A Cmr cassette was inserted into this site, and this construction was then introduced into E. chrysanthemi and recombined into the chromosome. The resulting strain, A3884, did not produce RhiE, indicating that the EcoRI site is within the rhiE gene. DNA of strain A3884 was extracted, digested with BglII or NsiI (two enzymes that do not cut the Cmr cassette), and ligated with pBluescript digested with BamHI or PstI. After the transformation of E. coli, Cmr clones were selected. The plasmids obtained with BglII contained the largest inserts, with more than 6 kb of chromosomal DNA (Fig. 1). A partial digestion with EcoRI allowed for the excision of the Cmr cassette of the plasmid and restored the wild-type allele of rhiE. A 2,420-bp DNA fragment surrounding the EcoRI site was sequenced. It contained a single open reading frame beginning at nucleotide 369 and ending at nucleotide 2159 and was followed by a sequence that could act as a Rho-independent terminator. However, the putative GTG initiation codon was not preceded by a classical Shine-Dalgarno sequence (Fig. 2). To check whether this GTG was the initiation codon, it was modified by site-directed mutagenesis to GTA. Synthesis of RhiE, checked by exclusive labeling of the product of the gene cloned under control of the T7 φ10 promoter, was not modified by this mutation (Fig. 2). It is possible that one of two in-frame ATG codons (positions 426 and 432), preceded by a putative Shine-Dalgarno sequence (AAAGA), could be the initiation codon. Mutation of either one of them (to ATC and ATT, respectively) did not abolish RhiE synthesis, indicating that both act as initiation codons (Fig. 2). We showed that both of them had to be mutated to prevent RhiE synthesis (Fig. 2). Since RhiE is a secreted protein, we looked for the presence of a signal sequence. The N-terminal part of the protein is hydrophobic and contains a putative signal peptidase cleavage site (ATG/A) at position 27 from the first methionine residue (position 426) (Fig. 2). This cleavage site was confirmed by sequencing the N terminus of the RhiE protein purified from rhamnose-induced E. chrysanthemi culture supernatant. Labeling of RhiE expressed in E. coli showed two forms of the protein (Fig. 2). Treatment of the cells with carbonyl cyanide m-chlorophenylhydrazone, which blocks signal sequence processing, led to the synthesis of only the larger form (Fig. 2). The smaller form was extractable from the E. coli periplasm (data not shown). Thus, the larger form corresponds to the nonmatured form of RhiE. The inefficient cleavage of the RhiE signal sequence in E. coli could result in the unusual presence of three prolyl residues in this sequence. Mature RhiE is a 551-amino-acid protein with a calculated molecular mass of 61,940 Da.
FIG. 1.
Cloning of rhiE. The thick line represents MudI1681 DNA, and the thin line represents E. chrysanthemi chromosomal DNA. The hatched box represents the Cmr cassette.
FIG. 2.
Determination of the RhiE initiation codon. (A) Wild-type rhiE or rhiE mutated at putative initiation codons was cloned in the pT7-5 vector. The proteins expressed were labeled with [35S]cysteine-[35S]methionine and separated by SDS-PAGE, and the gels were autoradiographed. Lanes 1 and 3, BL21/T7-RhiE; lane 2, BL21/T7-RhiE treated with carbonyl cyanide m-chlorophenylhydrazone; lane 4, BL21/T7-RhiE369 (M369V); lane 5, BL21/T7-RhiE426 (M426I); lane 6, BL21/T7-RhiE432 (M432I); lane 7, BL21/T7-RhiE426-432 (M426I M432I). (B) Nucleotide sequence of the rhiE translation initiation region. Putative initiations codons are in boldface. The Shine-Dalgarno sequence is underlined. The protein sequence is indicated above the nucleotide sequence. The signal sequence cutting site is indicated by a slash. The numbering is that for the nucleotide sequence deposited under accession number AJ438339.
The sequence of RhiE was compared with sequences in databases. The best homologies were found with several proteins of unknown function in Arabidopsis thaliana (35). These proteins have been classified in family 4 of the polysaccharide lyases by Coutinho and Henrissat (10). This family contains two other proteins. Only one of them, produced by A. aculeatus, has been biochemically characterized as a rhamnogalacturonate lyase (18). Homology between these proteins was restricted to their C-terminal parts.
RhiE is secreted by the Out pathway.
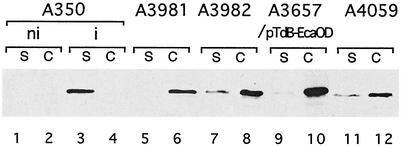
The presence of an N-terminal signal sequence suggested that RhiE secretion occurs via the type II Out pathway, since secretion through type I (Prt) or III (Hrp) secretion pathways is Sec independent (29). The absence of RhiE in the supernatant of an OutC mutant confirmed this hypothesis (Fig. 3). Two of the Out proteins, OutC and OutD, were shown to contain a secretion targeting signal for proteins secreted by the Out secretion machinery (5). The presence of the C-terminal PDZ domain of OutC is required for the secretion of most of the Out-secreted proteins, except for Cel5, PelI, and PemA. Introduction into a ΔoutD E. chrysanthemi mutant of an OutD protein from Erwinia carotovora or a hybrid containing the N-terminal domain of E. carotovora OutD and the C-terminal domain of E. chrysanthemi OutD allowed for the secretion of only PemA and PelI, indicating that the N-terminal domain of E. chrysanthemi OutD contains the information necessary for secretion of most of the Out-secreted proteins. Ten percent of RhiE was secreted from the mutant A3981 (outC ΔPDZ), and no RhiE was secreted by the outD mutant strain A3657 containing plasmid pTdB-EcaOD encoding E. carotovora OutD (Fig. 3). This indicates that RhiE secretion is dependent on the presence of the PDZ domain of OutC and requires the E. chrysanthemi OutD N-terminal domain. Thus, RhiE specificity towards the Out machinery components is similar to that of most of the Out-secreted proteins.
FIG. 3.
Cellular localization of RhiE in various E. chrysanthemi strains. Bacteria were grown in LB medium in the presence of rhamnose (except for lanes 1 and 2). Proteins of the cell fraction (lanes C) and the supernatant (lanes S) were separated by SDS-PAGE and blotted onto a membrane, and the presence of RhiE was detected with anti-RhiE antibodies. Lanes 1 and 2, noninduced A350 (ni); lanes 3 and 4, rhamnose-induced A350 (i); lanes 5 and 6, A3981(outC); lanes 7 and 8, A3982 (outCΔPDZ); lanes 9 and 10, A3657(pTdB-EcaOD); lanes 11 and 12, A4059 (dsbA dsbC).
Several studies have shown the necessity of disulfide bond formation in secreted proteins for their recognition by the secretion machinery (22, 31). However, these studies, performed with dsb mutants, have been weakened by the facts that the absence of disulfide bonds in secreted protein makes them unstable and that dsb mutations could also affect the secretion machinery. RhiE is the first characterized E. chrysanthemi protein which is secreted by the Out pathway but contains no disulfide bond. We studied RhiE secretion in a dsbA dsbC mutant, in which disulfide bonds are not formed in periplasmic proteins. In this mutant, RhiE was stable and accumulated in the cells (Fig. 3). This indicates that the Out machinery is not functional in the dsbA dsbC mutant.
Regulation of rhiE expression.
To study rhiE expression, a transcriptional fusion was constructed. A uidA-kan cassette was introduced into the unique EcoRI site of rhiE. After introduction into E. chrysanthemi, the construction was recombined into the chromosome by successive cultures in low-phosphate medium. The absence of RhiE in the culture supernatant confirmed the replacement of the wild-type gene by the uidA mutant allele in strain A3946. Expression of the fusion was strongly induced in the presence of rhamnose (>200-fold) (Table 3). However, the concentration of rhamnose necessary to reach the fully induced level was very high (>2 g/liter), which suggests that rhamnose does not enter the bacteria by a high-affinity transport system but rather by passive diffusion or by a nonspecific transport system. The induction of rhiE by other sugars present in pectin was tested. None of the sugars (galacturonate, galactose, xylose, fucose, ribose, and arabinose) was found to be an inducer of rhiE expression. The effect of regulatory mutations that affect pectinase gene expression was analyzed. While a mutation in kdgR or pecS did not modify rhiE expression, a mutation in pecT, which controls expression of all of the pectate lyase genes, led to a 1.6-fold increase in rhiE expression (Table 3). Mutations in these three regulators did not change the rhamnose-induced induction level (data not shown). In the presence of glucose as the sole carbon source, rhiE expression is reduced, which suggests a possible role of CRP as an activator of rhiE. This was confirmed when rhiE-uidA was introduced into a crp mutant: the fusion expression level was reduced 100-fold in the presence of rhamnose. rhiE expression increased twofold at the end of the exponential growth phase but was not modified in an expI mutant that no longer synthesizes the E. chrysanthemi major acyl-homoserine lactone (data not shown). The effects of various environmental conditions were tested. In contrast to what is observed for the pectate lyase genes, rhiE expression was not reduced at high temperature (37°C). However, it was reduced at high or low osmolarity of the medium.
TABLE 3.
rhiE expression in various backgrounds and under various induction and growth conditions
| Growth conditionsa | Additional mutation | Carbon source | Inducerb | β-Glucuronidase activityc (mean ± SD) |
|---|---|---|---|---|
| Standard | None | Glycerol | − | 7 ± 2 |
| + | 1,195 ± 190 | |||
| Glucose | − | <1 | ||
| + | 202 ± 41 | |||
| crp | Glucose | − | <1 | |
| + | 10 ± 2 | |||
| kdgR | Glycerol | − | 8 ± 1 | |
| pecS | Glycerol | − | 7 ± 1 | |
| pecT | Glycerol | − | 12 ± 2 | |
| Low osmolarity | None | Glycerol | + | 654 ± 108 |
| High osmolarity | None | Glycerol | + | 825 ± 49 |
| 37°C | None | Glycerol | + | 1,203 ± 94 |
Standard growth conditions were obtained by cultivation in M63 medium at 30°C with shaking at 200 rpm. Low-osmolarity medium was four fold-diluted M63 medium. High osmolarity was obtained by adding 0.3 M NaCl to M63 medium.
The inducer was 0.2% rhamnose.
β-Glucuronidase activity is expressed as nanomoles of p-nitrophenol formed per minute per milligram (dry weight) of bacteria.
Occurrence of RhiE in other Erwinia strains.
The supernatants of rhamnose-induced cultures of 21 strains of E. chrysanthemi and other Erwinia species (E. rhapontici, E. cypripedi, E. carnegiani, E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, E. carotovora subsp. odorifera, and E. carotovora subsp. betavasculorum) were examined for the presence of RhiE. Material reacting with the anti-RhiE antibody was detected in one-third of the E. chrysanthemi strains and in none of the other Erwinia strains, indicating that a homologue of rhiE is not present, or is not expressed, under these conditions in these strains (Table 2). No correlation could be found between the expression of RhiE, the geographical or plant origin of the Erwinia strains, their RecA restriction fragment length polymorphism (RFLP) group (37), and their ability to grow with rhamnose as the sole carbon source.
Characterization of RhiE activity.
The homology of RhiE with a family of proteins which includes a rhamnogalacturonate lyase led us to suppose that this protein could possess the same activity. We tried to detect lyase activity by measuring an increase in absorbance at 230 nm with several substrates (polygalacturonate, pectin, RG-I, and arabinogalactan) in the presence of an extract of E. coli overproducing RhiE. We could detect activity only with RG-I, confirming that RhiE is a rhamnogalacturonate lyase which cleaves the RG-I backbone between a rhamnose and a galacturonate and creates an unsaturated bond between carbons 4 and 5 of the galacturonate residue. RhiE activity was not increased by the addition of 0.2 mM Ca2+, Mn2+, Co2+, Cu2+, or Zn2+ and was not modified by the addition of 2 mM EDTA. The optimum pH of activity was 6.0. A possible role of RhiE in E. chrysanthemi pathogenicity was investigated by comparing the length of rotted tissue produced by the wild-type strain and by a rhiE mutant on chicory leaves. A significant decrease in the length of rotted tissue (13 ± 5 versus 26 ± 8 cm) showed that rhiE is required for the full virulence of E. chrysanthemi strain 3937. The purified enzyme was not able to provoke maceration of potato tubers or chicory leaves.
DISCUSSION
The plant-pathogenic bacterium E. chrysanthemi synthesizes and secretes a full range of enzymes that are able to degrade the smooth region of pectin, whose synthesis is induced by PGA or galacturonate. To identify enzymes able to depolymerize the backbone of RG-I, we analyzed the supernatants of rhamnose-induced cultures. The rhamnogalacturonate backbone of pectin can be cleaved by rhamnogalacturonate rhamnohydrolases and rhamnogalacturonate lyases, which cleave the α-1,4 linkage, or by rhamnogalacturonate galacturonohydrolases, which cleave the α-1,2 bond. Several of these enzymes can be produced in the same organism, as described for A. aculeatus (16, 17, 18). Only one major protein, RhiE, which belongs to the rhamnogalacturonate lyase family, was detected in E. chrysanthemi supernatants. Thus, in contrast to the case for the multiprotein family of pectate lyases, only one rhamnogalacturonase seems to be produced by this bacterium to degrade the RG-I backbone. Other possible explanations for the absence of detection of other rhamnose-induced secreted proteins is that they are synthesized in small amounts or that the induction conditions used did not allow for their synthesis. The gene rhiE does not seem to be present in all of the strains tested. The absence of rhamnose metabolism by E. chrysanthemi could render useless, and energetically costly, the complete degradation of the rhamnogalacturonate part of pectin into monosaccharides, which cannot subsequently be used as a carbon source. The absence of rhiE in strain A350 reduces the virulence of the bacterium on chicory leaves. Even if RhiE is not useful in providing carbon sources for the bacterium, its enzymatic activity could be required to liberate pectin smooth regions and increase their accessibility to pectate lyase action. E. chrysanthemi pathogenicity is not due to a single determinant but results from the addition of several factors that lead to bacterial cell wall degradation. The presence of RhiE in some strains could complete their arsenal of degrading enzymes and increase their virulence.
RhiE regulation was compared with that of other pectinases. The main inducing condition seems to be the presence of rhamnose, which does not induce the synthesis of the pectate lyases. Among the specific regulators of the pectate lyase genes tested, only pecT weakly controls rhiE expression. Besides the genes of all of the pectate lyases, pecT also regulates the expression of other virulence factors, such as the synthesis of exopolysaccharides (7). rhiE is also activated by CRP, the main activator of virulence factor genes in E. chrysanthemi (24). Thus, in addition to a specific regulation by rhamnose, rhiE is included in the regulatory network that controls E. chrysanthemi virulence factors.
In contrast to other Out-secreted proteins studied up to now, RhiE does not contain any cysteine residue. This led to a reexamination of the role of disulfide bond formation in Out secretion. RhiE accumulated in the periplasm of a dsbA dsbC mutant, unable to form disulfide bonds in the periplasmic proteins. This nonfunctionality of the Out machinery could result from the absence of disulfide bond formation in two proteins, OutS and OutK (reference 22 and unpublished results). However, results obtained with the cellulase Cel5 mutated at cysteine residues showed that disulfide bond formation is required for the secretion of Cel5 by E. chrysanthemi (4). Thus, the formation of disulfide bonds is required to stabilize not only the secreted proteins but also the secretion machinery.
RhiE does not require Ca2+ or other divalent cations for its activity, and its optimum pH is around 6.0. These properties are similar to those of A. aculeatus rhamnogalacturonate lyase and of pectin lyases (18, 36) and differ from those of P. cellulosa rhamnogalacturonate lyase and of pectate lyases, which have an absolute requirement for divalent cations (most frequently Ca2+) and are active at basic pH (14, 34). Rhamnogalacturonate lyases are found in two families of polysaccharide lyases in the classification of Coutinho and Henrissat (10). P. cellulosa rhamnogalacturonate lyase belongs to family PL11 (14), which contains only bacterial proteins. RhiE is the first bacterial rhamnogalacturonate lyase characterized in the PL4 family, which contains the A. aculeatus rhamnogalacturonate lyase (18). The A. thaliana MYST proteins, which also belong to family PL4, were thought to be dicotyledon-specific proteins and have no characterized activity (35). These proteins are probably also rhamnogalacturonate lyases. Thus, PL4 could be a widespread family of proteins with rhamnogalacturonate lyase activity found in plants, fungi, and bacteria. Rhamnogalacturonate lyases of PL4 and PL11 differ in their catalytic properties (pH and ion requirements), and further analysis is required to understand the basis of these differences.
Acknowledgments
We thank E. Bonnin for providing RG-I, E. Lojkowska for providing a collection of Erwinia strains, V. E. Shevchik for advice, and N. Cotte-Pattat, S. Reverchon, and V. James for reading the manuscript.
This work was supported by grants from the Centre National de la Recherche Scientifique and from the Ministère de l'Education Nationale et de la Recherche.
REFERENCES
- 1.Albersheim, P., A. G. Darvill, M. A. O'Neill, H. A. Schols, and A. G. J. Voragen. 1996. An hypothesis: the same six polysaccharides are components of the primary cell walls of all higher plants, p. 47-53. In J. Visser and A. G. J. Voragen (ed.), Pectins and pectinases. Elsevier, Amsterdam, The Netherlands.
- 2.Bauer, D. W., A. J. Bogdanove, S. V. Beer, and A. Collmer. 1994. Erwinia chrysanthemi hrp genes and their involvement in soft rot pathogenesis and elicitation of the hypersensitive response. Mol. Plant-Microbe Interact. 7:573-581. [DOI] [PubMed] [Google Scholar]
- 3.Bonnin, E., M. Brunel, Y. Gouy, L. Lesage-Meesen, M. Asther, and J.-F. Thibault. 2001. Aspergillus niger I-1472 and Pycnoporus cinnabarinus MUCL39533, selected for the biotransformation of ferulic acid to vanillin, are also able to produce cell wall polysaccharide-degrading enzymes and feruloyl esterases. Enzyme Microb. Technol. 28:70-80. [DOI] [PubMed] [Google Scholar]
- 4.Bortoli-German, I., E. Brun, B. Py, M. Chippaux, and F. Barras. 1994. Periplasmic disulfide bond formation is essential for cellulase secretion by the plant pathogen Erwinia chrysanthemi. Mol. Microbiol. 11:545-553. [DOI] [PubMed] [Google Scholar]
- 5.Bouley, J., G. Condemine, and V. E. Shevchik. 2001. The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II Out pathway of Erwinia chrysanthemi. J. Mol. Biol. 308:205-219. [DOI] [PubMed] [Google Scholar]
- 6.Castilho, B. A., P. Olfson, and M. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condemine, G., A. Castillo, F. Passeri, and C. Enard. 1999. The PecT repressor coregulates synthesis of exopolysaccharides and virulence factors in Erwinia chrysanthemi. Mol. Plant-Microbe Interact. 12:45-52. [DOI] [PubMed] [Google Scholar]
- 8.Condemine, G., and J. Robert-Baudouy. 1987. 2-Keto-3-deoxygluconate transport system in Erwinia chrysanthemi. J. Bacteriol. 169:1972-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condemine, G., and J. Robert-Baudouy. 1987. Tn5 insertion in kdgR, a regulatory gene of the polygalacturonate pathway in Erwinia chrysanthemi. FEMS Microbiol. Lett. 42:39-46. [Google Scholar]
- 10.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes. [Online.] http://afmb.cnrs-mrs.fr/CAZY/index.html.
- 11.Hugouvieux-Cotte-Pattat, N., and J. Robert-Baudouy. 1985. Lactose metabolism in Erwinia chrysanthemi. J. Bacteriol. 162:248-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugouvieux-Cotte-Pattat, N., and J. Robert-Baudouy. 1987. Hexuronate catabolism in Erwinia chrysanthemi. J. Bacteriol. 169:1223-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.McKie, V. A., J. P. Vincken, A. G. J. Voragen, L. A. M. van den Broek, E. Stimson, and H. J. Gilbert. 2001. A new family of rhamnogalacturonan lyases contains an enzyme that binds to cellulose. Biochem. J. 355:167-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Mutter, M., G. Beldman, S. M. Pitso, H. A. Schols, and A. G. J. Voragen. 1998. Rhamnogalacturonan α-d-galactopyranosyluronohydrolase, an enzyme that specifically removes the terminal nonreducing galacturonosyl residue in rhamnogalacturonan regions in pectin. Plant Physiol. 117:153-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mutter, M., G. Beldman, H. A. Schols, and A. G. J. Voragen. 1994. Rhamnogalacturonan α-l-rhamnopyranohydrolase, a novel enzyme specific for the terminal nonreducing rhamnosyl unit in rhamnogalacturonan regions of pectin. Plant Physiol. 106:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mutter, M., I. J. Colquhoun, H. A. Schols, G. Beldman, and A. G. J. Voragen. 1996. Rhamnogalacturonase B from Aspergillus aculeatus is a rhamnogalacturonan α-l-rhamnopyranosyl-(1-4)-α-d-galactopyranosyluronide lyase. Plant Physiol. 110:73-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasser, W., M. Bouillant, G. P. C. Salmond, and S. Reverchon. 1998. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl homoserine lactone signal molecules. Mol. Microbiol. 29:1391-1405. [DOI] [PubMed] [Google Scholar]
- 20.Nelson, M. A., S. T. Merino, and R. L. Metzenberg. 1997. A putative rhamnogalacturonase required for sexual development of Neurospora crassa. Genetics 146:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novel, G., M.-L. Didier-Fichet, and F. Stoeber. 1974. Inducibility of β-glucuronidase in wild-type and hexuronate-negative mutants of Escherichia coli K-12. J. Bacteriol. 102:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pugsley, A. P., N. Bayan, and N. Sauvonnet. 2001. Disulfide bond formation in secreton component PulK provides a possible explanation for the role of DsbA in pullulanase secretion. J. Bacteriol. 183:1312-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Résibois, A., M. Colet, M. Faelen, E. Schoonejans, and A. Toussaint. 1984. Phi-EC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology 137:102-112. [DOI] [PubMed] [Google Scholar]
- 24.Reverchon, S., D. Expert, J. Robert-Baudouy, and W. Nasser. 1997. The cyclic AMP receptor protein is the main activator of the pectinolysis genes in Erwinia chrysanthemi. J. Bacteriol. 179:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1994. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol. Microbiol. 11:1127-1139. [DOI] [PubMed] [Google Scholar]
- 26.Robert-Baudouy, J., W. Nasser, G. Condemine, S. Reverchon, V. E. Shevchik, and N. Hugouvieux-Cotte-Pattat. 2000. Pectic enzymes of Erwinia chrysanthemi, regulation and role in pathogenesis, p. 221-268. In G. Stacey and N. T. Keen (ed.), Plant-microbe interactions, vol. 5. APS Press, St. Paul, Minn.
- 27.Roeder, D. L., and A. Collmer. 1985. Marker exchange mutagenesis of a pectate lyase isoenzyme in Erwinia chrysanthemi. J. Bacteriol. 164:51-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto, M., Y. Shirane, I. Naribayashi, K. Kimura, N. Morishita, T. Sakamoto, and T. Sakai. 1994. Purification and characterization of a rhamnogalacturonase with protopectinase activity from Trametes sanguinea. Eur. J. Biochem. 226:285-291. [DOI] [PubMed] [Google Scholar]
- 29.Salmond, G. P. C., and P. J. Reeves. 1993. Membrane traffic wardens and protein secretion in Gram-negative bacteria. Trends Biochem. Sci. 18:7-12. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Shevchik, V. E., I. Bortoli-German, J. Robert-Baudouy, S. Robinet, F. Barras, and G. Condemine. 1995. Differential effect of dsbA and dsbC mutations on extracellular enzyme secretion in Erwinia chrysanthemi. Mol. Microbiol. 16:745-753. [DOI] [PubMed] [Google Scholar]
- 32.Surgey, N., J. Robert-Baudouy, and G. Condemine. 1996. The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J. Bacteriol. 178:2503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabor, S., and C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tardy, F., W. Nasser, J. Robert-Baudouy, and N. Hugouvieux-Cotte-Pattat. 1997. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J. Bacteriol. 179:2503-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavares, R., S. Aubourg, A. Lecharny, and M. Kreis. 2000. Organization and structural evolution of four multigene families in Arabidopsis thaliana: AtLCAD, AtLGT, AtMYST and AtHD-GL2. Plant Mol. Biol. 42:703-717. [DOI] [PubMed] [Google Scholar]
- 36.Vitali, J., B. Schick, H. C. Kester, J. Visser, and F. Jurnak. 1998. The three-dimensional structure of Aspergillus niger pectin lyase B at 1.7-A resolution. Plant Physiol. 116:69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waleron, M., K. Waleron, A. J. Podhajska, and E. Lojkowska. 2002. Genotyping of bacteria belonging to the former Erwinia genus by PCR-RFLP analysis of a recA gene fragment. Microbiology 148:583-595. [DOI] [PubMed] [Google Scholar]