Abstract
Protein folding in the living cell begins cotranslationally. To analyze how it is influenced by the ribosome and by the translocon complex during translocation into the endoplasmic reticulum, we expressed a mutant influenza hemagglutinin (a type I membrane glycoprotein) with a C-terminal extension. Analysis of the nascent chains by two-dimensional SDS-PAGE showed that ribosome attachment as such had little effect on ectodomain folding or trimer assembly. However, as long as the chains were ribosome bound and inside the translocon complex, formation of disulfides was partially suppressed, trimerization was inhibited, and the protein protected against aggregation.
INTRODUCTION
Protein folding in the living cell is a complex process. It is affected by a variety of milieu factors such as pH, calcium ions, and the redox environment; it is influenced by co- and posttranslational modifications; and it relies on the assistance of numerous molecular chaperones. Furthermore, it is now generally accepted that folding begins cotranslationally, i.e., while the polypeptide is still being synthesized by the ribosome (Fedorov and Baldwin, 1997; Netzer and Hartl, 1998; Hardesty et al., 1999).
For a typical protein, this means that folding occurs vectorially from the N terminus toward the C terminus. It also means that the ribosome and associated structures such as the translocon complex in the endoplasmic reticulum (ER) membrane are part of the immediate environment within which cotranslational folding takes place. The initial folding of soluble secretory proteins and membrane proteins synthesized in the ER are thus likely to take place in the aqueous, protein-lined channel connected to the exit tunnel of the large ribosomal subunit. Only after chain termination, which depending on the size of the protein may occur minutes after initiation, is the newly synthesized polypeptide chain released from the ribosome and the translocon complex and thus free to fold independently.
In this paper, we have for the first time analyzed to what extent the ribosome and the translocon complex affect the folding of a protein in the ER. We determined how far folding could proceed while the protein was still bound to the ribosome and the translocation complex and assessed the effects the translocon complex (composed of the sec61 protein channel, TRAM, signal peptidase, oligosaccharyl transferase, and other accessory proteins) (Rapoport et al., 1996) and the ribosomes on folding, oligomerization, and aggregation.
To be able to analyze folding in living cells, we made use of chimeric polypeptide probes containing the N-terminal ectodomain and transmembrane domain of influenza hemagglutinin (HA) and a C-terminal domain derived from receptor-associated protein (RAP) (Strickland et al., 1991). The fate of these probes in the ribosome-bound cycloheximide-arrested state was compared with that of the puromycin-released state using a two-dimensional SDS-PAGE technique recently developed in our laboratory (Chen et al., 1995). The results indicated that attachment to the ribosome per se did not affect the folding of the translocated ectodomain, nor did it interfere with formation of HA trimers. However, the translocon complexes to which the ribosomes were attached clearly played a role in restricting the folding of the ectodomain and preventing oligomerization as well as nonproductive intermolecular interactions.
MATERIALS AND METHODS
Generation of Recombinant Semliki Forest Virus (SFV)
In-frame fusion between the cDNA of HA (X31 strain) and the cDNA of the 298 amino residues (residues 25–322) of RAP (Strickland et al., 1991) was performed by PCR and verified by sequencing. The HA-RAP and HA-RAP (Tm−) constructs were cloned from Bluescript (Stratagene, La Jolla, CA) into the XbaI site of pSFV-1. Because HA has several SpeI sites within the coding region, we used an NruI variant pSFV-1 expression vector (a gift from Dr. Henrik Garoff, Karolinska Institute, Stockholm, Sweden). The pSFV (Nru1)-HA-RAP and the pSFV (Nru1)-HA-RAP (Tm−) were linearized with Nru1. The linearized DNA was used for in vitro transcription. The in vitro-transcribed RNA along with SFV helper RNA were electroporated into BHK-21 cells to generate recombinant SFV virus stocks as described (Liljeström and Garoff, 1991).
35S Labeling, Immunoprecipitation, and Two-dimensional Gel Analysis
BHK-21 cells were infected with the recombinant HA-RAP or HA-RAP (Tm−) SFV in serum-free medium at a multiplicity of infection of 5 for 1 h. The cells were pulse labeled with [35S]methionine and [35S]cysteine 9 h after infection, and sometimes chased in various conditions. The cells were then alkylated with 20 mM N-ethylmaleimide to prevent further oxidation before cell lysis and immunoprecipitation (Braakman et al., 1991). Immunoprecipitation with α-N-terminal peptide of HA (α-NHA), α-N2, α-CNX, and α-CRT were performed as previously described (Braakman et al., 1991; Hammond et al., 1994; Peterson et al., 1995). α-FLAG monoclonal antibody (M2) was purchased from Eastman Kodak (Rochester, NY). In a typical immunoprecipitation reaction with this antibody, 15 μg of α-FLAG antibody was added to 0.5 ml of cell lysate from a 60-mm dish.
The immunoprecipitates were analyzed using a two-dimensional SDS-PAGE system (nonreduced in the first dimension and reduced in the second dimension), which was previously developed in our laboratory to study nascent chains in live cells (Chen et al., 1995). 5.5% gels were used for the analysis of nascent chains of glycosylated HA-RAP and HA-RAP (Tm−); 7.5% gels were used to analyze unglycosylated HA-RAP nascent chains.
RESULTS
Constructing HA-RAP Chimeras
To study the folding of growing nascent chains in the ER of living cells, we made use of influenza HA constructs that had at their C terminus a sequence extension of ∼300 extra amino acid residues (Figure 1). The extension allowed us to study whether the attachment of the cytosolic C terminus to the ribosome affected the fate of the nascent chain. For the extension, a sequence from a lumenal ER protein, RAP, was chosen because it was polar and unlikely to interfere with folding of the lumenal and transmembrane domains of the HA molecule when added to the cytoplasmic C terminus. Moreover, because it was devoid of cysteines and methionines, it would not incorporate radioactivity during metabolic labeling with [35S]Translabel. This had the desired effect that after a radioactive pulse of 1 min, only nascent chains would be 35S labeled. Moreover, because the full-length molecules would not have any label, any degradation products derived from them would not contaminate the 35S autoradiograms. The HA portion of the fusion protein had, in contrast to the tail extension, 20 methionines and cysteines evenly spaced throughout. Therefore, the 35S label was expected to be highest in nascent chains of intermediate length and to trail off toward shorter and longer chains.
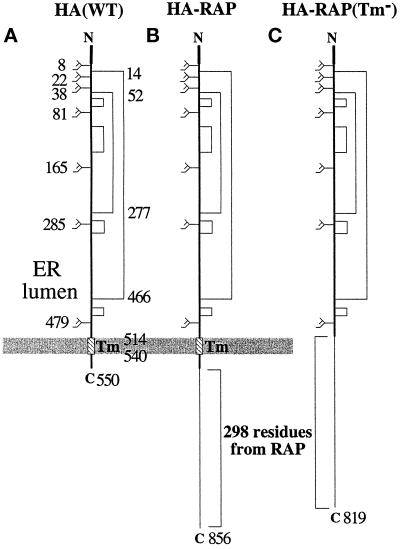
Figure 1.
HA-RAP and HA-RAP (Tm−) chimeras. (A) Linear structure of X31 HA(WT). The positions of the two major disulfide bonds (C52–277C and C14–466C) and the transmembrane domain (514–540) are numbered on the right of the molecule, the positions of the glycosylation sites are numbered on the left of the molecule. The mature HA contains a total of 550 amino acid residues. (B) Structure of HA-RAP fusion protein. Two hundred ninety-eight amino residues (residues 25–322) of RAP were fused to the cytosolic tail of HA. There is a FLAG tag at the very C-terminal end. The mature HA-RAP protein contains 856 amino acid residues. (C) Structure of HA-RAP (Tm−). This protein differs from the HA-RAP fusion protein in that the transmembrane domain and cytosolic tail from wild-type HA are absent. The mature HA-RAP (Tm−) contains 819 amino acid residues.
The RAP sequence (residues 25–322) was fused either to the C-terminal cytoplasmic tail of HA, generating a construct called HA-RAP (Figure 1B), or to the HA ectodomain, to form HA-RAP (Tm−) (Figure 1C). Because the membrane-spanning sequence of HA was preserved, HA-RAP was a transmembrane protein with the HA ectodomain in the ER lumen and with the authentic 10-residue C-terminal sequence plus the RAP sequences in the cytosol. Lacking the “stop-transfer sequence” composed by the transmembrane domain of HA, the C-terminal RAP portion of HA-RAP (Tm−) was, in contrast, expected to follow the HA ectodomain into the lumen of the ER. To the C-terminal ends of the RAP sequences in both constructs, a FLAG tag (eight amino acid residues) was added to allow distinction between full-length and incomplete chains by immunoprecipitation with anti-FLAG antibodies.
Cotranslational Folding of HA-RAP
For efficient expression of the fusion proteins, we used the recombinant SFV system (Liljeström and Garoff, 1991). BHK cells were infected with the recombinant viruses encoding HA-RAP or HA-RAP (Tm−), and 9 h after infection, the cells were pulse labeled for 1 min with [35S]Translabel. After the pulse, or after additional periods of chase, the cells were treated with 20 mM N-ethylmaleimide to prevent further disulfide formation, and lysed with detergent (Braakman et al., 1991). Postnuclear supernatants were immunoprecipitated using α-NHA and analyzed by two-dimensional SDS-PAGE.
The two-dimensional SDS-PAGE system used has been specifically designed to monitor the folding of growing nascent chains in the ER of live cells (Chen et al., 1995). It takes advantage of the observation that HA and other proteins with intrachain disulfides tend to have a faster electrophoretic mobility when oxidized than when reduced. When electrophoresed, nonreduced in the first dimension, and reduced in the second dimension, such proteins are characteristically offset from the diagonal of the two-dimensional gel. This also applies to oxidized nascent chains present in the ER of cells, except that they appear as lines rather than single spots in the gel because of their heterogeneous molecular size.
During co- and posttranslational folding of wild-type HA, three differentially oxidized forms are resolved by SDS PAGE depending on whether they have acquired disulfide bonds C52–277C and C14–466C (Braakman et al., 1991; Braakman and Helenius, unpublished results). Intermediate 1 (IT1) lacks both, intermediate 2 (IT2) has the former but not the latter, and the fastest migrating form, NT, has both. These two disulfide bonds generate large covalent loops in the polypeptide chains and cause easily detected shifts in SDS-PAGE mobility. It is noteworthy that gaps in the lines formed by nascent chains arise whenever the addition of N-linked glycans results in a molecular weight increase in the nascent chain (Chen et al., 1995).
The two-dimensional gel pattern obtained for nonreduced HA-RAP after 1 min of pulse labeling and no chase is shown in Figure 2A. Electrophoresis was performed in 5.5% polyacrylamide gels that allowed satisfactory resolution of proteins in the 60- to 120-kDa molecular mass range. As expected, no radioactivity was present in the position of full-length HA-RAP (compare with Figure 2B). The HA portion of the fusion protein had 20 methionines and cysteines evenly spaced throughout; therefore, the 35S label was highest in nascent chains of intermediate length and trailed off toward shorter and longer chains. The gap shown by an arrow corresponded to the addition of the most C-terminal N-linked glycan in position N479 (Chen et al., 1995). The location of this gap is close to the 80-kDa full-length wild-type HA, one of the molecular mass markers run in the second dimension. This is because translation of HA-RAP must proceed to residue 544 (almost full-length wild-type HA) before glycosylation site N479 reaches the active site of the oligosaccharide transferase in the lumen of the ER (Whitley et al., 1996).
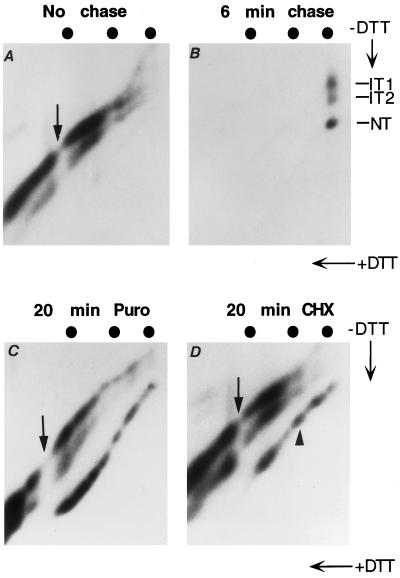
Figure 2.
Role of ribosome and translocon complex on nascent chain folding. BHK cells infected by recombinant SFV virus expressing HA-RAP were labeled for 1 min and chased in various conditions. The lysates were immunoprecipitated by α-NHA and analyzed by 5.5% two-dimensional gels. The markers are wild-type HA (80 kDa), muscle phosphorylase (97 kDa). and Escherichia coli β-galactosidase (115 kDa). (A) One-minute pulse for HA-RAP. (B) One-minute pulse followed by 6-min chase with 1 mM cysteine and 1 mM methionine. Shown is a very short exposure to demonstrate clear separation between IT1 and IT2. (C) One-minute pulse followed by 20-min chase in the presence of 1 mM puromycin. (D) One-minute pulse followed by 20-min chase in the presence of 1 mM cycloheximide.
The labeled HA-RAP nascent chains were present in three separate, nearly parallel lines (Figure 2A). If the sample was reduced, they all merged into a single line along the diagonal (Chen et al., 1995). Without previous reduction, the strongest of the nascent chain lines corresponded to IT1 running close to the diagonal. Approximately 20% of the nascent HA-RAP were present in the IT2 form that formed a distinct line below the diagonal. It started at an approximate molecular mass of 63 kDa, which indicated that the C52–277C disulfide bond could form well before the HA portion of the chimera had been fully synthesized. The x-ray crystal structure shows that this disulfide bond closes off the top domain of the HA molecule (Wilson et al., 1981).
The third line, corresponding to NT and barely visible below the IT2 line, contained <5% of the labeled nascent chains. Judging from its starting position, one could conclude that the HA-RAP chains involved in forming the NT line were all longer than wild-type HA. This indicated that, although the ectodomain can close the largest disulfide loop (C14–466C) cotranslationally, this was a rare event.
If the 1-min pulse was followed by a 6-min chase (Figure 2B), almost all the label in the nascent chains was replaced by full-length HA-RAP, which was present as three spots corresponding to the full-length IT1, IT2, and NT forms. This showed that once completed and dissociated from the ribosomes, the ectodomains of the HA-RAP chains continued to fold normally.
Effect of Ribosome and the Translocon Complex on Nascent Chain Folding
Two protein synthesis inhibitors, puromycin and cycloheximide, were used to further analyze the folding of ribosome-bound and free HA-RAP chains. These inhibitors differ in their effects: puromycin induces premature chain release (Nathans, 1964), whereas cycloheximide causes elongation arrest without chain release (Wettstein et al., 1964; Beckmann et al., 1990). When a 1-min pulse was followed by 20 min of chase in the presence of puromycin or cycloheximide, folding proceeded further than during normal cotranslational folding (Figure 2, C and D).
Several differences were observed between the puromycin-released and the cycloheximide-arrested HA-RAP chains. In puromycin-treated cells, the position of the gap (shown by an arrow) caused by the addition of N-linked glycan to N479 shifted toward lower molecular mass. The reason was that, when associated with the ribosome, a polypeptide chain must have ∼65 additional amino acid residues at its C terminus for the consensus glycosylation site to reach the oligosaccharide transferase located on the lumenal side of the translocon (Whitley et al., 1996). Thus, although still bound to the ribosome, nascent chains with a C-terminal end between residues 479 and 543 are not glycosylated in the N479 site. Only ribosome-bound nascent chains longer than 544 residues get glycosylated. This leaves a molecular mass gap (∼3 kDa) between nascent chains with 543 residues and six glycans and nascent chains with 544 residues and seven glycans. Because puromycin releases the nascent chains, which slide out of the ribosomes and through the translocon complexes, the consensus glycosylation acceptor sites of peptide chains between 479 and 543 residues become exposed to the oligosaccharide transferase. As a result, there is no longer a gap between chain 543 (now with seven glycans) and chain 544 (with seven glycans), but there is a gap between chain 478 (with six glycans) and chain 479 (now with seven glycans). The shift of the glycosylation gap also confirmed that the labeled chains analyzed in Figure 2A were indeed authentic nascent chains and that cycloheximide efficiently blocked the release of the chains from the ribosomes (Figure 2D).
The degree of folding was different for puromycin-released and cycloheximide-arrested HA-RAP chains. More than half of the chains with molecular masses between 80 and 97 kDa reached the NT form when they were released form the ribosome with puromycin (Figure 2C). When arrested on the ribosomes by cycloheximide, less than one-fourth of these chains reached the NT form (Figure 2D). This difference implied that although C14–466C can form in chains that are associated with the ribosomes and the translocon, the efficiency is much lower than for the same chains released from the complex. In other words, the ribosomes and/or the translocon complex impose constraints on the folding of the ectodomain of the HA-RAP chains of intermediate length.
For HA-RAP chains of ≥97 kDa, folding of ribosome-bound nascent chains was as effective as for the puromycin-released chains (see Figure 2D, arrowhead). This showed that ribosome binding per se does not inhibit efficient folding of the ectodomain of HA-RAP. But to fold efficiently, the bound HA-RAP chains had to possess a cytosolic tail longer than ∼160 amino acid residues. If the cytoplasmic tail was shorter, folding of the ectodomain was largely inhibited. Previous in vitro microsome experiments have shown that when the cytosolic tail is sufficiently long, the Tm segment of ribosome-bound nascent chains can move laterally out of the translocon (Do et al., 1996) and integrate into the lipid-containing part of the ER membrane (Martoglio et al., 1995; Mothes et al., 1997). Our results indicated that such cotranslational lateral exit of the Tm segment also occurred in vivo and demonstrated that once it occurred, the translocon complex ceased to impose constraints on the ectodomain folding.
Effect of Ribosome and the Translocon Complex on Nascent Chain Oligomerization
Wild-type HA is known to undergo efficient trimerization soon after it has reached the NT form (Braakman et al., 1991). To test whether NT of HA-RAP fusion proteins had the capacity to trimerize, cells were pulse labeled for 1 min and chased in the presence of puromycin for 20 min to release the HA-RAP nascent chains. The cell lysate was then immunoprecipitated with a trimer-specific monoclonal antibody called N2 (Braakman et al., 1991), and the precipitates were subjected to two-dimensional SDS-PAGE.
It was found that puromycin-released HA-RAP chains of variable lengths that had reached the NT form were in fact precipitated with trimer-specific antibodies (Figure 3A). The trimerization efficiency was independent of the length of the cytosolic tail, indicating that the C-terminal RAP extension did not interfere with trimerization. As with wild-type HA, chains that were still incompletely oxidized (Braakman et al., 1991), the IT1 and IT2 forms did not trimerize. When the HA-RAP (Tm−) was used instead of HA-RAP, no trimer formation was detected (Figure 3B), consistent with previous findings that not only must the HA be fully oxidized to trimerize, but it requires the presence of the transmembrane domain (Singh et al., 1990; Tatu et al., 1995).
Figure 3.
Effect of ribosome and the translocon complex on nascent chain oligomerization. BHK cells infected by recombinant SFV expressing HA-RAP (A, C, and G) or HA-RAP (TM−) (B, D–F, and H) were labeled for 1 min followed by a 20-min chase with either 1 mM puromycin or 1 mM cycloheximide. The lysate was immunoprecipitated with α-NHA, trimer-specific monoclonal antibody α-N2, or α-FLAG (M2) and analyzed by 5.5% two-dimensional gels. The markers are wild-type HA (80 kDa), muscle phosphorylase (97 kDa), and E. coli β-galactosidase (115 kDa). (A) HA-RAP: 1-min pulse followed by 20-min chase with puromycin, immunoprecipitated with α-N2. (B) Same as A, except HA-RAP (Tm−) virus was used. (C) HA-RAP: 1-min pulse followed by 20-min chase with cycloheximide, immunoprecipitated with α-N2. (D) Same as C, except HA-RAP (Tm−) virus was used. (E) HA-RAP (Tm−): 1-min pulse followed by 20-min chase with puromycin, immunoprecipitated with α-NHA, (F) HA-RAP (Tm−): 1-min pulse followed by 20-min chase with cycloheximide, immunoprecipitated with α-NHA. (G) HA-RAP: 1-min pulse, followed by 20-min chase with cycloheximide, immunoprecipitated with α-FLAG. (H) Same as G, except HA-RAP (Tm−) virus was used.
Next, we tested whether ribosome-associated nascent chains of HA-RAP and HA-RAP (Tm−) could form trimers. In the case of the ribosome-bound HA-RAP (Tm−), no trimerization was observed (Figure 3D). This inability was not due to a folding defect, because anti-NHA immunoprecipitations showed that the ectodomain of HA-RAP (Tm−) chains folded to NT after a 20-min chase in the presence of puromycin (Figure 3E) and cycloheximide (Figure 3F). However, precipitation with N2 showed that a sizable fraction of nascent HA-RAP chains in the form of NT was present in trimers (Figure 3C). The trimerization was hardly detectable for the nascent chains <97 kDa (Figure 3C), indicating that the ribosomes and the translocon complex inhibited trimerization of nascent chains with short cytosolic tails. The efficiency of trimer formation exceeded 50% for ribosome-bound nascent chains with molecular masses of ≥97 kDa (Figure 3C). Therefore, ribosome binding per se did not inhibit trimer formation. Inhibition apparently occurred when the cytoplasmic tail was too short to permit the protein from escaping the constraints imposed by the translocon complex.
The origin of the trimerization partners was addressed using the FLAG tag at the C terminus of HA-RAP to distinguish between full-length and incomplete chains. When α-FLAG immunoprecipitation was performed with the same lysate as that used in Figure 3C, NT nascent chains were precipitated (Figure 3G). This showed that full-length HA-RAP molecules were associated with the pulse-labeled, incomplete HA-RAP chains of the NT form. The radioactivity seen along the diagonal line across the gel represented nonspecific background: it was also found in the HA-RAP (Tm−) control, which did not show any precipitation of labeled HA forms (Figure 3H). The full-length version of HA-RAP (Tm−) was, however, efficiently precipitated by anti-FLAG antibody. Thus, the ribosome-associated HA-RAP nascent chains with cytosolic tails longer than ∼160 amino acid residues moved into a location in the ER membrane where they had access to previously synthesized, full-length HA-RAP molecules, and a large fraction formed trimers with them.
Effect of Ribosome and the Translocon Complex on Nascent Chain Aggregation
Previous studies have shown that unglycosylated HA misfolds and enters large aggregates and disulfide cross-linked complexes in the ER (Hurtley et al., 1989). Misfolding of the chains starts already cotranslationally (Chen et al., 1995). To determine how the ribosome and the translocon complex affect the aggregation, cells expressing HA-RAP were treated with tunicamycin and pulse labeled for 1 min. They were then chased for 5 or 10 min in the presence of either puromycin or cycloheximide. Anti-NHA immunoprecipitates were analyzed with 7.5% two-dimensional SDS-PAGE.
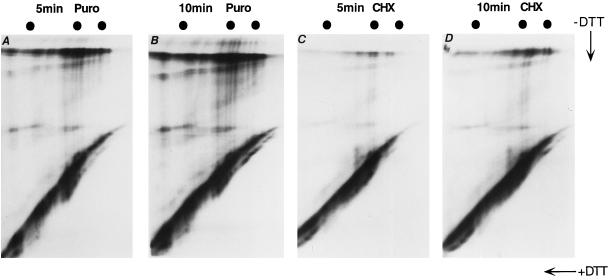
The gaps in the HA-RAP lines were missing (Figure 4, A–D), indicating that glycosylation was efficiently blocked. Within 5 or 10 min of chase, the fates of unglycosylated, puromycin-released nascent chains were shown in Figure 4, A and B. The intrachain disulfides were not those observed in the normal IT1 and IT2 but generated a random smear below the diagonal, diagnostic of misfolding. A large fraction of the labeled chains was found at the top of the gel in disulfide-bonded aggregates. These aggregates may also contain proteins other than HA. When the cycloheximide-treated samples were analyzed (Figure 4, C and D), similar misfolding was observed, but aggregate formation were strongly inhibited. Clearly, although the ribosome and the translocon complex did not prevent the unglycosylated nascent chains from misfolding, it protected them from aggregation. Interestingly, the protection was strong regardless of the length of the cytosolic tail.
Figure 4.
Misfolded ribosome-associated nascent chains are inhibited from aggregation. BHK cells infected by recombinant SFV virus expressing HA-RAP were pretreated with 5 μg/ml tunicamycin for 45 min in cysteine- and methionine-free medium. After a 1-min pulse in tunicamycin, the cells were chased with either 1 mM puromycin or 1 mM cycloheximide for 5 or 10 min. The lysate was immunoprecipitated by α-NHA and analyzed by 7.5% two-dimensional gels. The markers are egg white albumin (45 kDa), BSA (65 kDa), and muscle phosphorylase (97 kDa). (A) One-minute pulse followed by 5-min chase with 1 mM puromy-cin. (B) One-minute pulse followed by 10-min chase with 1 mM puromycin. (C) One-minute pulse followed by 5-min chase with 1 mM cycloheximide. (D) One-minute pulse followed by 10-min chase with 1 mM cycloheximide.
DISCUSSION
A nascent chain that emerges from a membrane-bound ribosome in the ER encounters an environment different from what is seen by a nascent chain in the cytosol. Because the most recently synthesized C-terminal part is moving through a passage in the large ribosomal subunit, previously synthesized sequences may already be folding inside the ER lumen. Alternatively, they may be in the process of moving through the translocon complex, they may be trapped inside the translocon complex as a Tm sequence, they may be present as a Tm sequence already extruded from the translocon into the ER membrane, or they may be part of a cytosolic loop between the translocon and the ribosome. In this study, we have investigated how the ribosome and the translocon complex affects the maturation of nascent influenza HA chains in the ER of the living cell.
One of the main conclusions was that association of nascent HA with the translocon complex causes partial suppression of ectodomain folding. This was particularly evident when one followed formation of the C14–466C disulfide in HA-RAP. Although the top domain could fold and acquire its disulfide C52–277C bond without problems cotranslationally, the stem domain that contains disulfide bond C14–466C close to the membrane surface was oxidized very slowly if at all. This suggested that the closer to the membrane and thus deeper inside the translocon complex a polypeptide segment, the less efficient its cotranslational oxidization and folding.
The slower folding of the stem domain was most likely caused by crowded conditions inside the translocon complex. The translocon complex is a large structure composed of numerous protein components (Rapoport et al., 1996). In addition to the Sec61 complex that forms the actual transmembrane channel through which the polypeptide enters the ER lumen, many accessory proteins are present. These include TRAM, the signal peptidase complex, the oligosaccharide transferase complex, glucosidase I, calnexin, and probably a number of other proteins. Some of the proteins present form the protein-conducting channel, whereas others are likely to extend quite far into the lumen of the ER. Although the transmembrane channel itself is thought to be rather wide (up to 40–60 Å in diameter for the open functional channel; Hamman et al., 1997), it may not be wide enough to allow proteins to fold efficiently. In the case of HA it is possible that the heavily glycosylated N-terminal segment containing Cys-14 may find it difficult to loop back into the translocon complex once the signal sequence has been cotranslationally cleaved, and to interact with the C-terminal segment inside the complex. The three N-linked glycans flanking Cys-14 are, moreover, known to interact with calnexin, a lectin-like chaperone that binds to HA already during translation (Chen et al., 1995; Hebert et al., 1997). This may further inhibit the free movement of the C-terminal segment.
Alternatively, the inefficient formation of the C14–466C disulfide in the translocon complex may be caused by inaccessibility of the cysteines to lumenal thiol oxidoreductases such as protein disulfide isomerase or ERp57 (Freedman, 1989) within the translocon complex. There is increasing evidence that these enzymes are directly involved in the oxidation of newly synthesized proteins in the ER (Frand and Kaiser, 1998). That thiol oxidases such as ERp57 do associate with growing nascent chains has been recently observed for viral glycoproteins in living cells (Molinari and Helenius, 1999). However, this interaction may only be possible once the cysteines have moved out of the narrow part of the translocon complex.
By restricting access to large segments of a nascent growing polypeptide chain, the translocon complex may serve a function similar to that of GroEL, another multimeric, complex that houses incompletely folded proteins within a polar cavity (for review, see Bukau and Horwich, 1998). GroEL is a member of the HSP60 molecular chaperone family that interacts with incompletely folded proteins in the cytosol of bacteria, in mitochondria, and in chloroplasts. Like GroEL, we observed that the translocon complex prevented nonproductive and premature interactions between nascent chains and other proteins in the ER. We found evidence for the latter by observing the fate for nonglycosylated misfolded nascent chains of HA. As long as these were associated with the ribosome and the translocon complex, their entry into disulfide cross-linked aggregates was greatly suppressed. Although the translocon complex helps prevent growing nascent chains from interacting with other incompletely folded proteins in the ER, ribosome binding as such is likely to prevent interactions between nascent chains by preventing close contacts. The center-to-center distance between ribosomes in ER-bound polysomes is ∼20–35 nm. In comparison, the length of the folded HA ectodomain is ∼34 nm.
That HA-RAP chains with a cytosolic loop longer than ∼160 amino acid residues were able to fold efficiently and to form trimers with full-length HA-RAP molecules confirmed the observation obtained in microsomes that the transmembrane domain can exit the translocon complex when the cytosolic loop is long enough (Mothes et al., 1997). It also allowed us to conclude that binding to the ribosome per se did not affect folding or oligomerization of nascent chains. The restrictions observed on folding and oligomerization were thus primarily caused by association with the translocon complex.
In summary, the results showed that the translocon complex, in addition to its role in translocation, provides a protective and restrictive environment for initial folding of a growing nascent chain. Together with the associated enzymes and chaperons, it can regulate the folding of incoming polypeptide chains. Whether this is generally important for the proper maturation of proteins in the ER remains to be established.
ACKNOWLEDGMENTS
We thank Dr. Dudley K. Strickland for the RAP cDNA plasmid and Dr. Henrik Garoff for the NruI pSFV-1 expression plasmid. We thank Jani Simons, Matt Bui, Ineke Braakman, and other members of the Mellman Helenius group for advice and help. We also thank Henry Tan for photographic work. Grant support from National Institutes of Health and the Swiss National Research Foundation is acknowledged.
REFERENCES
- Beckmann RP, Mizzen LA, Welch WJ. Interaction of hsp70 with newly synthesized proteins: implications for folding and assembly. Science. 1990;248:850–853. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Braakman I, Hoover-Litty H, Wagner KR, Helenius A. Folding of influenza hemagglutinin in the endoplasmic reticulum. J Cell Biol. 1991;114:401–411. doi: 10.1083/jcb.114.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Chen W, Helenius J, Braakman I, Helenius A. Cotranslational folding and calnexin binding of influenza hemagglutinin in the endoplasmic reticulum. Proc Natl Acad Sci USA. 1995;92:6229–6233. doi: 10.1073/pnas.92.14.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Falcone D, Lin J, Andrews DW, Johnson AE. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Fedorov AN, Baldwin TO. Cotranslational protein folding. J Biol Chem. 1997;272:32715–32718. doi: 10.1074/jbc.272.52.32715. [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- Freedman RB. Protein disulfide isomerase: multiple roles in the modification of nascent secretory proteins. Cell. 1989;57:1069–1072. doi: 10.1016/0092-8674(89)90043-3. [DOI] [PubMed] [Google Scholar]
- Hamman BD, Chen JC, Johnson EE, Johnson AE. The aqueous pore through the translocon has a diameter of 40–60 A during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharides, glucose trimming and calnexin during glycoprotein folding in the endoplasmic reticulum. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardesty B, Tsalkova T, Kramer G. Cotranslational folding. Curr Opin Struct Biol. 1999;9:111–114. doi: 10.1016/s0959-440x(99)80014-1. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Zhang J-X, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtley SM, Bole DG, Hoover-Litty H, Helenius A, Copeland CS. Interactions of misfolded Influenza hemagglutinin with binding protein (BiP) J Cell Biol. 1989;108:2117–2126. doi: 10.1083/jcb.108.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeström P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology. 1991;9:1–7. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- Martoglio B, Hofmann MW, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Molinari M, Helenius A. Glycoproteins form mixed disulfides with oxidoreductases during folding in living cells. Nature. 1999;402:90–93. doi: 10.1038/47062. [DOI] [PubMed] [Google Scholar]
- Mothes W, Heinrich SU, Graf R, Nilsson I, von HG, Brunner J, Rapoport TA. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Nathans D. Puromycin inhibition of protein synthesis: incorporation of puromycin into peptide chains. Proc Natl Acad Sci USA. 1964;51:585–592. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- Peterson JR, Ora A, Nguyen Van P, Helenius A. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol Biol Cell. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Singh I, Doms RW, Wagner KR, Helenius A. Intracellular transport of soluble and membrane-bound glycoproteins: folding, assembly and secretion of anchor-free influenza hemagglutinin. EMBO J. 1990;9:631–639. doi: 10.1002/j.1460-2075.1990.tb08155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland DK, Ashcom JD, Williams S, Battey F, Behre E, McTigue K, Battey JF, Argraves WS. Primary structure of α2-macroglobulin receptor-associated protein. J Biol Chem. 1991;266:13364–13369. [PubMed] [Google Scholar]
- Tatu U, Hammond C, Helenius A. Folding and oligomerization of Influenza hemagglutinin in the ER and the intermediate compartment. EMBO J. 1995;14:1340–1348. doi: 10.1002/j.1460-2075.1995.tb07120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein FO, Noll H, Penman S. Effect of cycloheximide on ribosomal aggregates engaged in protein synthesis in vitro. Biochim Biophys Acta. 1964;87:525–528. doi: 10.1016/0926-6550(64)90131-8. [DOI] [PubMed] [Google Scholar]
- Whitley P, Nilsson I, von Heijne G. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J Biol Chem. 1996;271:6241–6244. doi: 10.1074/jbc.271.11.6241. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]