Escherichia coli chemotaxis is arguably the best understood of all biological behaviors, but even this relatively “simple” system continues to offer up new molecular insights from Evolution's bag of signaling tricks. The latest surprise comes from Silversmith et al. (15), who report in this issue how the X-ray structure of a mutant CheY protein led them to the discovery of a CheY residue that plays a special role in this response regulator's phosphorylation activities. Their story not only clarifies a mechanistically murky step in chemotactic signal transduction but also provides us with a nice example of molecular detective work.
THE CHEMOTAXIS PHOSPHORELAY
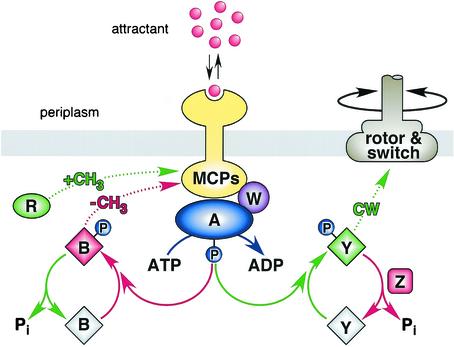
E. coli uses a signaling cascade of protein phosphorylation and dephosphorylation reactions to control its flagellar motors in response to environmental chemical changes (Fig. 1) (see references 1 to 3 for recent reviews). Some signaling reactions (depicted in green in Fig. 1) augment clockwise flagellar rotation, which promotes random directional turns during swimming. Other reactions (depicted in red) suppress clockwise signals to enhance counterclockwise rotation, the default state of the flagellar motors, which promotes forward swimming. The clockwise- and counterclockwise-enhancing elements of the signaling circuit are wired in opposition so that the two signals are balanced during steady-state conditions, producing episodes of forward movement punctuated by occasional random turns. Chemical stimuli transiently perturb the clockwise-counterclockwise balance to promote chemotactic movements.
FIG. 1.
Circuit elements and signaling logic of the E. coli chemotaxis pathway. Green, components and reactions that augment clockwise flagellar rotation; red, counterclockwise-enhancing elements. Cytoplasmic Che proteins are designated by single letters; MCPs (methyl-accepting chemotaxis proteins) are transmembrane chemoreceptors that modulate the autokinase activity of CheA in response to changes in ligand occupancy and methylation state. The CheB and CheY response regulators are inactive (gray) until phosphorylated. See text for additional details.
The flux of phosphates through the signaling cascade is governed by transmembrane chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs) (5). MCPs possess periplasmic ligand-binding domains for monitoring attractant compounds, such as serine or aspartate, and cytoplasmic signaling domains that undergo reversible methylation at 4 to 6 glutamic acid residues. CheA, a histidine kinase, and CheW, a still enigmatic coupling factor, bind to the cytoplasmic domains of MCP molecules to form ternary signaling complexes that oscillate between kinase-on and kinase-off activity states. The proportion of receptor signaling complexes in the kinase-off and kinase-on states reflects a dynamic interplay between chemoreceptor occupancy and methylation state. Ligand-binding changes shift the equilibrium distribution of signaling complexes to initiate motor responses and to set in motion a slower feedback process that adjusts the methylation states of the receptor population to restore the prestimulus proportions of the two signaling states.
The signaling cascade begins with CheA, which donates its phosphoryl groups to two competing response regulators, CheB and CheY, thereby activating them. Phospho-CheY binds to the FliM protein in the flagellar motors to augment clockwise rotation. Phospho-CheB, an MCP-specific methylesterase, demethylates MCP molecules to shift them to the counterclockwise (kinase-off) state. Its counterpart, CheR, an MCP-specific methyltransferase, operates at a constant rate that is not modulated by stimuli. Thus, net changes in MCP methylation state are brought about by changes in the relative rates of the methylation and demethylation reactions through feedback control of CheB activity.
TIMING IS EVERYTHING
The flux rates of phosphates through the CheY and CheB circuits are critical to proper chemotactic signaling. Motor responses occur within 200 ms of stimulus application, demonstrating that phospho-CheY levels change appreciably on this time scale (8, 14). Phospho-CheB levels presumably change on a comparable time scale, but sensory adaptation takes longer because receptor methylation and demethylation changes follow a slower time course. The disparity between rapid motor control and slower sensory adaptation endows E. coli with a few-second “memory” of its recent chemical past, enabling the cell to monitor temporal changes in chemoeffector levels as it swims about in spatial chemical gradients (12).
Phospho-CheY and phospho-CheB undergo rapid dephosphorylation, which enables the chemoreceptors to shift CheY and CheB phosphorylation states on a short time scale by modulating the production of signaling phosphates by CheA. Both response regulators undergo self-catalyzed dephosphorylation, but phospho-CheB has a much shorter half-life than phospho-CheY (17). To compensate, another E. coli signaling component, CheZ, is specifically dedicated to accelerating CheY dephosphorylation (6). Cells that lack CheZ are nonchemotactic and respond to stimuli with response latencies of seconds rather than milliseconds, demonstrating the critical importance of rapid phospho-CheY turnover (8, 13).
WHO'S THE CATALYST?
The nature of CheZ has long been a mystery. Is CheZ simply an allosteric effector that augments the rate of CheY autodephosphorylation, or does CheZ catalyze a novel CheY dephosphorylation reaction? CheZ is known to bind tightly to phospho-CheY and in this way might induce a conformational change that enhances the dephosphorylation reaction. However, the recently solved structure of a CheZ-CheY cocrystal revealed that at least one CheZ residue, Gln-147, plays a catalytic role in the dephosphorylation of phospho-CheY (21). Thus, the CheZ-mediated reaction is distinctly different from CheY autodephosphorylation.
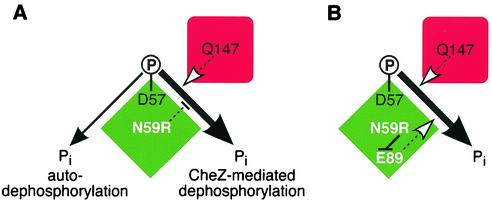
Does CheY also contribute novel catalytic determinants to the CheZ-mediated reaction? In an earlier study, Silversmith et al. (16) created a CheY mutant (CheY-N59R) that exhibited normal phosphorylation, autodephosphorylation, and CheZ binding but was highly “resistant” to CheZ augmentation of its dephosphorylation rate (Fig. 2A). At first glance, it might seem that Asn-59 must play a central role in the CheZ reaction, but this explanation was excluded by showing that other amino acid replacements at residue 59, for example, alanine, did not render the protein CheZ resistant (16). Perhaps the arginine side chain of CheY-N59R blocked CheZ access to the CheY active site or distorted the CheY active site through side chain interactions with another CheY residue. To test these possibilities, Silversmith et al. (15) determined the crystal structure of CheY-N59R complexed with a stable phosphoryl group analog. In a tightly reasoned series of experiments, they showed that Glu-89 of CheY, not Asn-59, was critical to the CheZ-mediated dephosphorylation reaction. The CheZ resistance of CheY-N59R proved to be due to formation of a salt bridge between the side chains of Arg-59 and Glu-89 that prevented the Glu-89 carboxyl group from participating in the CheZ-mediated dephosphorylation reaction (Fig. 2B).
FIG. 2.
Discovery of a CheY residue specifically needed for CheZ-mediated dephosphorylation. Green diamond, CheY; red square, CheZ. (A) Summary of CheY dephosphorylation reactions. CheY autophosphorylates at aspartate-57. The other active-site residues needed for that reaction are also involved in autodephosphorylation. CheZ residue Q147 plays an active role in accelerating the dephosphorylation of CheY. The CheY-N59R mutation has no effect on autodephosphorylation but disrupts CheZ-mediated dephosphorylation, even though N59 itself has no role in the CheZ-mediated reaction. (B) Mechanism of the CheY-N59R effect deduced by Silversmith et al. (15). The arginine side chain makes a salt bridge to the carboxyl group of CheY-E89, which prevents it from participating, as it normally does, in the CheZ-mediated reaction.
The curious case of CheY-N59R provides two cautionary notes for those of us who study structure-function relationships in proteins: (i) a lone mutant can give misleading molecular clues, and (ii) it's difficult to deduce molecular mechanisms without structure.
CONTROLLING CheZ
Activities that dephosphorylate response regulators are consistent features of signaling phosphorelays in bacteria. CheZ plays this role in the chemotaxis pathways of E. coli and other enteric bacteria. In the chemotaxis system of Sinorhizobium meliloti a second CheY protein serves as a phosphate sink to siphon phosphoryl groups from the motor-controlling CheY (11). Many other bacteria possess multiple CheY proteins that may serve a similar function (1). In simple two-component systems that control gene expression, the sensor kinase itself often acts as both a kinase and a phosphatase for its cognate response regulator (10, 18). By contrast, the complex pathway that negotiates sporulation decisions in Bacillus subtilis employs a number of dedicated phosphatases to provide checkpoint control over the phosphorylation states of key signaling components (4, 9).
Regulated dephosphorylation is a hallmark of gene-controlling phosphorelays. For example, in the EnvZ-OmpR system, medium osmolarity appears to control both the kinase and phosphatase activities of EnvZ (7, 19). Similarly, the phosphatases in the B. subtilis sporulation pathway respond to several sensory inputs (4). The signaling decisions in gene regulation pathways are probably made in a leisurely fashion compared to the millisecond response times needed for chemotactic behavior. It's surprising, therefore, that there is no compelling in vivo evidence for stimulus control of the phospho-draining activities in chemotaxis phosphorelays. Has Evolution really failed to exploit these opportunities for finer and faster control of response regulator activity?
To date, in vitro work on the E. coli signaling system has provided tantalizing hints that CheZ activity might be under stimulus control (20), but the field still awaits an experimental slam dunk that resolves the issue. Better understanding of the CheZ-mediated dephosphorylation reaction, including the role played by its CheY substrate, may provide useful clues to the molecular nature of that elusive control mechanism.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 2.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 3.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hess, J. F., K. Oosawa, N. Kaplan, and M. I. Simon. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79-87. [DOI] [PubMed] [Google Scholar]
- 7.Jin, T., and M. Inouye. 1993. Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. J. Mol. Biol. 232:484-492. [DOI] [PubMed] [Google Scholar]
- 8.Khan, S., K. Amoyaw, J. L. Spudich, G. P. Reid, and D. R. Trentham. 1992. Bacterial chemoreceptor signaling probed by flash photorelease of a caged serine. Biophys. J. 62:67-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perego, M., P. Glaser, and J. A. Hoch. 1996. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol. Microbiol. 19:1151-1157. [DOI] [PubMed] [Google Scholar]
- 10.Robinson, V. L., D. R. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7:626-633. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt, R. 2002. Sinorhizobial chemotaxis: a departure from the enterobacterial paradigm. Microbiology 148:627-631. [DOI] [PubMed] [Google Scholar]
- 12.Segall, J. E., S. M. Block, and H. C. Berg. 1986. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 83:8987-8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segall, J. E., A. Ishihara, and H. C. Berg. 1985. Chemotactic signaling in filamentous cells of Escherichia coli. J. Bacteriol. 161:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segall, J. E., M. D. Manson, and H. C. Berg. 1982. Signal processing times in bacterial chemotaxis. Nature 296:855-857. [DOI] [PubMed] [Google Scholar]
- 15.Silversmith, R. E., G. P. Guanga, L. Betts, C. Chu, R. Zhao, and R. B. Bourret. 2003. CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: a role for CheY glutamate 89. J. Bacteriol. 185:1495-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silversmith, R. E., J. G. Smith, G. P. Guanga, J. T. Les, and R. B. Bourret. 2001. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. J. Biol. Chem. 276:18478-18484. [DOI] [PubMed] [Google Scholar]
- 17.Stewart, R. C. 1993. Activating and inhibitory mutations in the regulatory domain of CheB, the methylesterase in bacterial chemotaxis. J. Biol. Chem. 268:1921-1930. [PubMed] [Google Scholar]
- 18.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 19.Tokishita, S., A. Kojima, and T. Mizuno. 1992. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of membrane-located protein kinase, EnvZ. J. Biochem. (Tokyo) 111:707-713. [DOI] [PubMed] [Google Scholar]
- 20.Wang, H., and P. Matsumura. 1996. Characterization of the CheAs/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol. Microbiol. 19:695-703. [DOI] [PubMed] [Google Scholar]
- 21.Zhao, R., E. J. Collins, R. B. Bourret, and R. E. Silversmith. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol. 9:570-575. [DOI] [PubMed] [Google Scholar]