Abstract
We recently identified a Bacillus anthracis glycoprotein which is a structural constituent of the exosporium filaments (P. Sylvestre, E. Couture-Tosi, and M. Mock, Mol. Microbiol. 45:169-178, 2002). This Bacillus collagen-like protein (BclA) contains an internal collagen-like region (CLR) of GXX repeats which includes a large proportion of GPT triplets. Here, we report that the polymorphic marker Ceb-Bams13, for which there are nine alleles (P. Le Flèche et al., BMC Microbiol. 1:2, 2001), maps within the open reading frame encoding BclA. The bclA gene in 11 B. anthracis strains representative of seven Ceb-Bams13 alleles was sequenced and compared to the Ames bclA gene sequence. The amino- and carboxy-terminal sequences surrounding the CLR are conserved. The CLR itself is highly polymorphic: it contains between 17 and 91 GXX repeats and one to eight copies of the 21-amino-acid sequence (GPT)5GDTGTT, named the BclA repeat. The length of the filament on the spore surface differed between the strains. We exchanged the bclA gene between strains with different CLRs and examined the spore surfaces by electron microscopy analysis. The length of the BclA CLR is responsible for the variation in filament length.
Bacillus anthracis, the etiological agent of anthrax, is an aerobic, gram-positive, spore-forming bacterium. B. anthracis spores can survive for several decades in the soil (23). Spores infect the host via intradermal inoculation, ingestion, or inhalation (6, 12, 14). After entry into the mammalian host, spores germinate and yield toxin-producing, capsulated bacilli. Toxemia and septicemia rapidly lead to death.
The exosporium is the outermost integument surrounding the mature B. anthracis spore. It is a loose-fitting, balloon-like structure covering the whole spore (5). Electron microscopy has revealed that the exosporium is composed of a paracrystalline basal layer with an hexagonal lattice structure and a hair-like outer region (2, 7, 9, 13, 22, 28). Recently, we identified a B. anthracis exosporium glycoprotein which is a structural constituent of the hair-like filaments on the outer layer of the exosporium (31). This glycoprotein contains a central region composed of GXX collagen-like repeats and was named BclA (for Bacillus collagen-like protein). It contains 70 GPT repeats and a 21-amino-acid sequence, (GPT)5GDTGTT, is repeated six times. We named this 21-amino-acid sequence the BclA repeat. Unlike the two closely related species Bacillus cereus and Bacillus thuringiensis, B. anthracis is considered to be an extremely monomorphic species. However, polymorphic markers containing tandem repeats have been found in the B. anthracis genome, and a multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA) has been developed for molecular typing of B. anthracis strains (11). It uses eight genetic loci on the chromosome and on the virulence plasmids pXO1 and pXO2. Five of the chromosomal markers (vrrA, vrrB1, vrrB2, vrrC1, and vrrC2) are found in open reading frames (ORFs), and variation in repeat number does not affect the translational reading frame. However, the protein products encoded by these five ORFs have not been identified. Five alleles were described for vrrA (1, 10). They differ in a 12-bp VNTR, with two to six copies present. The vrrA locus is found within an ORF that encodes a putative 30-kDa glutamine-rich protein (1, 10). For the vrrB locus, a total of 11 alleles were found within eight different size classes, resulting from combinations of nine nucleotide insertion-deletion polymorphisms (29). This ORF putatively codes for a 241- to 265-amino-acid protein, rich in glutamine, glycine, and histidine, with predicted molecular masses ranging from 24.9 to 27.8 kDa. The other markers (vrrC1 and vrrC2) have between 3 and 6 alleles (11).
MLVA provides good discrimination between different isolates. A worldwide B. anthracis collection (426 isolates) was classified into 89 distinct MLVA genotypes that fall into six major genetic groups which form two clusters themselves (A and B). Members of the A cluster are found worldwide and can be subdivided into four groups (A1, A2, A3, and A4). Cluster B is subdivided into two groups, B1 and B2. Only two genotypes are present in the B2 group. They are rare and have only been found in Europe (11).
Le Flèche and collaborators (15) described the use of a tandem repeat database to characterize polymorphic markers in B. anthracis. Fourteen markers were described, five of which are highly polymorphic. Here, we report that one of these five polymorphic markers, Ceb-Bams13, maps within bclA. We analyzed the bclA gene in various B. anthracis strains representative of different genotypes. The BclA polymorphism maps to the sequence encoding the internal collagen-like region (CLR), with a number of contiguous GXX triplets, varying from 17 to 91. We previously showed that BclA is a structural component of the filaments covering the outer layer of the exosporium (31). In an earlier study, Kramer and Roth (13) reported that the length of these hair-like projections was different in the Vollum and the Sterne B. anthracis strains. Here, we show that variations in the size of the CLR correlated to, and indeed are responsible for, the variation of filament length on the spore surface.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The 11 B. anthracis strains used in this work are listed in Table 1. The strains are from the following culture collections: Collection Institut Pasteur, strains 7702 Sterne, 8189, 53169, A2, and 5725; American Type Culture Collection, strains 6602 and 4229; and Agence Française de Sécurité Sanitaire des aliments, strain 6183. The other strains included in this study were previously described by Patra and collaborators (24) (strains RA3R and 7611R) and by Berthier and collaborators (3) (strain 9602). The strains A2R, 9602R, 6183R, and 5725R, derivatives for which R indicates a rough colony phenotype due to the spontaneous loss of pXO2, were isolated in our laboratory by growing the corresponding wild-type B. anthracis strains on chloramphenicol plates in an atmosphere of 5% CO2-95% air (30).
TABLE 1.
Analysis of exosporium filament length in parental strains and in complemented deletion mutants
| Strain | Plasmid content or bclA genotype | Ceb-Bams13 marker allele (reference) | Exosporium filament length (nm)a |
|---|---|---|---|
| 7702 | pXO1+ | 10 (15) | 60.84 ± 5.88 (81) |
| 7702b | pXO1+ | 10 (15) | 52.16 ± 5.83 (50) |
| 8189b | pXO1+, pXO2+ | 9 (15) | 47.29 ± 4.77 (70) |
| 53169b | pXO1+, pXO2+ | 7 (15) | 48.73 ± 2.62 (34) |
| 9602R | pXO1+ | 5 (15) | 29.20 ± 3.00 (52) |
| A2R | pXO1+ | 5 (15) | 27.69 ± 2.15 (45) |
| 6183R | pXO1+ | 5 (this work) | 28.14 ± 3.25 (57) |
| 6602 | pXO2+ | 4 (15) | 29.79 ± 3.71 (47) |
| RA3R | pXO1+ | 4 (this work) | 23.69 ± 2.67 (52) |
| 7611R | pXO1+ | 4 (this work) | 25.95 ± 4.11 (41) |
| 5725R | pXO1+ | 3 (15) | 25.12 ± 2.68 (75) |
| 4229 | pXO2+ | 1 (15) | 13.99 ± 3.18 (58) |
| PF31 | 7702 ΔbclA eag::bclA7702 | 61.30 ± 5.41 (51) | |
| PF34 | 7702 ΔbclA eag::bclA9602 | 25.76 ± 3.69 (45) | |
| PF32 | 9602R ΔbclA eag::bclA7702 | 60.23 ± 4.30 (52) | |
| PF33 | 9602R ΔbclA eag::bclA9602 | 24.85 ± 3.04 (30) | |
| PF35 | RA3R ΔbclA eag::bclA7702 | 59.76 ± 3.97 (60) | |
| PF36 | RA3R ΔbclA eag::bclA9602 | 24.09 ± 3.85 (54) | |
| PF37 | 7611R ΔbclA eag::bclA7702 | 59.09 ± 3.94 (44) | |
| PF38 | 7611R ΔbclA eag::bclA9602 | 25.41 ± 3.03 (30) |
Results are expressed in nanometers as means ± standard deviations with the number of measurements in parentheses.
Virulent spores were fixed in 4% formaldehyde before negative staining. When strain 7702 was analyzed under the same conditions, the filament length was reduced by about 14.3%.
Escherichia coli TG1 (18) and HB101(pRK212.1) (33) were used for cloning and mating experiments. E. coli was grown in L broth or on L agar (21), and B. anthracis was grown in brain heart infusion medium (Difco Laboratories) or on NBY medium for spore preparation (8). Antibiotic concentrations in the media were as follows: ampicillin, 100 μg/ml for E. coli; kanamycin, 40 μg/ml for E. coli; spectinomycin, 100 μg/ml for both E. coli and B. anthracis; erythromycin, 180 μg/ml for E. coli and 5 μg/ml for B. anthracis.
Preparation of spores.
B. anthracis spores were prepared as previously described (31).
PCR and sequencing.
PCR was performed with rTaq polymerase (Pharmacia) and synthetic oligonucleotides hybridizing to flanking or internal sequences of the bclA gene. Template DNA was prepared by boiling B. anthracis bacteria, grown on a brain heart infusion agar plate overnight, in 500 μl of sterile water for 15 min, followed by centrifugation at 10,000 × g for 5 min. A long segment including the entire bclA ORF was amplified by using the primers pex 1 (5′-CCGTTAGAATCCATTGCAAGATGATAAGGC-3′) and pex 2 (5′-CGACCAACCATACTGTGTGCAGCTCTTGGC-3′). The sequence encoding the CLR was amplified by using the primers pex 5 (5′-CCCTAATCTTGTAGGACCTACATTACCACC) and pex 6 (5′-CCCACCGGAGTTAAATGCATATAGTCCTGC). DNA was denatured at 94°C for 5 min. Thirty amplification cycles of 30 s of denaturation at 94°C, 30 s of annealing at 58°C, and 2 min of extension at 72°C were performed followed by one cycle of 7 min at 72°C. The PCR products were analyzed by agarose gel electrophoresis. The amplicons generated by primers pex 1 and pex 2 were sequenced by using the two primers flanking the bclA gene, pex 3 (5′-CGTGTCATTTTCTTTCGGTTTTGCATCTAC) and pex 4 (5′-GTGCCTCCTACGGAATGTCATACAAC), and the Taq-dyed dideoxy terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) with an ABI-377 instrument.
Plasmid constructions.
The DNA fragments encoding the bclA genes were amplified by PCR from chromosomal DNA of B. anthracis 7702 or 9602R as the template and pex 1 and pex 2 as the primers.
The 7702 amplicon was digested with BamHI and HincII and then inserted into pUC19 (39) previously digested with BamHI and Ecl136II, to give pUC19-bclA. The SmaI fragment from pUC1318 Erm, containing the erythromycin resistance cassette (35), was inserted into pUC19-bclA previously cut with SfiI, thus deleting an internal 369-bp fragment (31); the resulting plasmid was thus pUC19-bclA-Erm (pSP14). The EcoRI-HindIII fragment from pSP14 containing the disrupted bclA gene was then inserted into pFX113 (4) previously digested with EcoRI and HindIII to give pFX113-bclA-Erm (pSP15).
The 7702 and 9602 amplicons were digested with BamHI and HincII and inserted into pAT28 (34) digested with BamHI and Ecl136II to give pAT28-bclA7702 and pAT28-bclA9602, respectively. The bclA genes of strains 7702 and 9602 were then inserted into the eag gene as follows: the EcoRI-SalI fragments of pAT28-bclA7702 and pAT28-bclA9602 were blunt ended with Vent polymerase and ligated to pSAL322 (20), itself digested with BamHI and blunt ended, to give pSAL322-bclA7702 (pSP21) and pSAL322-bclA9602 (pSP22), respectively.
Construction of recombinant strains.
Recombinant plasmids were transferred from E. coli to B. anthracis by heterogramic conjugation (33). Allelic exchange was carried out as previously described by Pezard and collaborators (25).
pSP15 was introduced into strains 7702, 9602R, RA3R, and 7611R, and the recombinant strains PF09 (7702 ΔbclA), PF11 (9602R ΔbclA), PF13 (RA3R ΔbclA), and PF14 (7611R ΔbclA) were obtained after allelic exchange.
The recombinant strains PF31 (7702 ΔbclA eag::bclA7702), PF32 (9602R ΔbclA eag::bclA7702), PF35 (RA3R ΔbclA eag::bclA7702), PF37 (7611R ΔbclA eag::bclA7702), PF34 (7702 ΔbclA eag::bclA9602), PF33 (9602R ΔbclA eag::bclA9602), PF36 (RA3R ΔbclA eag::bclA9602), and PF38 (7611R ΔbclA eag::bclA9602) were obtained by allelic exchange between the mutated eag::bclA gene in pSP21 or pSP22 and the chromosomal eag locus in PF09, PF11, PF13, or PF14.
Immunoblot analysis and glycoprotein detection.
Spore extracts were prepared and Western blotting was performed as previously described (31) with the anti-BclA monoclonal antibody 35B8. Glycoprotein detection was performed as previously described (31).
Electron microscopy analysis.
Spores were analyzed after rehydratation and negative staining as previously described (31). Filament lengths were determined as the mean of at least 30 measurements at different points on the spore border of the distance from the edge of the exosporium crystal to the extremity of the filaments. For each strain, lengths were measured on a minimum of five spores and from at least three independent experiments. The results are expressed as the means of filament length (in nanometers) ± standard deviation. For strains 8189 and 53169 carrying the virulence plasmids pXO1 and pXO2, spores were fixed with 4% formaldehyde before negative staining.
Nucleotide sequence accession numbers.
The GenBank accession number for the bclA genes from B. anthracis strains listed in Table 1 are as follows (strains are given in parentheses): AJ516936 (Ames), AJ516937 (7702), AJ516938 (8189), AJ516939 (53169), AJ516940 (A2), AJ516941 (9602), AJ516942 (6183), AJ516943 (6602), AJ516944 (7611), AJ516945 (RA3), AJ516946 (5725), and AJ516947 (4229).
RESULTS
Distribution of bclA gene and variation of BclA protein.
The BclA glycoprotein was first described from spores of the B. anthracis Sterne strain, and we identified the corresponding bclA gene from the database genome sequence of the Ames strain (www.tigr.org) (31). Here, we found that the polymorphic marker Ceb-Bams13 identified by Le Flèche and collaborators (15) is indeed localized within the ORF encoding BclA. Nine alleles of this marker have been described in a total of 31 B. anthracis strains typed. We therefore analyzed the expression of bclA in these strains by testing for protein production with an anti-BclA monoclonal antibody. Spores of all strains were labeled in an indirect immunofluorescence test, indicating that all strains produce BclA (data not shown).
We used 11 strains to investigate the size of the bclA genes and of the corresponding proteins (Table 1): 8 strains were representative of the seven alleles described for the Ceb-Bams13 marker (15) and 3 French strains were representative of 3 different genotypes (genotypes 3, 79, and 80) of the 89 described by MLVA analysis (11). The DNA fragment encoding the CLR of bclA in each strain was amplified by PCR. The PCR products were about 240 to 900 bp (Fig. 1). The entire bclA gene from each of the 11 strains was sequenced and compared to that of bclA from the Ames sequence (www.tigr.org) (Table 2). The number of amino acids of the corresponding BclA protein varied from 223 in strain 4229 to 445 in Sterne strain 7702; the calculated molecular masses of the mature BclA proteins were between 19.7 and 38.7 kDa. The predicted pIs for all of the proteins were acidic (3.4 to 3.7).
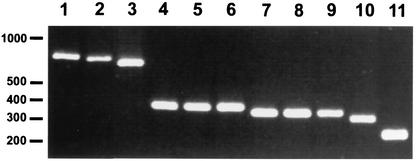
FIG. 1.
Differences in the size of the bclA gene in 11 different B. anthracis strains. The sequences encoding the CLR were amplified by PCR and analyzed by 2% agarose gel electrophoresis. Lane 1, strain 7702; lane 2, strain 8189; lane 3, strain 53169; lane 4, strain 9602R; lane 5, strain A2R; lane 6, strain 6183R; lane 7, strain 6602; lane 8, strain RA3R; lane 9, strain 7611R; lane 10, strain 5725R; lane 11, strain 4229. Molecular size markers (in base pairs) are indicated in the left margin.
TABLE 2.
Analysis of the BclA proteins in 12 B. anthracis strains
| Strain(s) | No. of amino acids in:
|
No. of GXX/GPT repeats in CLR | No. of BclA repeatsa in CLR | No. of GPT triplets before first BclA repeata in CLR | No. of GXX triplets after last BclA repeata in CLR | |
|---|---|---|---|---|---|---|
| BclA | CLR | |||||
| 7702 | 445 | 273 | 91/71 | 8 | 8 | 14 |
| 8189 | 391 | 219 | 73/57 | 6 | 5 | 13 |
| Ames | 382 | 210 | 70/54 | 6 | 5 | 13 |
| 53169 | 370 | 198 | 66/52 | 5 | 5 | 13 |
| 9602R, A2R, 6183R | 262 | 90 | 30/22 | 2 | 5 | 11 |
| 6602 | 253 | 81 | 27/21 | 1 | 6 | 14 |
| RA3R, 7611R | 253 | 81 | 27/21 | 1 | 9 | 11 |
| 5725R | 244 | 72 | 24/18 | 1 | 6 | 11 |
| 4229 | 223 | 51 | 17/11 | 1 | 0 | 10 |
The BclA repeat is defined as the 21-amino-acid sequence (GPT)5GDTGTT.
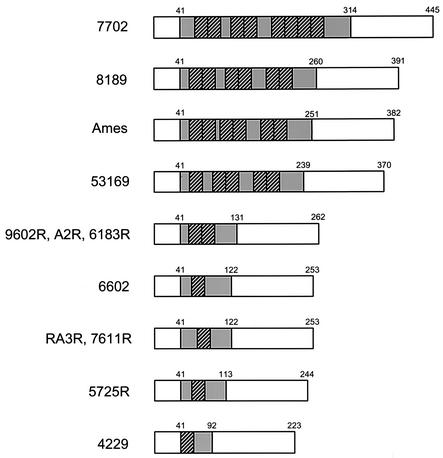
The differences in sequence were entirely restricted to the CLR; the 40 amino-terminal and 132 carboxy-terminal amino acids flanking the CLR were identical in all strains. All BclA sequences studied differed from the Ames BclA protein sequence (Fig. 2). The BclA proteins encoded by strains 9602R, A2R, and 6183R are identical, and those encoded by strains RA3R and 7611R are also identical to each other.
FIG. 2.
Schematic comparison of predicted BclA proteins in 12 B. anthracis strains. The area shaded in grey represents the CLR, consisting of a variable number of GXX triplet motifs. The amino acid positions of the start and end of the CLR are indicated. Within the CLR, the BclA repeats of 21 amino acids are represented with striped boxes. They are flanked by different numbers of GPT triplets.
The BclA proteins produced by the different strains were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting of spore extracts as previously described (31). A high-molecular-weight immunoreactive band was detected in each strain (Fig. 3) but with different sizes in different strains. Three additional reactive bands were detected in both RA3R and 7611R spore extracts. All of the immunoreactive bands were glycosylated (data not shown).
FIG. 3.
Analysis of spore extracts from 11 B. anthracis strains. Proteins were separated by SDS-7% PAGE and analyzed by immunoblotting with anti-BclA monoclonal antibody. Lane 1, strain 7702; lane 2, strain 8189; lane 3, strain 53169; lane 4, strain 9602R; lane 5, strain A2R; lane 6, strain 6183R; lane 7, strain 6602; lane 8, strain RA3R; lane 9, strain 7611R; lane 10, strain 5725R; lane 11, strain 4229. The sizes of the molecular mass markers are given in kilodaltons in the left margin.
Analysis of the BclA CLR.
The CLRs of the different BclA proteins varied from 51 to 273 amino acids in length and are composed of GXX units, including a large proportion of GPT triplets (Table 2). The proline and threonine contents of this region are 23.5 to 27.2% and 31.4 to 35.5%, respectively. The abundance of these residues probably contributes to the slow migration of BclA proteins in SDS-PAGE (31). Within the CLRs there were one to eight copies of the 21-amino-acid sequence (GPT)5GDTGTT, named the BclA repeat (Fig. 2). In all strains carrying at least two BclA repeats, they were grouped in pairs, and in strain 7702, four motifs were clustered together. There were diverse numbers of GPT triplets upstream from the first BclA repeat, downstream from the last one, and between BclA repeat groups. In the CLRs of three strains (strains RA3R, 7611R, and 6602) containing 81 residues with one BclA repeat, two different organizations of the GXX triplets were observed. Thus, the CLR displays diverse numbers and organizations of the GXX triplets, resulting in this segment of BclA being highly polymorphic.
Comparative analysis of the exosporium filaments.
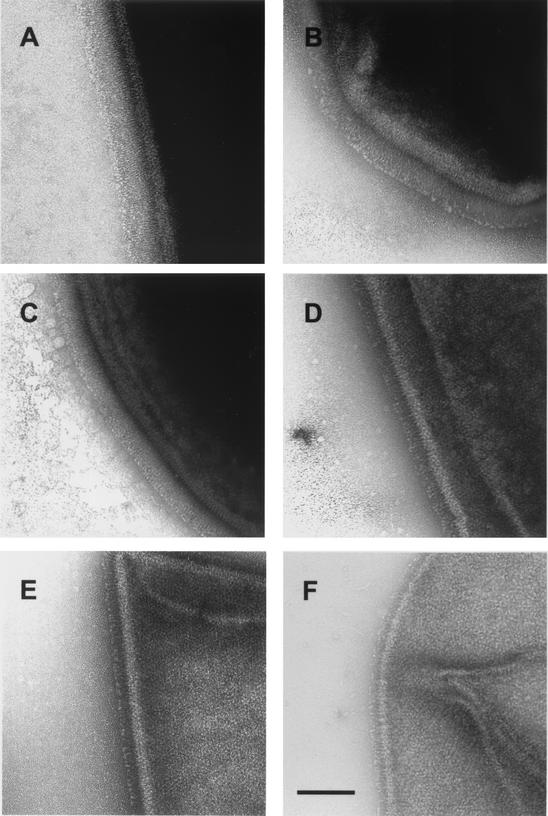
The exosporium from the different strains of B. anthracis was examined by electron microscopy for the presence of filaments of which BclA is the structural component (31). The main difference between strains was the length of filaments (Table 1). Strain 7702 had the longest projections (Fig. 4A), and strain 4229 had the shortest projections (Fig. 4F). The filament lengths were directly proportional to the lengths of the CLRs. The exosporium of strains RA3R, 7611R, and 6602, which contain a CLR of 81 residues with two different organizations, were ultrastructurally indistinguishable.
FIG. 4.
Electron microscopy analysis of the spore surface after negative staining. (A) Strain 7702; (B) strain 8189; (C) strain 53169; (D) strain 6183R; (E) strain 5725R; (F) strain 4229. Bar, 100 nm.
To determine whether CLR length is directly responsible for the length of the filaments, four complementation assays were performed. bclA deletion mutants of strains 7702 and 9602R were constructed. Deletion of bclA leads to the loss of filaments (31). A copy of one of the genes (bclA7702 or bclA9602) together with its promoter region was then inserted into the chromosome of strains 7702 ΔbclA (PF31 and PF34) and 9602R ΔbclA (PF32 and PF33). The genes were inserted into the eag gene encoding the S-layer protein EA1, a locus that does not affect B. anthracis sporulation. The recombinant strains were tested for the production of the BclA protein. Spore extracts of the parentals, the ΔbclA mutants, and the complemented strains were subjected to SDS-PAGE and Western blot analysis (Fig. 5). Production of BclA was restored in all complemented strains. In strains carrying the bclA7702 gene (Fig. 5, lanes D and G) the apparent molecular mass of BclA was identical to that of strain 7702 (Fig. 5, lane A), and in strains carrying the bclA9602 gene (Fig. 5, lanes E and H), the apparent molecular mass of BclA was identical to that of strain 9602R (Fig. 5, lane B). The surfaces of the spores were examined under the electron microscope. Complementation restored the production of filaments (Fig. 6). The lengths of the filaments on strains PF31 and PF32 carrying the bclA7702 gene (Fig. 6D and G) were the same as those on strain 7702 (Fig. 6A). Similarly, the lengths of the filaments on strains PF34 and PF33 carrying the bclA9602 gene (Fig. 6E and H) were the same as those on strain 9602R (Fig. 6B). Thus, the bclA gene and the length of the protein determine the filament length.
FIG. 5.
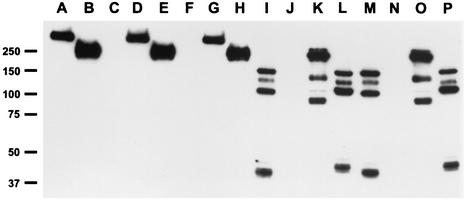
Analysis of spore extract from strains 7702, 9602R, RA3R, and 7611R and their mutants. Proteins were separated by SDS-7% PAGE and analyzed by immunoblotting with anti-BclA monoclonal antibody. Lane A, parental strain 7702; lane B, parental strain 9602R; lane C, strain 7702 ΔbclA (PF09); lane D, strain 7702 ΔbclA eag::bclA7702 (PF31); lane E, strain 7702 ΔbclA eag::bclA9602 (PF34); lane F, strain 9602R ΔbclA (PF11); lane G, strain 9602R ΔbclA eag::bclA7702 (PF32); lane H, strain 9602R ΔbclA eag::bclA9602 (PF33); lane I, parental strain RA3R; lane J, strain RA3R ΔbclA (PF13); lane K, strain RA3R ΔbclA eag::bclA7702 (PF35); lane L, strain RA3R ΔbclA eag::bclA9602 (PF36); lane M, parental strain 7611R; lane N, strain 7611R ΔbclA (PF14); lane O, strain 7611R ΔbclA eag::bclA7702 (PF37); lane P, strain 7611R ΔbclA eag::bclA9602 (PF38). The sizes of the molecular mass markers are given in kilodaltons in the left margin.
FIG. 6.
Electron microscopy analysis of the spore surface of parental strains and complemented deletion mutants after negative staining (see the legend to Fig. 5 for details). (A) Parental strain 7702; (D) strain 7702 ΔbclA eag::bclA7702 (PF31); (E) strain 7702 ΔbclA eag::bclA9602 (PF34); (B) parental strain 9602R; (G) strain 9602R ΔbclA eag::bclA7702 (PF32); (H) strain 9602R ΔbclA eag::bclA9602 (PF33); (I) parental strain RA3R; (K) strain RA3R ΔbclA eag::bclA7702 (PF35); (L) strain RA3R ΔbclA eag::bclA9602 (PF36). Bar, 100 nm.
The anti-BclA antibody gave several bands (40 to 150 kDa) in the immunoblots of spore extracts of strains RA3R and 7611R (Fig. 3, lanes 8 and 9, and 5, lanes I and M) but not in those of the other strains. However, no significant differences in length or ultrastructure organization were detected by electron microscopy between these two strains and strain 6602, which also contains a CLR of 81 residues but which presents a different GXX triplet organization. To test whether the CLR organization or the genetic background of these strains is responsible for the different immunoblot findings, gene exchange experiments were performed. The bclA7702 and bclA9602 genes were separately inserted into ΔbclA mutants of strains RA3R and 7611R. Complementation restored the production of filaments to RA3R ΔbclA (Fig. 6K and L) and 7611 ΔbclA (data not shown). Immunoblotting with the anti-BclA antibody revealed several bands in the complemented strains. These proteins migrated with apparent molecular masses of 90 to 250 kDa (Fig. 5, lanes K and O) for the strains complemented with a copy of the bclA7702 gene and of 42 to 140 kDa (Fig. 5, lanes L and P) for the strains complemented with a copy of the bclA9602 gene. These profiles differed from those of the parental strains (Fig. 5, lanes I and M) and from those of the BclA proteins in strains 7702 (Fig. 5, lane A) and 9602R (Fig. 5, lane B). This suggests that the genetic backgrounds of the RA3R and 7611R strains contribute to the multiplicity of BclA bands. The lengths of exosporium filaments in these strains were nevertheless dependent on which bclA genes they carried (Table 1 and Fig. 6K and L).
DISCUSSION
The bclA gene encoding the structural constituent of exosporium filaments is prevalent in B. anthracis strains, and BclA was produced by all strains studied. We also report that the polymorphic marker Ceb-Bams13 (15) encompasses the DNA sequence encoding the CLR of BclA. A substantial diversity in the sizes of the BclA genes was identified in strains representative of eight Ceb-Bams13 alleles. These differences are due to different numbers of GXX amino acid repeats in the CLR, and consequently, this segment of BclA is highly polymorphic. All strains studied carried at least one 21-amino-acid sequence of (GPT)5GDTGTT (the BclA repeat), suggesting that it is an essential feature of BclA; it may be essential for the structural organization of BclA on the spore surface. The number of BclA repeats is between 1 in strain 4229 and 8 in Sterne strain 7702. No strains with 3, 4, or 7 repeats were found. The length of the CLR was directly correlated to the length of filaments on the spore surface. CLRs of the same length, but with different CLR organizations, as found for allele 4, gave filaments of indistinguishable length and ultrastructure. The size of the filaments of the Sterne strain spores we report is in agreement with that reported in earlier studies of Sterne spores (7, 13).
The presence of an extensive CLR in the BclA sequence suggests that it is capable of forming a collagen-like structure. Mammalian collagen molecules form a triple-helix domain of variable length which consists of many GXY triplet repeats. Posttranslational hydroxylation of proline residues and glycosylation of the polypeptide are believed to be essential for the assembly of a stable triple helix. The presence of glycosylated threonine residues in the Y position have been described as an alternative to hydroxyproline in the triple-helix-stabilizing function (19). All BclA proteins were glycosylated, and their CLRs were rich in threonine residues (31.4 to 35.5%). We have previously proposed that threonine residues may be glycosylated and that glycosyl residues contribute to the stability of the exosporium filaments (31).
The ability of bacterial collagen-like proteins to form a triple helix has been recently reported in an in vitro model (38) with the streptococcal collagen-like proteins (Scl). Both Scl proteins have a similar organization (16, 17, 26, 27, 37). Their putative extracellular part consists of a variable amino-terminal noncollagenous region and a central highly variable CLR. The carboxy-terminal part contains characteristic cell-wall-associated regions. Recombinant polypeptides derived from Scll and Scl2 proteins, which contain 50 and 79 GXY repeats in their CLRs, respectively, can form collagen-like triple helices in vitro, despite the absence of hydroxyproline residues (38). Moreover, the length of the collagenous tail was dependent on the CLR length, and theoretical predictions (32) for a collagen triple helix were confirmed by experimental measurement (38). The Scl proteins are translocated to the cell surfaces of group A streptococci but have not been visualized as a cell surface structure. For B. anthracis, it is likely that exosporium filaments are organized in a triple-helix structure formed by three BclA chains. Experimental measurements of filaments on the spore surface correlate to theoretical estimations (32), and their lengths depend on the CLR size.
BclA is the first identified B. anthracis protein containing a VNTR region where this variation has been associated with phenotypic variation. Nine different alleles of Ceb-Bams13 were observed among the 31 diverse strains (15). It is likely that there are additional alleles. The fact that two different organizations of CLR were found for allele 4 (one in strains RA3R and 7611R and another in strain 6602) reveals one of the limitations of phylogenetic studies based only on size variations of polymorphic markers in the absence of prior characterization of polymorphism. Interestingly, multiple organizations for a given allele allow discrimination between strains otherwise appearing indistinguishable. We do not know whether other Ceb-Bams13 alleles present several CLR organizations.
The BclA proteins extracted from strains RA3R and 7611R gave a multiple-glycosylated-band pattern. The other strains each gave only a single band. Such multiple-band patterns were also observed for the ΔbclA mutants of RA3R and 7611R complemented with heterologous bclA genes. This suggested that posttranslational modifications, probably implicated in the stability or glycosylation of BclA on the spore surface or both, depending on the genetic background of the strains, were responsible for the phenomenon. Strain RA3 is of genotype 79 and strain 7611 is of genotype 80, and both are in the B2 cluster defined in the MLVA analysis of Fouet and collaborators and Keim and collaborators (5a, 11). Among the 8 MLVA markers, only the pXO2 marker differs between genotypes 79 and 80, indicating a similar chromosomal background as far as can be defined. These two genotypes are the most common in France but are extremely rare elsewhere in the world (5a). Strains RA3 and 7611 originated from anthrax outbreaks in two different mountain regions of France: the Pyrenees and the Alps. The hydrogeological characteristics of the soil could be relevant to the prevalence of the B2 cluster in certain regions of France. The exosporium filaments are the outermost structure of the spore, and therefore, particular variants of BclA may contribute to the properties of the spores in response to various environments. The identification of VNTR variation within a surface structural gene of the infectious form suggests that B. anthracis may use this variation to modulate its properties, as reported for other pathogens (36). Functional analysis of genes carrying VNTR variations, revealed by the B. anthracis genome data, should bring further insights into the biology of this otherwise monomorphic organism. Nevertheless, the significance of BclA polymorphism remains currently unresolved.
Acknowledgments
We wish to thank A. Fouet for interest and helpful discussions during this work and for critical reading of the manuscript.
P.S. was supported by DGA.
REFERENCES
- 1.Andersen, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaman, T. C., H. S. Pankratz, and P. Gerhardt. 1971. Paracrystalline sheets reaggregated from solubilized exosporium of Bacillus cereus. J. Bacteriol. 107:320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthier, M., J. L. Fauchère, J. Perrin, B. Grignon, and D. Oriot. 1996. Fulminant meningitis due to Bacillus anthracis in 11-year-old girl during Ramadan. Lancet 347:828.. [DOI] [PubMed] [Google Scholar]
- 4.Brossier, F., M. Weber-Levy, M. Mock, and J.-C. Sirard. 2000. Role of toxin functional domains in anthrax pathogenesis. Infect. Immun. 68:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DesRosier, J. P., and J. C. Lara. 1984. Synthesis of the exosporium during sporulation of Bacillus cereus. J. Gen. Microbiol. 130:935-940. [Google Scholar]
- 5a.Fouet, A., K. L. Smith, C. Keys, J. Vaissaire, C. Le Doujet, M. Lévy, M. Mock, and P. Keim. 2002. Diversity among French Bacillus anthracis isolates. J. Clin. Microbiol. 40:4732-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedlander, A. M., S. L. Welkos, M. L. Pitt, J. W. Ezzell, P. L. Worsham, K. J. Rose, B. E. Ivins, J. R. Lowe, G. B. Howe, P. Mikesell, and W. B. Lawrence. 1993. Postexposure prophylaxis against experimental inhalation anthrax. J. Infect. Dis. 167:1239-1242. [DOI] [PubMed] [Google Scholar]
- 7.Gerhardt, P., and E. Ribi. 1964. Ultrastructure of the exosporium enveloping spores of Bacillus cereus. J. Bacteriol. 88:1774-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hachisuka, Y., K. Kojima, and T. Sato. 1966. Fine filaments on the outside of the exosporium of Bacillus anthracis spores. J. Bacteriol. 91:2382-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Andersen, K. H. Wilson, M. E. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein, F., J. S. Walker, D. F. Fritzpatrick, R. E. Lincoln, B. G. Mahlandt, W. I. Jones, Jr., J. P. Dobbs, and K. J. Hendrix. 1966. Pathophysiology of anthrax. J. Infect. Dis. 116:123-138. [DOI] [PubMed] [Google Scholar]
- 13.Kramer, M. J., and I. L. Roth. 1968. Ultrastructural differences in the exosporium of the Sterne and Vollum strains of Bacillus anthracis. Can. J. Microbiol. 14:1297-1299. [DOI] [PubMed] [Google Scholar]
- 14.Laforce, F. M., F. H. Bumford, J. C. Feeley, S. L. Stokes, and D. B. Snow. 1969. Epidemiologic study of a fatal case of inhalation anthrax. Arch. Environ. Health 18:798-805. [DOI] [PubMed] [Google Scholar]
- 15.Le Flèche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, R. M. Ireland, S. D. Reid, G. G. Adams, and J. M. Musser. 2000. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun. 68:6542-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, B. J. Shelvin, E. A. Graviss, and J. M. Musser. 2001. Identification and characterization of a second extracellular collagen-like protein made by group A Streptococcus: control of production at the level of translation. Infect. Immun. 69:1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Mann, K., D. E. Mechling, H. P. Bachinger, C. Eckerskorn, F. Gaill, and R. Timpl. 1996. Glycosylated threonine but not 4-hydroxyproline dominates the triple helix stabilizing positions in the sequence of a hydrothermal vent worm cuticle collagen. J. Mol. Biol. 261:255-266. [DOI] [PubMed] [Google Scholar]
- 20.Mesnage, S., E. Tosi-Couture, M. Mock, P. Gounon, and A. Fouet. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23:1147-1155. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Moberly, B. J., F. Shafa, and P. Gerhardt. 1966. Structural details of anthrax spores during stages of transformation into vegetative cells. J. Bacteriol. 92:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 24.Patra, G., J. Vaissaire, M. Weber-Levy, C. Le Doujet, and M. Mock. 1998. Molecular characterization of Bacillus strains involved in outbreaks of anthrax in France in 1997. J. Clin. Microbiol. 36:3412-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pezard, C., P. Berche, and M. Mock. 1991. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect. Immun. 59:3472-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen, M., and L. Björck. 2001. Unique regulation of SclB-a novel collagen-like surface protein of Streptococcus pyogenes. Mol. Microbiol. 40:1427-1438. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen, M., A. Edén, and L. Bjorck. 2000. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 68:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth, I. L., and R. P. Williams. 1963. Comparison of the fine structure of virulent and avirulent spores of Bacillus anthracis. Texas Rep. Biol. Med. 21:394-399. [PubMed] [Google Scholar]
- 29.Schupp, J. M., A. M. Klevytska, G. Zinser, L. B. Price, and P. Keim. 2000. vrrB, a hypervariable open reading frame in Bacillus anthracis. J. Bacteriol. 182:3989-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirard, J. C., M. Mock, and A. Fouet. 1995. Molecular tools for the study of transcriptional regulation in Bacillus anthracis. Res. Microbiol. 146:729-737. [DOI] [PubMed] [Google Scholar]
- 31.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 32.Traub, W., and K. A. Piez. 1971. The chemistry and structure of collagen. Adv. Protein Chem. 25:243-352. [DOI] [PubMed] [Google Scholar]
- 33.Trieu-Cuot, P., C. Carlier, P. Martin, and P. Courvalin. 1987. Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol. Lett. 48:289-294. [Google Scholar]
- 34.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and Gram-positive bacteria. Nucleic Acids Res. 18:4296.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whatmore, A. M. 2001. Streptococcus pyogenes sclB encodes a putative hypervariable surface protein with a collagen-like repetitive structure. Microbiology 147:419-429. [DOI] [PubMed] [Google Scholar]
- 38.Xu, Y., D. R. Keene, J. M. Bujnicki, M. Höök, and S. Lukomski. 2002. Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J. Biol. Chem. 277:27312-27318. [DOI] [PubMed] [Google Scholar]
- 39.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]