Abstract
Glucose performs key functions as a signaling molecule in the yeast Saccharomyces cerevisiae. Glucose depletion is known to regulate gene expression via pathways that lead to derepression of genes at the transcriptional level. In this study, we have investigated the effect of glucose depletion on protein synthesis. We discovered that glucose withdrawal from the growth medium led to a rapid inhibition of protein synthesis and that this effect was readily reversed upon readdition of glucose. Neither the inhibition nor the reactivation of translation required new transcription. This inhibition also did not require activation of the amino acid starvation pathway or inactivation of the TOR kinase pathway. However, mutants in the glucose repression (reg1, glc7, hxk2, and ssn6), hexose transporter induction (snf3 rgt2), and cAMP-dependent protein kinase (tpk1w and tpk2w) pathways were resistant to the inhibitory effects of glucose withdrawal on translation. These findings highlight the intimate connection between the nutrient status of the cell and its translational capacity. They also help to define a new area of posttranscriptional regulation in yeast.
INTRODUCTION
The ability to sense and respond to changes in the nutritional environment is essential for the flexibility of growth exhibited by the yeast Saccharomyces cerevisiae. The signal transduction pathways that control this flexibility can be induced by levels of specific nutrients such as amino acids and glucose. In general, these pathways bring about changes in gene expression (most commonly transcription) or posttranslational modifications (Thevelein, 1994).
Nutrients also control the level of protein synthesis in cells. Starvation for amino acids or purines is known to cause a general inhibition of translation (summarized in Figure 1A) as well as an activation of many genes involved in amino acid biosynthesis (Hinnebusch, 1984; Tzamarias et al., 1989; Rolfes and Hinnebusch, 1993). A detailed model to account for this has been presented (Hinnebusch, 1996). It proposes that the accumulation of uncharged tRNAs activates the Gcn2p protein kinase, which phosphorylates the α-subunit (Sui2p) of the translation initiation factor eIF2 (Dever et al., 1992; Ramirez et al., 1992). This phosphorylation traps eIF2 in an inactive GDP-bound form via sequestration of the eIF2B guanine nucleotide exchange factor. As eIF2-GTP is required for translation initiation, the accumulation of eIF2-GDP results in a general inhibition of translation (Trachsel, 1996).
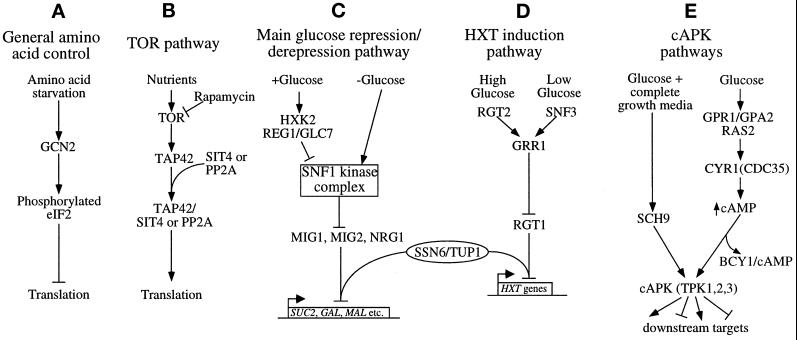
Figure 1.
Summary of the signaling pathways involving translational or glucose-mediated controls in S. cerevisiae. Arrows represent activating signals, and blunt-ended lines represent inhibitory signals.
Another example of signal-mediated translational control in yeast involves the inhibition of the TOR kinase pathway (Figure 1B). This pathway is inhibited by both nutrient starvation and the drug rapamycin, leading to the inhibition of translation (Barbet et al., 1996). The targets of rapamycin, Tor1p and Tor2p, are phosphatidylinositol kinase homologues. Mutations in these kinases generate a rapamycin-resistant phenotype. Tor1p and Tor2p are thought to control translation initiation by stimulating the association of type 2A (PP2A) and type 2A–related (Sit4p) phosphatases with Tap42p via phosphorylation of Tap42p (Di Como and Arndt, 1996; Jiang and Broach, 1999). A mutant form of Tap42p, Tap42-11p, confers partial rapamycin resistance to cells by maintaining its association with the phosphatases even when Tor1p and Tor2p are inactivated. The mechanism by which Tap42p controls translation initiation is not currently understood, although rapamycin treatment has been shown to lead to the degradation of eIF4G (a component of the eIF4F cap-binding complex) (Berset et al., 1998). More recent studies have also shown that rapamycin treatment affects both amino acid transporters and transcription of the ribosomal protein genes (Schmidt et al., 1998; Powers and Walter, 1999).
Glucose is one of the major signaling nutrients for S. cerevisiae as well as its preferred carbon source. Several well-characterized signal transduction pathways (summarized in Figure 1, C–E) allow yeast to perceive the level of glucose, and collectively these pathways initiate the appropriate response to this level (Thevelein, 1994; Gancedo, 1998). In general, many of these responses involve alterations in programs of gene expression, and the majority of these alterations occur at the level of mRNA transcription.
In this paper, we have made use of a preliminary observation from our laboratory and from others (Martinez-Pastor and Estruch, 1996) that protein synthesis in yeast is inhibited by removal of the carbon source from the growth medium, whereas transcription is unaffected. We have expanded on this observation to show that the translational inhibition is rapid, reversible, and specific for glucose or fructose removal. The inhibition and its reversal do not require new transcription and do not make use of previously described nutrient-based translational control pathways. However, the inhibition is overcome in glucose repression, hexose transporter (HXT) induction, and cAMP-dependent protein kinase (cAPK) mutants. These results suggest that the yeast translational apparatus is tightly controlled by the nutritional status of the cell, and they indicate the existence of a previously undescribed pathway that can lead to the rapid inhibition of protein synthesis in response to environmental changes.
MATERIALS AND METHODS
Strains and Growth Conditions
The strains used in this study are listed in Table 1. They are listed as yAS numbers with their more common names in parentheses. The strain yAS2568 (W3031A) was used as the wild-type strain unless stated otherwise. Strains were grown on either standard yeast extract/peptone medium (YP) or synthetic complete (SC) yeast nitrogen base/ammonium sulfate/amino acid medium supplemented with 2% carbon source, as indicated (Guthrie and Fink, 1991). Strains were grown at 30°C except for temperature-sensitive (ts) mutants, which were grown at 26°C. The identities of most mutant strains were confirmed with the use of growth phenotypes specific to each strain. The gcn2::URA3 mutant (yAS2569) was generated with the use of standard methods by transformation of yAS2568 with a BstEII–SnaBI fragment from the plasmid p781 (Wek et al., 1992). The SNF1::G418 strains (yAS2605 [reg1Δ snf1Δ], yAS2606 [hxk2Δ snf1Δ], and yAS2602 [snf3Δ rgt2Δ snf1Δ]) were generated by transformation of strains yAS2576, yAS2578, and yAS2537 with a PCR product containing 40 base pairs found upstream and downstream of the SNF1 ORF surrounding the KanMX2 gene (Wach et al., 1994). Insertions were verified by Southern blotting. yAS2570 and yAS2571 were generated by five backcrosses of the H1645 strain (Dever et al., 1992) to yAS2568 followed by replacement of the resident SUI2 URA3 plasmid with either the wild-type or mutant SUI2 LEU2 plasmid (strains and plasmids used here were kindly provided by Tom Dever, National Institutes of Health). yAS2566 and yAS2567 were generated by transformation of yAS2531 (yM4127) with the plasmids pBM3259 (SNF3-1) and pBM3270 (RGT2-1), respectively (Özcan et al., 1998) (strains and plasmids used here were kindly provided by Mark Johnston, Washington University).
Table 1.
Yeast strains used in this study
| Strain name | Genotype | Source |
|---|---|---|
| yAS306 (W3031A) | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Sachs strain collection |
| yAS879 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 rpb1-1 | Sachs strain collection |
| yAS956 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 bcy1∷URA3 | Sachs strain collection |
| yAS2568 (W3031A) | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Sachs strain collection |
| yAS2569 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1gcn2∷URA3 | This study |
| yAS2570 | MATa ade2 his3-11,15 leu2-3,112 trp1 ura3 sui2Δ p[SU12 LEU2 CEN] | This study |
| yAS2571 | MATa ade2 his3-11,15 leu2-3,112 trp1 ura3 sui2Δ p[SU12 S51A LEU2 CEN] | This study |
| yAS2410 (CY4907) | MATa ade2 his3 leu2 trp1 ura3 SSD1-v tap42∷TRP1 p[TAP42 LEU2 CEN] | Di Como and Arndt (1996) |
| yAS2411 (CY4908) | MATa ade2 his3 leu2 trp1 ura3 SSD1-v tap42∷TRP1 p[tap42-11 LEU2 CEN] | Di Como and Arndt (1996) |
| yAS2531 (yM4127) | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 | Özcan et al. (1996a) |
| yAS2527 (yM4509) | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 rgt1∷hisG | Özcan et al. (1996b) |
| yAS2533 (yM4817) | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 rgt2∷HIS3 | Özcan et al. (1996a) |
| yAS2534 (yM6175) | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 snf3∷HIS3 | Özcan et al. (1998) |
| yAS2537 (yM6212) | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 rgt2∷HIS3 snf3∷HIS3 | Özcan et al. (1998) |
| yAS2602 | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 rgt2∷HIS3 snf3∷HIS3 snf1∷G418 | This study |
| yAS2532 (yM4767) | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 ura3-52 RGT2-1 | Özcan et al. (1996a) |
| yAS2529 (yM4553) | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 mig1∷HIS3 | Özcan and Johnston (1996) |
| yAS2566 | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 p[SNF3-1 URA3 CEN] | This study |
| yAS2567 | MATa ade2-101 his3-Δ200 leu2-3,112 lys2-801 trp1-901 tyr1-501 ura3-52 p[RGT2-1 URA3 CEN] | This study |
| yAS2572 (FY250) | MATα his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 | Sherwood and Carlson (1997) |
| yAS2573 (FY251) | MATa his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 | Sherwood and Carlson (1997) |
| yAS2574 (MCY2616) | MATα his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 glc7-T152K | Tu and Carlson (1994) |
| yAS2575 (MCY1595) | MATα his3-Δ200 his4-539 lys2-801 ura3-52 snf1-Δ3 | Celenza et al. (1989) |
| yAS2576 (MCY3278) | MATα his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 reg1∷URA3 | Tu and Carlson (1995) |
| yAS2605 | MATα his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 reg1∷URA3 snf1∷G418 | This study |
| yAS2577 (PS7050-1D) | MATα his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 grr1∷URA3 | Sherwood and Carlson (1997) |
| yAS2578 (PS7851-2A) | MATα his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 hxk2∷URA3 | Sherwood and Carlson (1997) |
| yAS2606 | MATα his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 hxk2∷URA3 snf1∷G418 | This study |
| yAS2579 (PS3851-3A) | MATa his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 gsf1-1 | Sherwood and Carlson (1997) |
| yAS2580 (PS5204-1C) | MATα his3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 gsf2-Δ1∷TRP1 | Sherwood and Carlson (1997) |
| yAS2504 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 hxk1∷URA3 hxk2∷LEU2 glk1∷LEU2 HXK2∷TRP1 | Randez-Gil et al. (1998) |
| yAS2505 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 hxk1∷URA3 hxk2∷LEU2 glk1∷LEU2 HXK2A15∷TRP1 | Randez-Gil et al. (1998) |
| yAS2506 (JC821A/pKC886) | MATα his3-11,15 leu2 Δtrp1 ura3 glc7∷HIS3 p[GLC7 TRP1 CEN] | Ramaswamy et al. (1998) |
| yAS2507-26 (JC821A/pKC866-x) | MATα his3-11,15 leu2 Δtrp1 ura3 glc7∷HIS3 p[GLC7-x TRP1 CEN] | Ramaswamy et al. (1998) |
| yAS2497 (SJ17) | MATα leu2-3,112 gal4 ura3-52 | Tung and Hopper (1995) |
| yAS2498 (SJ17-ND) | MATα leu2-3,112 gal4 ura3-52 ssn6-Δ6∷URA3 | Tung and Hopper (1995) |
| yAS2501 (LRA85) | MATα leu2 his4-539 ura3-52 cdc35-11 | Tung and Hopper (1995) |
| yAS2539 (MLY41a) | MATa ura3-52 | Pan and Heitman (1999) |
| yAS2540 (MLY132a) | MATa ura3-52 gpa2∷G418 | Pan and Heitman (1999) |
| yAS2541 (MLY232a) | MATa ura3-52 gpr1∷G418 | Pan and Heitman (1999) |
| yAS2542 (MLY265a) | MATa ura3-52 sch9∷G418 | J. Heitman |
| yAS962 (SP1) | MATa ade8 his3 leu2 trp1 ura3 | Nikawa et al. (1987) |
| yAS963 (S18-3B) | MATa ade8 his3 leu2 trp1 ura3 tpk1w1 tpk2∷HIS3 tpk3∷TRP1 | Nikawa et al. (1987) |
| yAS964 (S15-5B) | MATa ade8 his3 leu2 trp1 ura3 tpk1∷URA3 tpk2w1 tpk3∷TRP1 | Nikawa et al. (1987) |
| yAS965 (S22-5D) | MATa ade8 his3 leu2 trp1 ura3 tpk1∷URA3 tpk2∷HIS3 tpk3w1 | Nikawa et al. (1987) |
| yAS982 (TK161-R2V) | MATa ade8 his3 leu2 trp1 ura3 RAS2val19 | Nikawa et al. (1987) |
| yAS2599 (ASY62) | MATa ade8 his3 leu2-3,112 trp1 ura3-52 tpk1∷ADE8 tpk2∷HIS3 tpk3∷TRP1 msn2∷HIS3 msn4∷LEU2 | Smith et al. (1998) |
Analysis of Ribosomal Distribution on Sucrose Density Gradients
Yeast cultures were grown to an OD600 of 0.4, and 200 ml was harvested at 8000 × g for 10 min at 30°C in a Sorvall (Newtown, CT) GSA rotor. Cells were resuspended in 200 ml of medium either with or without a carbon source. After a specific time (10 min in the majority of experiments), the culture was added to cold 200-ml centrifuge bottles containing 2 ml of 10 mg/ml cycloheximide. All subsequent steps were performed at 4°C. The cells were centrifuged at 8000 × g for 10 min in a Sorvall GSA rotor and washed in 50 ml of lysis buffer (20 mM HEPES, pH 7.4, 2 mM MgOAc, 100 mM KOAc, 100 μg/ml cycloheximide, 0.5 mM DTT). Cells were pelleted for 3 min with the use of a clinical centrifuge, resuspended in 800 μl of lysis buffer, and transferred to 1.5-ml microcentrifuge tubes. After pelleting for 5 min at 7000 rpm in an Eppendorf (Westbury, NY) microcentrifuge, cells were resuspended in an equal volume of lysis buffer and lysed in the presence of 1 volume of glass beads by vortexing six times for 20 s at 40-s intervals. Lysates were cleared briefly at 10,000 rpm for 5 min in an Eppendorf microcentrifuge, followed by a 20-min 10,000-rpm centrifugation to give the final lysate. The A260 was measured, and 9 A260 units were loaded onto 15–50% linear sucrose gradients as described by Luthe (1983). The gradients were centrifuged in a SW41 rotor (Beckman Instruments, Palo Alto, CA) for 2.5 h at 40,000 rpm, after which the gradients were collected from the bottom. The A254 was measured continuously to generate the traces shown in the figures.
[35S]Methionine Incorporation Assays
These were performed as described by Sachs and Deardorff (1992). Briefly, yeast were grown in SCD lacking methionine (SCD-met) to an OD600 of 0.4, and two 7.5-ml aliquots were pelleted in a clinical centrifuge for 3 min. The two cell pellets were resuspended immediately in 20 ml of SCD-met and SC-met, respectively. Methionine was added to a final concentration of 60 ng/ml, of which 0.5 ng/ml was [35S]methionine (cell-labeling grade, 1175 Ci/mmol; New England Nuclear, Boston, MA). One-milliliter samples were added to 1 ml of 20% trichloroacetic acid (TCA) at the various time points and heated at 95°C for 20 min in glass tubes. The protein precipitate was collected on GFC filters (Whatman, Clifton, NJ), washed with 10 ml of 10% TCA and then with 10 ml of 95% ethanol, and counted in Scintiverse (Fisher, Pittsburgh, PA) scintillation fluid. This process was scaled down and fewer time points were taken for screening through the glc7 mutants.
Northern Blotting
Yeast (yAS2568) were grown in YPD to an OD600 of 0.5. One 15-ml aliquot plus two 30-ml aliquots were centrifuged in a clinical centrifuge. The 15-ml aliquot cell pellet was immediately frozen in liquid N2. The two 30-ml aliquot cell pellets were resuspended in 30 ml of YPD or YP. Fifteen-milliliter aliquots were pelleted, and the pellets were frozen after 5 and 10 min. RNA was extracted from the frozen pellets and used for Northern blots as described by Boeck et al. (1998). The following probes were used: ACT1, a 165-base pair EcoRI fragment from pAS340; PGK1, a 3.1-kilobase HindIII fragment from pAS214; CYH2, a 1.6-kilobase SalI fragment from pAS215; PAB1, a 785-base pair NdeI–SpeI fragment from pAS77; and MFA2, a 326-base pair EcoRI fragment from pAS225.
Immunoblotting of eIF4G
Yeast were grown in YPD to an OD600 of 0.5. Ten-milliliter aliquots were pelleted and resuspended in YPD or YP for various times. Cells were pelleted, frozen in liquid N2, and resuspended in 500 μl of TCA/lysis buffer (10% TCA, 10 mM Tris, pH 8.0, 100 mM NH4OAc, 1 mM EDTA, 1 mM PMSF, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin). Cells were lysed with 0.5 volume of glass beads at 4°C by vortexing (six times for 20 s at 40-s intervals). The TCA precipitates in the lysate were then pelleted and resuspended in loading buffer (3.5% SDS, 14% glycerol, 120 mM Tris base, 0.0025% bromphenol blue, 8 mM EDTA, 120 mM DTT). Samples were boiled and electrophoretically separated with the use of an 8% SDS polyacrylamide gel. Western blotting for eIF4G was performed as described by Tarun and Sachs (1996).
ATP Measurements
The protocol used was adapted slightly from that of Simpson and Hammond (1989). Yeast were grown in SCD to an OD600 of 0.5. As a control, a 20-μl sample was removed from the original culture and added to 20 μl of 10% TCA. This was further processed as described below and then used as the 100% standard. Two 15-ml culture aliquots were centrifuged in a clinical centrifuge for 3 min at 30°C. The two cell pellets were resuspended in 15 ml of SCD or SC, and 20-μl aliquots were removed after 1, 2.5, 5, 7.5, 10, 15, 20, and 30 min. Immediately after removal, these aliquots were mixed with 20 μl of 10% TCA. This was vortexed for 1 min, and 10 μl was removed and added to 990 μl of reaction buffer (25 mM HEPES, pH 7.75, 2 mM EDTA). This was mixed, and 100 μl was added directly to 100 μl of Enliten luciferin/luciferase reagent (Promega, Madison, WI). Luminescence was measured on a Turner Designs (Sunnyvale, CA) TD-20/20 luminometer with the use of a 10-s integration period. Standard curves were determined with the use of ATP standards, and these were used to ensure linearity under these conditions. Assays were performed at least twice for each strain, with maximal errors of ±5% of the value shown.
RESULTS
Glucose Withdrawal Results in a Loss of Polyribosomes
When extracts were prepared from yeast grown and washed for 10 min in standard glucose-containing medium (YPD), they exhibited a normal polyribosome profile (Figure 2A). Such a profile includes an accumulation of multiple ribosomes bound to single mRNAs (polysomes) toward the bottom of the gradient. When cells grown in YPD were washed for 10 min in YP lacking glucose, however, polysomes were lost and the level of single 80S ribosomes increased (Figure 2A). This change in polysome profile has been noted previously for many temperature-sensitive strains bearing mutations in key translation initiation factors upon transfer to the restrictive temperature and is indicative of an inhibition of translational initiation (Hartwell and McLaughlin, 1969; Mathews et al., 1996). We also found that polysomes could be maintained in cells washed in buffer provided that glucose and amino acids were present (our unpublished results).
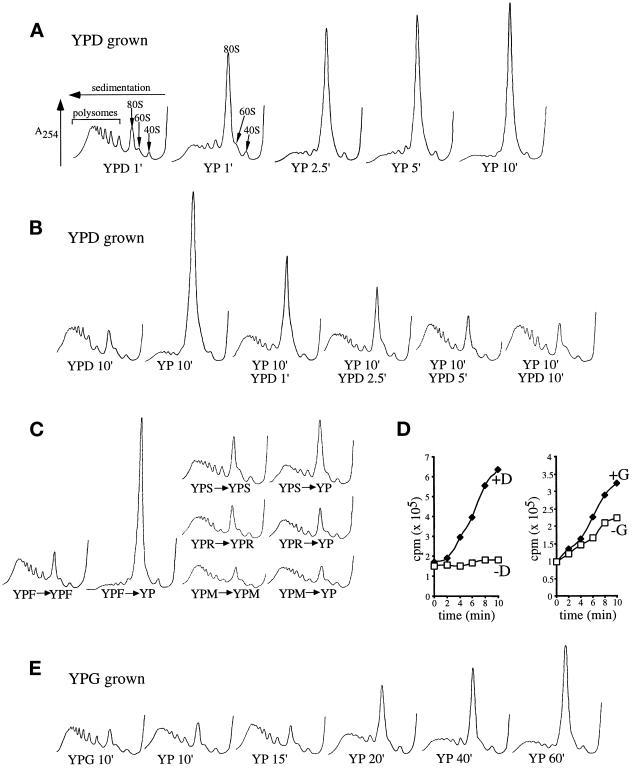
Figure 2.
Glucose or fructose removal from yeast medium specifically inhibits translation initiation. (A) Polyribosome traces from the wild-type strain (yAS2568). Yeast was grown in YPD and resuspended in YP medium lacking (YP) or containing (YPD) glucose for the indicated times (minutes). Polyribosomes were analyzed as described in MATERIALS AND METHODS. The peaks that contain the small ribosomal subunit (40S), the large ribosomal subunit (60S), and both subunits (80S) are indicated by arrows. The polysome peaks generated by 2, 3, 4, 5, etc. 80S ribosomes on a single mRNA are bracketed. (B) Polyribosome traces from wild-type yeast that were washed in the absence of glucose (YP) for 10 min and then incubated in medium containing glucose for the indicated times (minutes). (C) Polyribosome traces from wild-type yeast grown on YP medium with a variety of 2% carbon sources (fructose [F], sucrose [S], maltose [M], and raffinose [R]). Cells were washed for 10 min either in the presence (+) or absence (−) of the carbon source. (D) [35S]Methionine incorporation into proteins over time in wild-type yeast. Cells were grown in synthetic complete minus methionine medium, harvested, and resuspended in a labeling mixture in the presence (♦) or absence (□) of a carbon source (glucose [D] or galactose [G]). Aliquots were taken at the indicated times, and the levels of [35S]methionine incorporated into proteins were determined. (E) Polyribosome traces from wild-type yeast grown in galactose medium (YPG) and resuspended in YP or YPG medium for the indicated times (minutes).
Glucose Withdrawal Inhibits Translation Rapidly and Reversibly
The kinetics of polysome redistribution upon glucose withdrawal was investigated. Extracts from cells that had been washed for increasing amounts of time in the absence of glucose were examined on polysome gradients (Figure 2A). A redistribution of polysomes into the 80S peak was evident after 1 min of glucose depletion, and after 2.5 min this redistribution was almost complete.
The rate of protein synthesis was measured by [35S]methionine incorporation to confirm that the redistribution in polysomes was associated with an inhibition of protein synthesis. Cells were transferred to medium containing or lacking glucose and labeled with [35S]methionine for the next 10 min (Figure 2D). Yeast cells incorporated [35S]methionine linearly in the presence of glucose. However, upon transfer to medium lacking glucose, there was an almost complete inhibition of further [35S]methionine incorporation. These data, together with the nearly complete loss of polysomes in the absence of glucose, indicate that translation initiation is inhibited by glucose removal.
We also asked how quickly the polysome profile could be restored by the readdition of glucose to inhibited cells (Figure 2B). As expected, a 10-min wash without glucose (YP 10′) drastically reduced polysome levels. Subsequent addition of glucose-containing medium led to a marked increase in polysomes after 1–2.5 min and a complete restoration of polysomes after 5 min. Therefore, glucose removal causes a sudden inhibition of translation initiation that can be rapidly reversed.
The Polysome Redistribution Is Carbon Source Specific
We next investigated whether the loss of polyribosomes was specific to glucose withdrawal or a general consequence of carbon source depletion. Accordingly, yeast were grown on various carbon sources (glucose [D], fructose [F], galactose [G], maltose [M], sucrose [S], and raffinose [R]) and then washed for 10 min in the presence or absence of the carbon source. Only yeast growing on glucose or fructose before polysome analysis exhibited polysome loss upon carbon source depletion (Figure 2, B, C, and E). Yeast cells grown on other carbon sources had nearly normal levels of polysomes, even after carbon source removal. These polysomes are indicative of elongating ribosomes, because after galactose withdrawal the yeast continues to incorporate [35S]methionine at approximately half the rate observed when galactose is maintained (Figure 2D).
We also determined how long it took after galactose removal for a decrease in translation to become evident (Figure 2E). After 15 min, there was no major effect of galactose removal upon translation, but at times thereafter, a gradual decrease in the level of polysomes was observed. It is interesting to compare the polysome profile 1 min after glucose removal (Figure 2A) with that produced 40 or 60 min after galactose removal (Figure 2E); they have a very similar pattern, even though the length of time in the absence of a carbon source was vastly different.
We conclude from these data that the rapid inhibition of translation upon carbon source withdrawal is specific to the removal of either glucose or fructose. This carbon source specificity is identical to that of many previously described glucose-dependent signal transduction pathways (Thevelein, 1994). We also found that 2-deoxyglucose had a general inhibitory effect on translation and therefore could not be used to determine whether the absence of glucose or a glucose metabolite leads to translational inhibition (our unpublished results).
Translational Inhibition Is Not Caused by a Gross Decrease in mRNA Abundance
One possible explanation for the inhibition of translation initiation upon glucose withdrawal is that it occurs indirectly via a massive decay of mRNA. Therefore, we assessed the abundance of specific mRNAs with a range of half-lives (11 min for PAB1, 45 min for PGK1, 30 min for ACT1, 43 min for CYH2, and 2.5 min for MFA2 [Herrick et al., 1990]) by Northern blot analysis after glucose removal (Figure 3A). In general, the level of mRNAs did not decrease when glucose was withdrawn. In fact, the level of certain mRNAs (e.g., CYH2) increased when glucose was removed. This effect could be due to the inhibition of translation, which presumably has already taken place and leads to the stabilization of certain mRNAs (Peltz et al., 1992; Beelman and Parker, 1994). These results rule out the formal possibility that glucose withdrawal leads to translational inhibition as a result of increased mRNA instability.
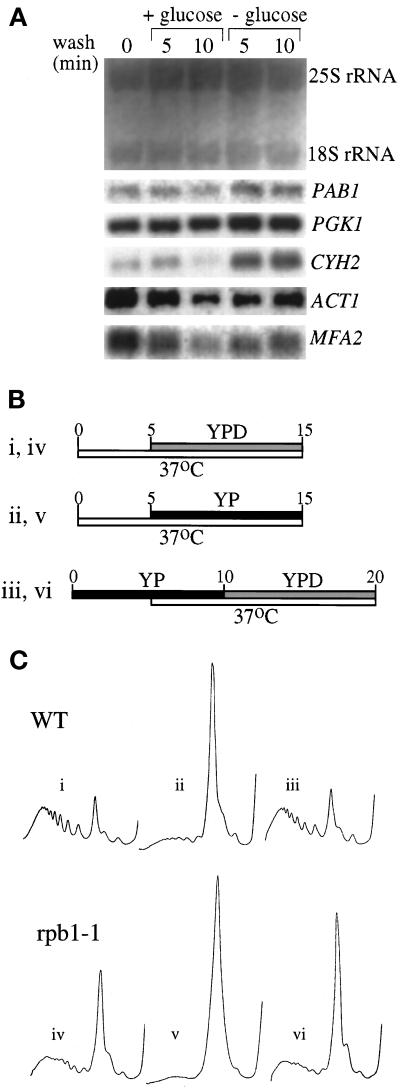
Figure 3.
Neither mRNA degradation nor transcription contributes to the inhibition of translation upon glucose withdrawal or the recovery after glucose readdition. (A) Northern blot stained with methylene blue to visualize the rRNA levels (upper panel). The same blot was probed for a variety of specific mRNAs (lower panels) (see MATERIALS AND METHODS for details). (B) Line diagram for the experimental protocol used in C. The Roman numerals refer to the polyribosome traces shown in C. The timing (minutes) and length of the temperature change from 30 to 37°C is depicted by the lower, white bars. The upper bars represent medium changes and length of washes (gray bars represent a change to YPD, and black bars represent a change to YP). (C) Polyribosome traces for the wild-type (yAS306) and RNA polymerase II mutant rpb1-1 (yAS879) strains. Roman numerals refer to B, which depicts the order and timing of YP or YPD washes and temperature changes that occurred before cells were harvested for polyribosome analysis.
Neither Translational Inhibition nor Recovery Requires New Transcription
The rpb1-1 temperature-sensitive mutant was used to assess whether an additional round of gene expression is a requirement for the process of translational inhibition. This strain harbors a mutation in an RNA polymerase II gene, and after a shift to the restrictive temperature, it is rapidly inhibited for the transcription of most mRNAs (Nonet et al., 1987; Herrick et al., 1990). rpb1-1 or its isogenic wild-type parent was shifted to 37°C 5 min before a 10-min incubation in medium with or without glucose (Figure 3B). In wild-type yeast, this temperature shift did not affect the polysome profile in the presence of glucose (Figure 3Ci), and the removal of glucose still inhibited translation (Figure 3Cii). For the rpb1-1 mutant, the 15-min 37°C incubation would be expected to lead to a substantial decrease in the quantity of mRNA and therefore polysomes, because the average half-life of a yeast mRNA is ∼15 min. Indeed, as shown in Figure 3Civ, after 15 min at 37°C the rpb1-1 strain contains approximately half the polysomes present in the wild-type parent. However, even allowing for this decrease in polysomes after transcriptional shutdown, there was still a rapid additional decrease in polysome levels when glucose was removed from the medium (Figure 3Cv).
A similar result was observed for the recovery of translational activity after the readdition of glucose. In these experiments, glucose was removed from yeast for 10 min to allow for the inhibition of translation. Halfway through this period, the yeast cells were shifted to 37°C to completely inhibit mRNA transcription in the rpb1-1 strain. The levels of polysomes were then assayed after readdition of glucose for another 10 min (Figure 3B). In the wild-type strain (Figure 3Ciii), the shift in temperature to 37°C did not affect its ability to recover polysomes after readdition of glucose. For the rpb1-1 strain (Figure 3Cvi), polysomes recovered to almost the same level as that seen when transcription was inhibited in the presence of glucose (Figure 3Civ). These results indicate that new transcription is not required for glucose withdrawal to inhibit translation or for glucose readdition to stimulate translation.
The Inhibition of Translation by Glucose Withdrawal Does Not Occur through Previously Described Translational Inhibitory Mechanisms
As described in the INTRODUCTION, some prominent examples of signal-mediated translational inhibition have been discovered. First, starvation for amino acids leads to a global inhibition of translation (Tzamarias et al., 1989). We confirmed this result (Figure 4A) with the use of a wild-type strain in which the withdrawal of amino acids reduced the level of polysomes. Surprisingly, glucose removal had a more severe effect than even amino acid depletion, because it generated a greater reduction in polysomes. The amino acid starvation pathway of translational inhibition requires the Gcn2p protein kinase, which phosphorylates the α-subunit of eIF2 (a translation initiation factor encoded by the SUI2 gene). The mutation of Ser51 to Ala in Sui2p disrupts this regulatory circuit (Dever et al., 1992). This mutation was used to investigate whether the translational inhibition by glucose withdrawal occurs by a similar mechanism. Consistent with previous results, the SUI2-S51A strain was resistant to the inhibition of translation caused by amino acid withdrawal (Figure 4A). However, the mutant was sensitive to the removal of glucose at the translational level (Figure 4A). Similar effects were found with a gcn2 null strain (our unpublished results). These results support the conclusion that glucose removal and amino acid starvation elicit translational inhibition via different mechanisms.
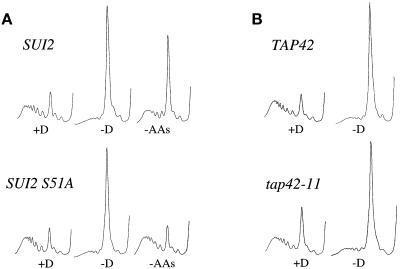
Figure 4.
Mutants in other translational control pathways are still inhibited for translation upon glucose withdrawal. (A) Polyribosome traces from the wild-type (yAS2570) and SUI2-S51A (yAS2571) strains. Yeast was grown in SCD-Leu medium and washed for 10 min in SCD-Leu (+D), SC-Leu (−D), or SCD minus all amino acids (−AAs). (B) Polyribosome traces from the wild-type (yAS2410) and tap42-11 (yAS2411) strains. Yeast was grown in SCD-Leu medium and then washed for 10 min in either SCD-Leu (+D) or SC-Leu (−D).
Another signal-mediated translational inhibitory response is induced by inactivation of the TOR signaling pathway (Barbet et al., 1996). For example, rapamycin or nutrient starvation inhibits the TOR proteins, which are normally capable of phosphorylating Tap42p. A specific mutation, tap42-11, confers rapamycin resistance (Di Como et al., 1996). If the inhibition of translation by glucose removal is brought about via inactivation of Tor1p and Tor2p, we reasoned that the tap42-11 mutant would be at least partially resistant to glucose withdrawal. However, the tap42-11 mutant exhibited the same level of translational inhibition on glucose withdrawal as its isogenic parent (Figure 4B). The kinetics and magnitude of the rapamycin-dependent inhibition of translation also bear little relation to those of the inhibition of translation by glucose depletion (Barbet et al., 1996; Powers and Walter, 1999). Previously, even 30 min after rapamycin treatment only a limited decrease in polysomes was observed (Powers and Walter, 1999), whereas there was almost complete loss of polysomes 2.5 min after glucose depletion in the present study. It has also been suggested that rapamycin leads to the selective degradation of the translation initiation factor eIF4G (Berset et al., 1998). When glucose was withdrawn from yeast cells, we found no evidence that eIF4G was degraded (our unpublished results). These results suggest that the inhibition of translation by glucose removal is unlikely to result from the inactivation of the TOR pathway.
Specific Glucose Repression Mutants Exhibit Resistance to Translational Inhibition by Glucose Withdrawal
Growth in the presence of glucose represses the expression of a wide variety of genes whose products are involved in the use of alternative carbon sources and oxidative metabolism (Gancedo, 1998). This process involves the signal transduction pathway summarized in Figure 1C. One of the most significant changes that occurs in yeast cells depleted of glucose is the activation of the glucose derepression pathway. The absence of glucose generates an active Snf1p protein kinase complex that inhibits the activity of several transcriptional repressors. This leads to the derepression of many glucose-repressed genes. We analyzed a series of mutants in the glucose repression/derepression pathway to test whether this pathway is involved in the inhibition of translation upon glucose removal. These mutants have been isolated and characterized during many years of study of this pathway (Entian, 1980; Celenza and Carlson, 1986; Niederacher and Entian, 1987; Schüller and Entian, 1987; Vallier and Carlson, 1994; Tu and Carlson, 1994, 1995; Devit et al., 1997; Sherwood and Carlson, 1997).
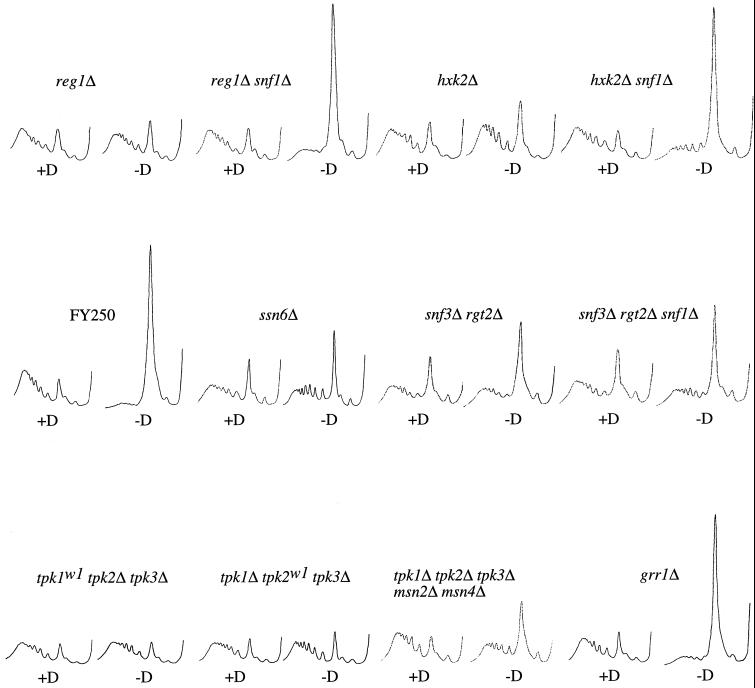
A number of mutants in the glucose repression pathway that give constitutive derepression were identified as translationally resistant to the removal of glucose. For example, as shown in Figure 5 and Table 2, reg1Δ, hxk2Δ, and ssn6-Δ6 were resistant to translational inhibition on glucose removal. However, other glucose repression mutations, such as glc7-T152K, mig1Δ, gsf1-1, and gsf2-Δ1, did not give resistance to the translational inhibition by glucose withdrawal, even though these mutants are derepressed.
Figure 5.
Polyribosome analyses for a selection of strains used in this study. Examples of polyribosome traces generated with the use of the wild-type strain (yAS2572 [FY250]) and various mutant strains (yAS2576 [reg1Δ], yAS2605 [reg1Δ snf1Δ], yAS2578 [hxk2Δ], yAS2606 [hxk2Δ snf1Δ], yAS2498 [ssn6Δ], yAS2537 [rgt2Δ snf3Δ], yAS2602 [rgt2Δ snf3Δ snf1Δ], yAS963 [tpk1w1 tpk2Δ tpk3Δ], yAS964 [tpk1Δ tpk2w1 tpk3Δ], yAS2599 [tpk1Δ tpk2Δ tpk3Δ msn2Δ msn4Δ], and yAS2577 [grr1Δ]) washed for 10 min in the presence (+D) or absence (−D) of glucose.
Table 2.
A summary of the sensitivity/resistance of various mutant yeast strains to glucose removal at the translational level
| Relevant genotype | Resistance to translational inhibition on glucose withdrawala | Ratio of [35S]-methionine incorporation with or without glucoseb | Doubling time (min)c | Repressed (R) or derepressed (DR) on glucosed |
|---|---|---|---|---|
| Wild typee | − | >16 | 80–145e | R |
| snf1-Δ3 (yAS2575) | − | N | 105 | R |
| reg1Δ (yAS2576) | +++ | 2.29 | 110 | DR |
| reg1Δ snf1Δ (yAS2605) | − | N | 95 | R |
| hxk2Δ (yAS2578) | +++ | 1.89 | 100 | DR |
| hxk2Δ snf1Δ (yAS2606) | − | N | 240 | R |
| HXK2 hxk1Δ glk1Δ (yAS2504) | − | N | 85 | R |
| hxk2S15A hxk1Δ glk1Δ (yAS2505) | − | N | 85 | DR |
| glc7-T152K (yAS2574) | − | N | 100 | DR |
| glc7-Q48K (yAS2511) | + | 5.76 | 100 | DR |
| gsf1-1 (yAS2579) | − | N | 115 | DR |
| gsf2-Δ1 (yAS2580) | − | N | 85 | DR |
| mig1Δ (yAS2529) | − | N | 100 | DR |
| ssn6-Δ6 (yAS2498) | ++ | N | 215 | DR |
| rgt1Δ (yAS2527) | − | N | 100 | N |
| RGT2-1f (yAS2532/yAS2567) | − | N | 90/200f | N |
| SNF3-1 (yAS2566) | − | N | 200 | N |
| rgt2Δ (yAS2533) | − | N | 120 | N |
| snf3Δ (yAS2534) | − | N | 120 | N |
| snf3Δ rgt2Δ (yAS2537) | ++ | 2.15 | 225 | DR |
| snf3Δ rgt2Δ snf1Δ (yAS2602) | ++ | 2.81 | 230 | N |
| grr1Δ (yAS2577) | − | N | 110 | DR |
| tpk1w1 tpk2Δ tpk3Δ (yAS963) | +++ | 1.67 | 220 | R |
| tpk1Δ tpk2w1 tpk3Δ (yAS964) | +++ | 1.72 | 260 | R |
| tpk1Δ tpk2Δ tpk3w1 (yAS965) | − | N | 145 | R |
| tpk1Δ tpk2Δ tpk3Δ | ||||
| msn2Δ msn4Δ (yAS2599) | ++ | 1.94 | 139 | N |
| bcy1Δ (yAS956) | − | N | 90 | N |
| RAS2val19 (yAS982) | − | N | 135 | N |
| cdc35-11 (yAS2501) | −/+ | N | 155 | N |
| gpa2Δ (yAS2540) | − | N | 100 | N |
| gpr1Δ (yAS2541) | − | N | 100 | N |
| sch9Δ (yAS2542) | −/+ | N | 125 | N |
Cells were generally grown on YPD and then transferred to YPD or YP. It was noted that certain resistant strains grown on SCD gave lower levels of translational resistance to glucose withdrawal.
Rates of incorporation were measured after transfer to medium with or without glucose. N, not determined.
Approximate doubling time of the strain on YPD unless plasmid maintenance required the use of SCD-minus medium.
Measured by growth on YP sucrose plus 2-deoxyglucose or taken from previous work in a number of laboratories. N, the strains were not tested.
Wild-type backgrounds to all the strains used were tested.
The dominant RGT2-1 allele was tested either on a plasmid in a wild-type strain (grown on SCD) or in the RGT2-1 strain.
Reg1p is a regulatory subunit of the Glc7p protein phosphatase 1. It is thought to directly maintain Snf1p in an inactive state via its dephosphorylation (Tu and Carlson, 1995; Ludin et al., 1998). Therefore, it was unexpected that a reg1 mutant was resistant to glucose removal but the glc7-T152K mutant (which has been shown to be defective in Reg1p binding) was not. To further examine this discrepancy, we analyzed a series of strains containing mutations in the GLC7 gene (see Ramaswamy et al. [1998] for a full listing of the mutants used). One of these mutations, glc7-Q48K, conferred mild resistance to the inhibition of translation by glucose withdrawal by both the polysome and [35S]methionine-labeling assays (Table 2). This mutant also exhibited constitutive derepression, as judged by its growth on sucrose in the presence of 2-deoxyglucose. Interestingly, the Gln48 residue lies in a similar region of the modeled structure of PP1A (based on the rabbit protein phosphatase type 1) as Thr152 and many other residues involved in glucose repression (see Baker et al., 1997). The identification of a glc7 mutation that confers resistance to glucose withdrawal is consistent with current models of Reg1p action (Ludin et al., 1998; Alms et al., 1999). As such, the resistance of the reg1Δ mutant would be via the inactivation of Glc7p toward specific substrates.
It has been shown that deletion of Hxk2p hexokinase, but not Glk1p and Hxk1p (which are also responsible for phosphorylating glucose to give glucose-6-phosphate), leads to constitutive derepression (Entian, 1980). This result has been interpreted to indicate that further metabolism of glucose to glucose-6-phosphate is not the primary signaling event for glucose repression. We tested a mutant strain deleted for both hxk1 and glk1 and found that protein synthesis was still inhibited upon glucose removal (Table 2). This finding shows that resistance to the inhibition of translation follows the same hexokinase specificity as the glucose repression pathway (i.e., only hxk2Δ has a phenotype). It was also suggested recently that Reg1p/Glc7p can dephosphorylate Hxk2p and that this event is involved in glucose repression (Randez-Gil et al., 1998; Alms et al., 1999). Therefore, we tested a strain containing a mutation at the site of Hxk2p phosphorylation (Ser15 to Ala15) that has been reported to be derepressed (Randez-Gil et al., 1998). This mutation did not lead to resistance to glucose withdrawal at the translational level (Table 2). We conclude from this result that maintenance of Hxk2 Ser15 phosphorylation in a reg1Δ strain is not the reason why cells are resistant to the translational inhibition induced by glucose removal.
As only a subset of glucose repression mutants exhibited resistance to translational inhibition by glucose withdrawal, it seemed possible that these mutations either had common effects outside glucose repression or generated a higher level of derepression than the mutants that had no effect. If the glucose repression pathway is involved in the resistance phenotype, then a SNF1 null mutation in the reg1Δ or hxk2Δ mutant (in which constitutively active Snf1p is normally maintained [Jiang and Carlson, 1996; Ludin et al., 1998]) would be expected to reestablish sensitivity to translational inhibition on glucose removal. As shown in Figure 5 and Table 2, translation was inhibited in both the hxk2Δ snf1Δ and reg1Δ snf1Δ mutants upon glucose removal to the same extent as in the wild type (FY250). This result demonstrates that the constitutive Snf1p activity associated with the reg1Δ and hxk2Δ mutants is required for their resistance to glucose removal at the translational level. In combination with the involvement of proteins throughout the repression pathway (from Hxk2p at the cell membrane to the transcriptional repressor Ssn6p), this suggests that derepression of the glucose repression pathway leads to the observed resistance phenotype. Thus, it seems likely that the level of constitutive derepression in a mutant is critical in determining whether it is resistant or sensitive to glucose removal at the translational level.
Our observation that a transcriptional repression mutant (ssn6Δ) is resistant to the translational inhibition upon glucose removal is difficult to integrate with the finding that new transcription is not required for the inhibition of translation or its recovery after glucose readdition (Figure 3C). This contradiction suggests that mutants in the glucose repression pathway could be acting in an indirect way to confer resistance to the effects of glucose withdrawal upon translation (see DISCUSSION).
The snf3Δ rgt2Δ Mutant Is Resistant to the Translational Inhibition upon Glucose Withdrawal
A different pathway has been described in which the presence of glucose is sensed by two integral membrane proteins, Rgt2p and Snf3p. These are high- and low-affinity glucose sensors, respectively, and they signal the presence of glucose to allow for the transcription of the HXT genes (Figure 1D) (Özcan et al., 1996a,b, 1998). This pathway involves Grr1p, which forms part of the SCF ubiquitin-conjugating complex that is involved in protein degradation. It has been proposed that this complex targets the transcriptional repressor Rgt1p for degradation and thus allows HXT gene expression (Li and Johnson, 1997). We chose to investigate whether these glucose sensors were responsible for signaling changes in glucose levels in the medium to the translational apparatus.
The constitutive activation of this pathway via the deletion of rgt1 or the presence of dominant active SNF3-1 or RGT2-1 mutations did not prevent the rapid inhibition of translation induced by glucose depletion (Table 2). Inactivation of the pathway via deletion of grr1, snf3, or rgt2 also did not prevent the response to glucose. However, the snf3Δ rgt2Δ double mutant did exhibit resistance (Figure 5 and Table 2). This strain, however, exhibits complex phenotypes. For instance, the strain grows poorly on glucose, perhaps explaining the lower level of polysomes even in the presence of glucose. In addition, like the reg1Δ and hxk2Δ mutants, this strain is constitutively derepressed. One explanation for this is that this mutant reacts as if glucose were limiting as a result of the decreased glucose transport caused by the constitutive transcriptional repression of the HXT genes (Schmidt et al., 1999). Interestingly, the grr1Δ mutant (which is also constitutively derepressed for a similar reason [Flick and Johnston, 1991; Vallier et al., 1994]) is sensitive to glucose removal at the translational level. Because Grr1p lies downstream of the Rgt2p and Snf3p glucose sensors, it seems unlikely that the resistance observed in the snf3Δ rgt2Δ mutant is a consequence of its derepression phenotype. However, to test this directly, we deleted SNF1 in the snf3Δ rgt2Δ background to abolish derepression (Schmidt et al., 1999). In the snf3Δ rgt2Δ mutant, deletion of SNF1 does not reestablish the inhibition of translation upon glucose removal (Figure 5 and Table 2). This result demonstrates that, in contrast to the reg1Δ and hxk2Δ mutants, the snf3Δ rgt2Δ mutant is not resistant as a consequence of constitutively active Snf1p. This opens the possibility that the Snf3p and Rgt2p glucose sensors may be directly involved in signaling the levels of glucose in the medium to the translational machinery.
Low Levels of cAPK Activity Prevent the Inhibition of Translation on Glucose Removal
Other signal transduction pathways in which glucose has been implicated as a signaling nutrient converge on the cAPK (Figure 1E). Two tpkw mutants (tpk1w1 tpk2Δ tpk3Δ and tpk1Δ tpk2w1 tpk3Δ), in which two of the genes encoding cAPK were deleted and the third was severely impaired, were resistant to the effects of glucose removal at the translational level (Figure 5 and Table 2). These mutants have an undetectable level of cAPK activity and therefore represent an extreme inhibition (Cameron et al., 1988). Interestingly, another of these tpkw mutants (tpk1Δ tpk2Δ tpk3w1) was not resistant to the inhibition of translation caused by glucose depletion (Table 2). This mutant was also previously shown to have the highest cAPK activity of these three mutants (Nikawa et al., 1987).
In low-cAPK mutants such as the tpkw mutants, the transcription factors Msn2p and Msn4p are constitutively nuclear, and as a result, the stress-controlled genes are constitutively expressed. Interestingly, the removal of glucose has been shown to induce the stress response, and this is mediated via the relocalization of the Msn2p and Msn4p transcription factors from the cytoplasm to the nucleus. This relocalization is thought to require inactivation of cAPK upon glucose removal (Görner et al., 1998). We examined whether the resistance to glucose removal at the translational level in low-cAPK mutants is a result of the constitutive activity of the stress response pathway in these mutants. We used a strain that is deleted for MSN2 and MSN4 as well as all of the cAPK genes (TPK1, TPK2, and TPK3). In this strain, the deletion of the Msn2p and Msn4p transcription factors abolishes the constitutive stress response that is characteristic of low-cAPK mutants (Smith et al., 1998). However, this strain is still largely resistant to the translational inhibition caused by glucose removal (Figure 5 and Table 2). In addition, when glucose is removed from a wild-type strain in which the stress response genes are preinduced (via heat, salt, or ethanol stress), translation is still largely inhibited (our unpublished results). Therefore, the activity of the stress response pathway does not explain the translational resistance to glucose withdrawal that is evident in low-cAPK mutants.
In [35S]methionine incorporation assays, the rate of incorporation in the presence of glucose for the tpkw mutants was significantly lower than that for the wild-type parent (our unpublished results). This correlates with their slow growth (Table 2). However, it was clear that in the absence of glucose these mutants incorporated [35S]methionine, whereas the wild-type strain showed impaired incorporation (Table 2). In addition, the tpk1Δ tpk2Δ tpk3Δ msn2Δ msn4Δ strain has only a slightly impaired growth rate compared with wild-type strains and yet is still resistant to glucose withdrawal at the translational level (Table 2).
Two pathways have been described that regulate cAPK activity in response to glucose. These are the RAS/cAMP pathway and the fermentable growth medium–induced pathway. The RAS/cAMP pathway is involved in the metabolic switch that occurs when cells are transferred onto glucose (Jiang et al., 1998), and the fermentable growth medium–induced pathway is involved in the maintenance of high cAPK activity during growth on glucose (Thevelein, 1991; Crauwels et al., 1997). Neither activating nor inhibitory mutations in various components of these pathways (Bcy1Δ, RAS2val19, gpa2Δ, gpr1Δ, cdc35-11, and sch9Δ) were found to have a significant effect on the level of translational inhibition caused by glucose removal (Table 2). It seems likely that the inhibitory mutations in these pathways do not reduce the cAPK activity sufficiently to allow resistance to glucose removal at the translational level.
An Analysis of ATP Levels after Removal of the Carbon Source
Our identification of yeast mutants resistant to the inhibitory effects of glucose withdrawal on translation supports the hypothesis that this translational inhibition is a signal-mediated event. However, our interpretations are complicated by the fact that glucose is also the main energy source of the cells (Thevelein, 1994). Therefore, an alternative possibility is that upon glucose removal the energy levels in the cell decrease to such an extent that translation can no longer proceed, because translation is one of the major energy-consuming processes in the cell.
To investigate whether energy depletion explains the translational inhibition, we measured ATP levels with the use of a luciferin/luciferase assay. This assay has the advantage that it can be used to assess the ATP levels quickly with a minimum of cellular manipulations (Simpson and Hammond, 1989). Figure 6 shows the results of time-course experiments whereby yeast was incubated in the presence or absence of glucose, with the amount of ATP expressed as a percentage of the starting ATP level of the culture. The precise starting ATP levels for all the strains tested were remarkably similar, falling in a range between 250 and 350 amol/cell. This correlates well with previous measures of the intracellular ATP level in yeast (Simpson and Hammond, 1989).
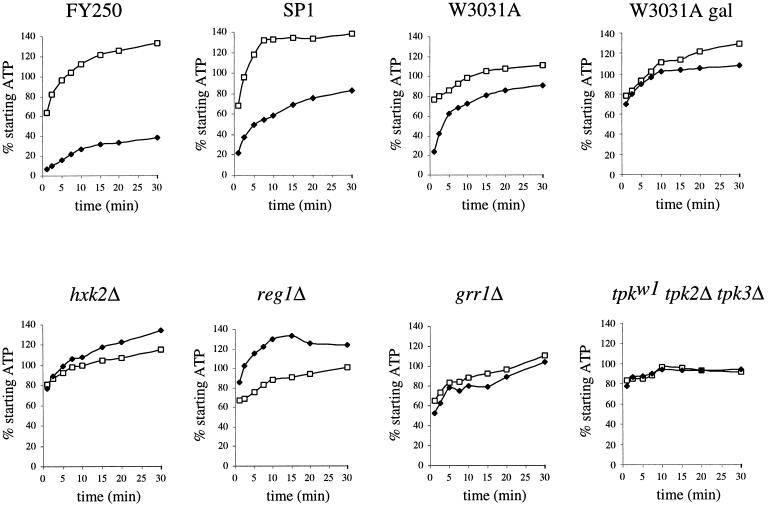
Figure 6.
Measurement of intracellular ATP levels after glucose withdrawal from the growth medium. The percentage of the starting intracellular ATP level for 30 min after transfer of yeast to medium with glucose (□) or without glucose (♦) is plotted. The W3031A gal graph is identical to the others except that the strain was grown in galactose medium and was transferred to medium with (□) or without (♦) galactose. Strains used are the wild-type strains yAS2572 (FY250), yAS962 (SP1), and yAS2568 (W3031A) and the mutant strains yAS2578 (hxk2Δ), yAS2576 (reg1Δ), yAS2577 (grr1Δ), and yAS963 (tpk1w1 tpk2Δ tpk3Δ).
We found that after glucose removal, wild-type strains (FY250 [yAS2572], SP1 [yAS962], and W3031A [yAS2568]) exhibited a rapid (after 1 min or less) decrease in ATP level to ∼20% of the starting value (Figure 6). This is consistent with the results of Wilson et al. (1996), who showed that the AMP/ATP ratio increases immediately after glucose removal. A decrease to ∼70% of the starting level of ATP also was observed in the presence of glucose. It is not clear why this decrease occurred, but it may relate to the cellular manipulations during the experiment. The decrease in ATP level that was observed upon glucose removal coincides with the translational inhibition described above. However, there was a variation in the rate of recovery of the ATP level (Figure 6, compare FY250, SP1, and W3031A). In some wild-type strains, the ATP level quickly recovered to >70% of that found before glucose withdrawal after 10 min (Figure 6, W3031A) (our unpublished results). Only in one wild-type background, FY250, did the low ATP level persist in the absence of glucose (Figure 6). The recovery of ATP levels is consistent with the results of Ditzelmüller et al. (1983), who found that ATP levels remained remarkably constant under a range of different growth conditions. Even though there was a wide variation between strain backgrounds in the level of ATP reduction and recovery after glucose removal, all of these strains were equally inhibited at the translational level. Therefore, it is possible that the decrease in ATP is either required for or generated by the inhibition of translation. However, it does not seem likely that reduced energy levels account directly for the translational inhibition upon glucose removal, because ATP levels recover rapidly to near normal levels in some inhibited strains.
The removal of galactose from a wild-type culture grown on galactose (W3031A gal) or the removal of glucose from the reg1Δ, hxk2Δ, and tpk1w1 tpk2Δ tpk3Δ mutants growing on glucose did not result in an initial decrease in ATP. As under these conditions strains are resistant to the translational inhibition upon carbon source removal, this might suggest that the initial decrease in ATP is involved in the translational inhibition. However, some of the glucose repression mutants that were not resistant to the translation inhibition exhibited no decrease in ATP after glucose removal. The most convincing example of this involves the grr1Δ strain, which exhibited a complete inhibition of translation on glucose removal (Figure 5) but did not show a significant decrease in ATP when glucose was withdrawn (Figure 6). This result supports the conclusion that the decrease in ATP resulting from glucose removal is not required to generate the inhibition of translation.
Since GTP hydrolysis is also required for both the initiation and the elongation steps of translation, the levels of GTP could affect the translational capacity of the cell after glucose removal. We do not favor this hypothesis for several reasons. First, our identification of yeast mutants that have no connection with the regulation of GTP levels and yet are resistant to the inhibitory effects of glucose withdrawal on translation does not support this hypothesis. In addition, the “runoff” of polysomes observed after glucose removal requires that translational elongation continues while initiation is inhibited (Mathews et al., 1996). Translational elongation of a polypeptide requires at least two GTP molecules per amino acid added, whereas initiation requires only one or two GTP molecules per polypeptide chain (Merrick and Hershey, 1996). The fact that translational elongation continues after glucose removal and allows for the runoff of polysomes is highly suggestive that GTP levels are not limiting. Finally, the level of GTP is intrinsically linked to the level of ATP, because GDP is converted to GTP by the enzyme nucleoside diphosphate kinase, which uses ATP as the phosphate donor (Parks and Argawal, 1973). As with ATP levels, the levels of GTP have been shown to remain relatively constant as long as cell viability is maintained (Ditzelmüller et al., 1983). As the ATP levels do not correlate with the inhibition of translation upon glucose removal, it seems likely that GTP levels will also fail to correlate.
DISCUSSION
In S. cerevisiae, the action of glucose as a signaling molecule affects a diverse number of biochemical pathways. Here we extend this diversity by showing that glucose depletion leads to an almost complete inhibition of translation. This inhibition is specific to either glucose or fructose as the carbon source, and it occurs very rapidly after carbon source removal. Readdition of glucose causes a rapid reversal of this inhibition. The inhibition does not come about via a gross decay of mRNA, and neither the inhibition nor its reversal by readdition of glucose requires transcription of new mRNAs. We found that translational inhibition by glucose removal does not require activation of the amino acid starvation pathway or inactivation of the TOR pathway, and it is unlikely to occur via a direct effect of ATP depletion. Furthermore, an investigation of a large number of mutants previously implicated in glucose-related signal transduction pathways highlighted possible roles for components of the main glucose repression, HXT induction, and cAPK pathways in the regulation of a cell's translational response to rapid glucose removal.
The glucose repression mutants reg1Δ and hxk2Δ, which are constitutively derepressed for the glucose-repressible genes, are resistant to the translational inhibition. This resistance requires Snf1p kinase activity. A mutant in the Ssn6p transcriptional repressor is also derepressed and translationally resistant. Therefore, it seems that when the glucose derepression pathway is constitutively active, the yeast are resistant to glucose removal at the translational level. This could explain why cells growing on galactose are resistant to carbon source removal, because these cells are also derepressed. Mutants with constitutively low cAPK activity are also resistant to the removal of glucose at the translational level. It has been shown previously that these mutants are not derepressed for the glucose-repressible genes (Hubbard et al., 1992). The translational phenotypes of the glucose repression and cAPK mutants are consistent with previous observations that the Snf1p kinase and cAPK act antagonistically to affect a similar set of cellular functions (Thompson-Jaeger et al., 1991; Hubbard et al., 1992).
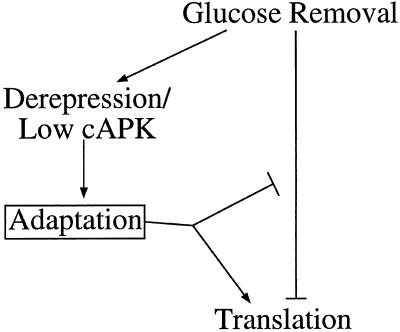
Translation is rapidly inhibited when glucose is replaced with an alternative carbon source such as galactose, and this inhibition is maintained for >2 h (our unpublished results). It seems likely that the recovery of translational activity under these circumstances is different from the rapid recovery seen when glucose is added back soon after glucose removal. We favor a model (Figure 7) in which glucose removal leads to activation of the derepression pathway and to cAPK inactivation. This would allow for the expression or modification of a factor(s) that could slowly overcome the inhibitory effect on translation caused by glucose removal. In this model, mutations in pathways that result in constitutive expression or modification of the factor(s) in cells growing in glucose would confer resistance to the translational inhibition caused by glucose removal. Such cells would be preadapted to the inhibitory effects of glucose removal upon translation.
Figure 7.
Multiple pathways may be involved in glucose-dependent translational control. Glucose removal is shown to inhibit translation. Translation resumes after switching carbon sources via a process termed adaptation. Adaptation might either directly reverse the translational inhibition or somehow overcome the inhibition by activating translation. Adaptation appears to be induced by the derepression of glucose-repressible genes and/or by low cAPK activity (both of which are also brought about by glucose removal).
The candidate gene strategy described here was adopted with the hope that we would identify the components of the pathway responsible for both sensing and transducing the signal for translational inhibition generated by glucose depletion. Intriguingly, the snf3Δ rgt2Δ mutant is partially resistant to the translational inhibition caused by glucose removal. This resistance is not affected by the reversal of the derepression phenotype associated with this mutant. Rgt2p and Snf3p are integral membrane proteins that are thought to sense the level of glucose. Clearly, this may indicate that the absence of glucose is somehow sensed and a signal is transmitted to the translational apparatus via these integral membrane proteins. However, this interpretation is complicated by results from dominant RGT2 or SNF3 mutants. These mutants constitutively signal the presence of glucose, as judged by the expression level of the HXT genes (Özcan et al., 1996a), and yet translation is still regulated by glucose removal. An alternative possibility is that the snf3Δ rgt2Δ mutant is adapted to glucose removal by an as yet uncharacterized mechanism.
This example highlights the importance of distinguishing whether a particular resistant mutant alters the sensing/transduction pathway or induces adaptation. The classification of resistant mutants will require a greater understanding of how translation is inhibited upon glucose removal and how the putative adaptation process occurs. Then, by understanding the nature of the mutations in each class, we imagine that it will be possible to generate a more complete view of how glucose removal leads to the inhibition of translation.
Although the precise physiological role of the translational inhibition described here is unknown, there are many possibilities. The regulation of translation could preserve energy until alternative carbon sources are available. Because protein synthesis is one of the major energy-consuming processes in the cell, its shutdown might allow for the maintenance of ATP levels in the cell. However, this explanation is difficult to reconcile with the result that the ATP levels of a series of resistant mutants stay high even though translation continues in these cells (Figure 6). Another possibility is that the translational shutdown could be involved in the efficient switch in gene expression that occurs when yeast is starved of glucose. This might be an indirect effect in which the absence of protein synthesis allows the existing mRNA and protein pools to turn over, thereby facilitating the efficient reprogramming of gene expression. In addition, it is also possible that the translational inhibition makes a direct contribution to the switch in gene expression. In this scenario, only those mRNAs that facilitate the switch to alternative metabolic or growth pathways would be translated. A precedent exists for specific translational activation under conditions of general translational repression in the amino acid starvation pathway. In this system, translation of a specific mRNA (GCN4) is actually activated via small upstream ORFs even though general mRNA translation is inhibited (Hinnebusch, 1996).
Yeast respond to glucose starvation by redirecting gene expression to alter metabolism. This reprogramming of the gene expression of the cell may involve both transcriptional and translational controls. The precise role of translational controls in allowing the efficient reprogramming of gene expression has been assessed only in a relatively small number of cases. It will be interesting in future work to determine how the regulation described here relates to other translational controls and whether specific mRNAs are immune to this regulation. Together with future work aimed at understanding the basic mechanism underlying the glucose effect on translation, these studies should help to elucidate how a cell can rapidly control its gene expression in response to rapid environmental changes.
ACKNOWLEDGMENTS
We thank Kim Arndt, John Cannon, Marian Carlson, Tom Dever, Anita Hopper, Mark Johnston, Kelly Tatchell, Stephen Garrett, and Jose Antonio Prieto for their generous and expeditious donations of strains and plasmids. We also thank Lucy Otero, Hilary Ashe, and Panda Hershey for their help in performing some of the time-course experiments. We thank Sandy Wells, Lucy Otero, and Ronald Boeck for their help and advice, along with the whole Sachs laboratory staff past and present for their helpful encouragement and discussion. This work was supported in part by National Institutes of Health grant GM50308 to A.B.S. and by a European Molecular Biology Organization long-term fellowship to M.P.A.
REFERENCES
- Alms GR, Sanz P, Carlson M, Haystead TAJ. Reg1p targets protein phosphatase 1 to dephosphorylate hexokinase II in Saccharomyces cerevisiae: characterizing the effects of a phosphatase subunit on the yeast proteome. EMBO J. 1999;15:4157–4168. doi: 10.1093/emboj/18.15.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SH, Frederick DL, Bloecher A, Tatchell K. Alanine-scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:615–626. doi: 10.1093/genetics/145.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S, Levin L, Zoller M, Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell. 1988;53:555–556. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Eng FJ, Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol Cell Biol. 1989;9:5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crauwels M, Donaton MCV, Pernambuco MB, Winderickx J, de Winde JH, Thevelein JM. The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable-growth-medium-induced (FGM) pathway. Microbiology. 1997;143:2627–2637. doi: 10.1099/00221287-143-8-2627. [DOI] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. Phosphorylation of initiator factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Devit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Ditzelmüller G, Wöhrer W, Kubicek CP, Röhr M. Nucleotide pools of growing, synchronized and stressed cultures of Saccharomyces cerevisiae. Arch Microbiol. 1983;135:63–67. doi: 10.1007/BF00419484. [DOI] [PubMed] [Google Scholar]
- Entian K-D. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol Gen Genet. 1980;178:633–637. doi: 10.1007/BF00337871. [DOI] [PubMed] [Google Scholar]
- Flick JS, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Hartwell LH, McLaughlin CS. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc Natl Acad Sci USA. 1969;62:468–474. doi: 10.1073/pnas.62.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996. pp. 199–244. [Google Scholar]
- Hubbard EJA, Yang X, Carlson M. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics. 1992;130:71–80. doi: 10.1093/genetics/130.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Davis C, Broach JR. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 1998;17:6942–6951. doi: 10.1093/emboj/17.23.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthe DS. A simple technique for the preparation and storage of sucrose gradients. Anal Biochem. 1983;135:230–232. doi: 10.1016/0003-2697(83)90755-8. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Estruch F. Sudden depletion of carbon source blocks translation, but not transcription, in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;390:319–322. doi: 10.1016/0014-5793(96)00683-7. [DOI] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB. Origins and targets of translational control. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996. pp. 1–30. [Google Scholar]
- Merrick WC, Hershey JWB. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996. pp. 31–70. [Google Scholar]
- Niederacher D, Entian K-D. Isolation and characterization of the regulatory HEX2 gene necessary for glucose repression in yeast. Mol Gen Genet. 1987;206:505–509. doi: 10.1007/BF00428892. [DOI] [PubMed] [Google Scholar]
- Nikawa J-I, Cameron S, Toda T, Ferguson KM, Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987;1:931–937. doi: 10.1101/gad.1.9.931. [DOI] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Dover J, Rosenwald AG, Wölfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996a;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Johnston M. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol Cell Biol. 1996;16:5536–5545. doi: 10.1128/mcb.16.10.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996b;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks RE, Jr, Argawal RP. Nucleoside diphosphokinases. In: Boyer PD, editor. The Enzymes. 3rd ed. Vol. 8. New York: Academic Press; 1973. pp. 307–333. [Google Scholar]
- Peltz SW, Donahue JL, Jacobson A. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5778–5784. doi: 10.1128/mcb.12.12.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy NT, Li L, Kahlil M, Cannon JF. Regulation of yeast glycogen metabolism and sporulation by Glc7p protein phosphatase. Genetics. 1998;149:57–72. doi: 10.1093/genetics/149.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M, Wek RC, Vazquez de Alana CR, Jackson BM, Freeman B, Hinnebusch AG. Mutations activating the yeast eIF2α kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol Cell Biol. 1992;12:5801–5815. doi: 10.1128/mcb.12.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randez-Gil F, Sanz P, Entian K-D, Prieto JA. Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast. Mol Cell Biol. 1998;18:2940–2948. doi: 10.1128/mcb.18.5.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfes RJ, Hinnebusch AG. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol Cell Biol. 1993;13:5099–5111. doi: 10.1128/mcb.13.8.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Deardorff JA. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signaling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MC, McCartney RR, Zhang X, Tillman TS, Solimeo H, Wölfl S, Almonte C, Watkins SC. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller HJ, Entian K-D. Isolation and expression analysis of two yeast regulatory genes involved in the derepression of glucose-repressible enzymes. Mol Gen Genet. 1987;209:366–373. doi: 10.1007/BF00329667. [DOI] [PubMed] [Google Scholar]
- Sherwood PW, Carlson M. Mutations in GSF1 and GSF2 alter glucose signaling in Saccharomyces cerevisiae. Genetics. 1997;147:557–566. doi: 10.1093/genetics/147.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson WJ, Hammond JRM. Cold ATP extractants compatible with constant light signal firefly luciferase reagents. In: Stanley PE, McCarthy BJ, Smither R, editors. ATP Luminescence: Rapid Methods in Microbiology. Oxford, UK: Blackwell Scientific; 1989. pp. 45–52. [Google Scholar]
- Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM. Fermentable sugars and intracellular acidification as specific activators of the Ras-adenylate cyclase signaling pathway in yeast: the relationship to nutrient-induced cell cycle control. Mol Microbiol. 1991;5:1301–1307. doi: 10.1111/j.1365-2958.1991.tb00776.x. [DOI] [PubMed] [Google Scholar]
- Thevelein JM. Signal transduction in yeast. Yeast. 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- Thompson-Jaeger S, François J, Gaughran JP, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel H. Binding of initiator methionyl-tRNA to ribosomes. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996. pp. 113–138. [Google Scholar]
- Tu J, Carlson M. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6789–6796. doi: 10.1128/mcb.14.10.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung K-S, Hopper AK. The glucose repression and RAS-cAMP signal transduction pathways of Saccharomyces cerevisiae each affect RNA processing and the synthesis of a reporter protein. Mol Gen Genet. 1995;247:48–54. doi: 10.1007/BF00425820. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Roussou I, Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989;57:947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Vallier LG, Carlson M. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 1994;137:49–54. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier LG, Coons D, Bisson LF, Carlson M. Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics. 1994;136:1279–1285. doi: 10.1093/genetics/136.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wek RC, Cannon JF, Dever TE, Hinnebusch AG. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2α kinase GCN2. Mol Cell Biol. 1992;12:5700–5710. doi: 10.1128/mcb.12.12.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]