Abstract
Background
Acetylcholinesterase is irreversibly inhibited by organophosphate and carbamate insecticides allowing its use in biosensors for detection of these insecticides. Drosophila acetylcholinesterase is the most sensitive enzyme known and has been improved by in vitro mutagenesis. However, its stability has to be improved for extensive utilization.
Results
To create a disulfide bond that could increase the stability of the Drosophila melanogaster acetylcholinesterase, we selected seven positions taking into account first the distance between Cβ of two residues, in which newly introduced cysteines will form the new disulfide bond and second the conservation of the residues in the cholinesterase family. Most disulfide bonds tested did not increase and even decreased the stability of the protein. However, one engineered disulfide bridge, I327C/D375C showed significant stability increase toward denaturation by temperature (170 fold at 50°C), urea, organic solvent and provided resistance to protease degradation. The new disulfide bridge links the N-terminal domain (first 356 aa) to the C-terminal domain. The quantities produced by this mutant were the same as in wild-type flies.
Conclusion
Addition of a disulfide bridge may either stabilize or unstabilize proteins. One bond out of the 7 tested provided significant stabilisation.
Background
Acetylcholinesterase (AChE, EC 3.1.1.7) is a serine hydrolase, which catalyzes the hydrolysis of acetylcholine. This enzyme is the target of organophosphate and carbamate insecticides which phosphorylate or carbamoylate the serine of the active site blocking the hydrolysis of the neurotransmitter acetylcholine. The post-synaptic membrane then remains depolarized and synaptic transmission cannot take place so the insect dies. These compounds are used to control proliferation of various agricultural pests: insects, acari and nematodes. One of the consequences is that pesticide residues remain in the environment and are potentially toxic for all animals, including humans since cholinergic transmission is well conserved. Insecticide residues can be detected with biosensors using AChE as biological element to detect low levels of contaminants in crops, soil, water or food samples [1,2].
Drosophila AChE (DmAChE) was found to be the most sensitive enzyme when compared to enzymes of non-insect origin and in-vitro-mutagenesis has permitted the selection of enzymes up to 300-fold more sensitive [3,4]. But like most enzymes from mesophilic organisms, DmAChE is not stable, and this instability precludes its utilization in biosensors. It can be stabilized by additives: proteins such as bovine serum albumin, reversible inhibitor, polyethylene glycol or by encapsulation in liposomes [5-8]. Another way to stabilize the enzyme is to use in vitro mutagenesis to modify the primary structure of the protein. Elimination of a free cysteine and mutation of the hydrophobic residues at the protein surface into hydrophilic residues have been used to increase the stability of DmAChE [9,10]. Here we focused on another method: engineering new disulfide bridges.
Disulfide bonds are present in most extracellular proteins, where they presumably stabilize the native conformation by lowering the entropy of the unfolded form [11] or by decreasing the unfolding rate of irreversibly denatured proteins [12,13]. This stabilizing property makes disulfide bond cross-linking an attractive strategy for engineering additional conformational stability into proteins by site-directed mutagenesis [14].
DmAChE is a dimer linked to membrane via a GPI anchor. There are eight cysteines in each monomer [15]. Six are involved in intrachain disulfide bonds, they are highly conserved in the protein family and their mutations result in inactivation of the protein. One cysteine is involved in an interchain disulfide bond and one, at position 290 (328 using precursor numbering) remains free [16,17]. The aim of this work was to stabilize DmAChE by introducing new disulfide bonds.
Results
Mutation
There are 35 potential disulfide bridges in DmAChE if we consider that every distance between two Cβ of 3.6 to 4 Å is suitable to form a disulfide bridge following the mutation of the two residues in cysteines. Among them, we selected 7 using two criteria: the two amino-acids involved should not be conserved in the cholinesterase family and a serine at these positions is present in one of the available sequences [18]. All these 7 disulfide bonds were predicted by MODIP, automated software for modeling disulfide bonds in proteins [19] with grades A (ideal stereochemistry), B (geometrically suitable but with distorted stereochemistry) and C (sites close enough to allow the formation of a disulfide bond) [20]. We verified that the engineered disulfide bonds were formed by assaying free sulfhydryl groups with the Ellman reagent in the presence of 6 M urea. The results were consistent with the expected disulfide bonds. We verified that the new cysteines did not promote a higher degree of polymerisation. SDS-gel electrophoresis performed in non-reducing conditions showed that all mutants were dimeric proteins like the wild type: introduction of cysteines did not provide additional intersubunit interactions in the mutants.
In our conditions, production of wild type DmAChE in insect cells via the secretory network is 52 nmoles per liter, five bridges did not significantly affect this protein production; two, m3 and m4 decreased production and no mutation increased production (Table 1).
Table 1.
Production ratio of mutant in baculovirus compared to wild type enzyme. Reference is the wild type DmAChE which was produced at 52 nanomoles per liter of culture. *: significant difference, n: number of batches analyzed
| Relative production | ||||||
| Mutant code | Mutated amino-acids | Grade (MODIP) | distance (Cβ) in tertiary structure (Å) | n | mean | Standard error |
| m1 | R24/A169 | B | 3.65 | 5 | 1.43 | 0.71 |
| m2 | I327/D375 | C | 3.77 | 20 | 1.06 | 0.73 |
| m3 | L354/A456 | C | 3.56 | 19 | 0.22 * | 0.26 |
| m4 | T369/M476 | A | 3.62 | 15 | 0.04 * | 0.07 |
| m5 | L388/Q427 | C | 3.70 | 16 | 0.96 | 0.63 |
| m6 | A452/S533 | B | 3.91 | 7 | 0.73 | 0.45 |
| m7 | T464/S543 | D | 3.96 | 8 | 0.77 | 0.47 |
Heat denaturation
We first analyzed denaturation with the most common method used to study protein denaturation: incubation at high temperature. The stability of the mutated protein was estimated by studying irreversible thermal inactivation at several temperatures (from 35 to 65°C) and plotted the first-order denaturation rate constant (kd) against the reciprocal of the absolute temperature (°K-1). It appeared that one bridge (m2) increased thermostability while one (m6) decreased it (Fig. 1).
Figure 1.
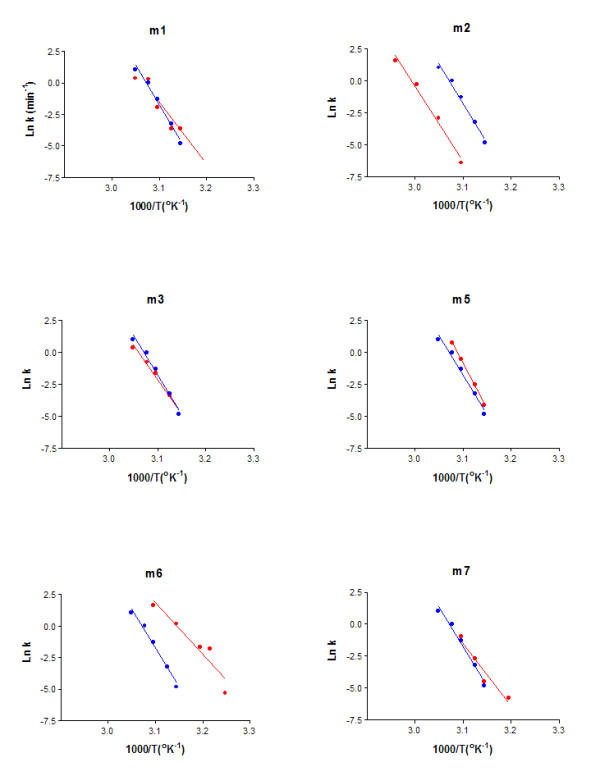
Arrhenius plots of thermal inactivation rate constants of mutated DmAChE (in red) compared to wild type (in blue). k: denaturation first order rate constant (in min-1).
Urea and organic solvent denaturation, protease sensitivity
Stability was assayed with three denaturing agents. In all cases, denaturation was irreversible and followed apparent first order kinetics. Stability was characterized by the half-life (t50), the time at which 50% of an initial enzymatic activity is preserved. The half life of the wild type protein was 13.6 min. in 4 M urea. Protease was used as a denaturant because a protein's resistance to proteolysis increases with its conformational stability due to the fact that the susceptibly to proteolysis reflects the rate of local unfolding [21,22]. The half life of wild type DmAChE was 13.9 min in 0.1 mg/ml pronase. Detection of insecticides in food requires their extraction with organic solvent. Although the solvent should be eliminated before the assay, low amounts may remain in solution and inactivate the enzyme. We used acetonitrile as model because it is soluble in water. The half life of the wild type protein was 1.7 min in 20% acetonitrile. The thermostability provided by bridge m2 is conserved for other denaturing agents (Table 2). Identically, the low stability provided by bridge m6 is found again. In addition, low stability was found for bridges m1 and m5.
Table 2.
Relative stability of mutated AChEs. For each mutation, the t50 ratio (t50 mutant/t50 wild type) was calculated for each denaturation agent (*: significant difference. n: number of independent batches analyzed)
| mutation | n | 50°C | 20% acetonitrile | 4 M urea. | 0.1 mg/mL pronase. |
| m1 | 1 | 2 | 0.43* | 0.40* | 0.27* |
| m2 | 10 | 170* | 2.11* | 12.35* | 1.60* |
| m3 | 9 | 1.4 | 0.85 | 1.87* | 2.37* |
| m5 | 6 | 0.47* | 0.13* | 0.05* | 0.17* |
| m6 | 2 | 0.05* | 0.33* | 0.26* | 0.71 |
| m7 | 2 | 0.73 | 0.94 | 0.71 | 0.79 |
Specific activity
The specific activity of the mutants, and the patterns of the pS curves, were not significantly changed with the introduction of new bridges (Fig. 2). This suggests that entrance of the substrate into the active site as well as the catalytic efficiency was not affected by the mutations.
Figure 2.
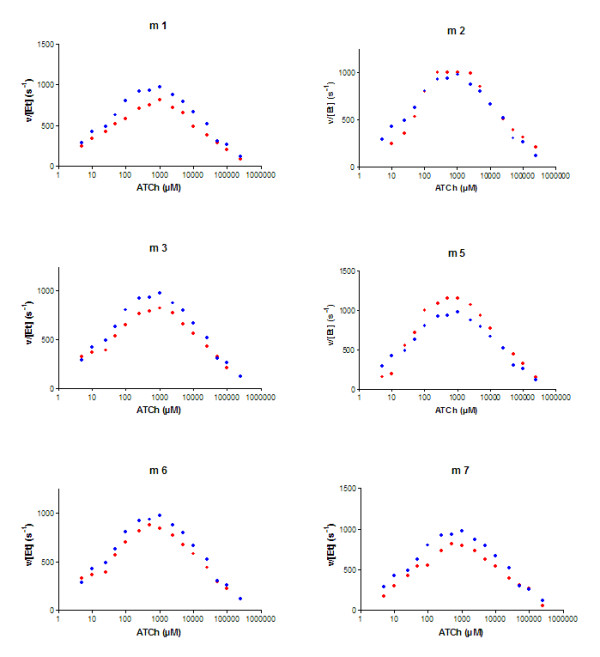
Effect of mutations on acetylthiocholine hydrolysis versus substrate concentration (log scale). (blue dots): wild type; (red dots): mutant, Acetylthiocholine concentration in micromoles per liter; v/[Et] specific activity in s-1.
Discussion
From the first works of Villafranca et al. [23] and Perry and Wetzel [24], introduction of non-native disufide bonds has been used to stabilize proteins [25-34]. These successes pushed us to use this technique to stabilize DmAChE.
The effect of addition of disulfide bridges was either stabilization or destabilization
Most new disulfide bonds introduced in DmAChE did not affect protein stability, one decreased stability. Destabilization has sometimes been reported [35,36]. This instability has been interpreted as the result of atypical sets of dihedral angles in newly formed disulfide bridges [37], from stabilization of the denatured state [38] or from reduction of disulfide bonds followed by disulfide exchange or chemical reaction of the SH groups formed [39,40]. Attempts to predict destabilization by modeling using MODIP failed, suggesting that selected positions were too flexible for a fulfilling prediction.
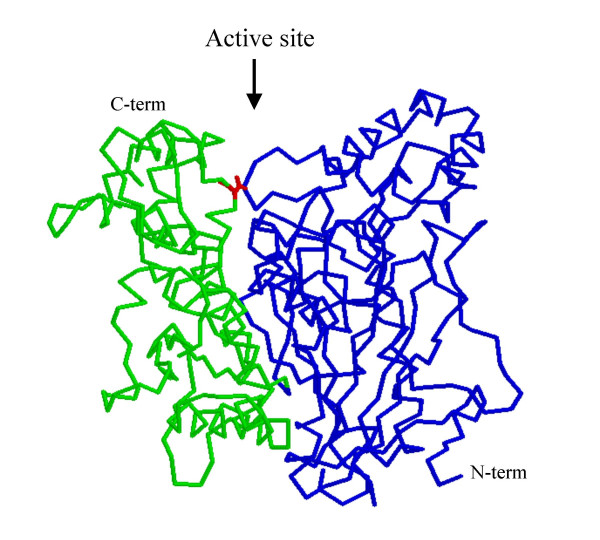
We found one mutation which stabilizes the protein (m2). Two subdomains forming the active site may be distinguished in cholinesterases and mutations decreasing interactions between them decrease protein stability [41]. Disulfide bridge m2 links the two subdomains of the enzyme (Fig. 3), strengthens subdomain interactions and increases overall stability. This suggests that the contact area of the two subdomains is the weakest site of the protein, taking into account the hypothesis that unfolding of a protein molecule starts at its weakest site, and local stabilization of this fragile region results in global stabilization of the whole molecule [42].
Figure 3.
Position of mutation m2 (I327/D375). The cross link has been colored in red. The disulfide bridge links two subdomains of the protein.
Addition of new disulfide bonds may impair protein production
Production is a key issue for application of the stable enzymes in biosensors. We found that addition of a disulfide bond may result in a decrease of protein production since two mutations out of the seven studied, affected protein production. Most probably, increasing the number of sulfhydryl groups in a protein decreases the folding efficiency by increasing the number undesirable disulfide bonds which results in a misfolded protein.
Conclusion
Addition of a disulfide bridge may either stabilize or unstabilize proteins.
Methods
Protein engineering
Possible sites for the introduction of disulfide bonds were located according to Wakarchuk et al. [23], by searching for pairs of residues for which the inter- Cβ distance was between 3.6 and 4 Å in the structure of DmAChE [17].
Protein production and purification
cDNA encoding DmAChE and mutants were expressed with the baculovirus system [43]. We expressed a soluble dimeric form deleted of the hydrophobic peptide at the C-terminal end which is exchanged for a glycolipid anchor. A 3 × histidine tag replaced the loop from amino-acids 103 to 136 to facilitate purification. This external loop is at the other side of the molecule with respect to the active site entrance and its deletion affects neither the activity nor the stability of the enzyme. Secreted AChE was purified to homogeneity using the following steps, ammonium sulfate precipitation, ultrafiltration with a 50 kDa cut off membrane, affinity chromatography with procainamide as ligand, NTA-nickel chromatography and anion exchange chromatography [7]. Residue numbering followed that of the mature protein.
Enzyme activity
The kinetics of substrate hydrolysis was followed at 25°C in 25 mM sodium phosphate buffer pH 7, containing 1 mg/ml BSA. Hydrolysis of acetylthiocholine, an analogue of the neurotransmitter allowing easy detection of the reaction product, was studied spectrophotometrically at 412 nm using the method of Ellman et al. [44], at substrate concentrations ranging from 2 μM to 300 mM, in 1 cm path-length cuvettes. Activity was measured for 1 minute after addition of the enzyme to the reaction mixture. The concentration of the enzymes was determined by active site titration using irreversible inhibitors with high affinity [45].
Denaturation
DmAChE is denatured irreversibly, ΔGd cannot be determined. Instead, the changes in the stability relative to a wild-type protein may be defined as the rate of enzymatic activity decrease [46]. All denaturation experiments were performed with 10 picomoles enzyme in 1 ml 25 mM phosphate buffer pH7 at 25°C. AChE was incubated in denaturing conditions, aliquots were taken out at regular times, diluted 10-fold in enzyme reaction mixture and remaining activity was measured, since residual enzymatic activity is related to the proportion of non-denatured protein. To analyze heat sensitivity, enzymes were incubated at 50°C and 1 mg/ml bovine serum albumin was added to the buffer. Aliquots were mixed with cold buffer chilled on ice and the solution was incubated at 25°C for ten minutes before recording the remaining activity. For urea denaturation, unfolding of DmAChE was induced by adding 4 M urea to the incubation buffer. The effect of organic solvent was followed by incubation of the enzyme in 20% acetonitrile. The effect of protease sensitivity was determined by incubation of AChE with 0.1 mg/ml pronase.
Abbreviations
DmAChE: Drosophila acetylcholinesterase, ATCh: acetylthiocoline, BSA Bovine Serum Albumin.
Authors' contributions
AL and LL performed in vitro mutagenesis, ORS and CL produced the protein and performed biochemical analysis. DF conceived and coordinated the study. All authors participated in the interpretation of the results, in the writing and revising of the manuscript, read and approved the final manuscript. This work has been supported by grants from CRSSA (Centre de recherche du Service de Santé de l'Armée) and the European contract n° QLK3-CT-2000-00650.
Contributor Information
Omid Ranaei Siadat, Email: ranaei@irnewideas.ir.
Andrée Lougarre, Email: Andree.Lougarre@ipbs.fr.
Lucille Lamouroux, Email: Lucille.Lamouroux@ipbs.fr.
Caroline Ladurantie, Email: Caroline.Ladurantie@ipbs.fr.
Didier Fournier, Email: Didier.Fournier@ipbs.fr.
References
- Marty JL, Sode K, Karube I. Biosensor for detection of organophosphate and carbamate insecticides. Electroanalysis. 1992;4:801–803. doi: 10.1002/elan.1140040217. [DOI] [Google Scholar]
- Bachmann TT, Leca B, Vilatte F, Marty JL, Fournier D, Schmid RD. Improved multianalyte detection of organophosphates and carbamates with disposable multielectrode biosensors using recombinant mutants of Drosophila acetylcholinesterase and artificial neural networks. Biosens Bioelectron. 2000;15:193–201. doi: 10.1016/S0956-5663(00)00055-5. [DOI] [PubMed] [Google Scholar]
- Villatte F, Marcel V, Estrada-Mondaca S, Fournier D. Engineering sensitive acetylcholinesterase for detection of organophosphate and carbamate insecticides. Biosens Bioelectron. 1998;13:157–164. doi: 10.1016/S0956-5663(97)00108-5. [DOI] [PubMed] [Google Scholar]
- Boublik Y, Saint-Aguet P, Lougarre A, Arnaud M, Villatte F, Estrada-Mondaca S, Fournier D. Acetylcholinesterase engineering for detection of insecticide residues. Protein Eng. 2002;15:43–50. doi: 10.1093/protein/15.1.43. [DOI] [PubMed] [Google Scholar]
- Payne CS, Saeed M, Wolfe AD. Ligand stabilization of cholinesterases. Biochim Biophys Acta. 1989;999:46–51. doi: 10.1016/0167-4838(89)90028-9. [DOI] [PubMed] [Google Scholar]
- Wilson EJ, Massoulie J, Bon S, Rosenberry TL. The rate of thermal inactivation of Torpedo acetylcholinesterase is not reduced in the C231S mutant. FEBS Lett. 1996;379:161–164. doi: 10.1016/0014-5793(95)01504-3. [DOI] [PubMed] [Google Scholar]
- Estrada-Mondaca S, Fournier D. Stabilization of recombinant Drosophila acetylcholinesterase. Protein Expr Purif. 1998;12:166–172. doi: 10.1006/prep.1997.0831. [DOI] [PubMed] [Google Scholar]
- Nasseau M, Boublik Y, Meier W, Winterhalter M, Fournier D. Substrate-permeable encapsulation of enzymes maintains effective activity, stabilizes against denaturation, and protects against proteolytic degradation. Biotechnol Bioeng. 2001;75:615–618. doi: 10.1002/bit.10074. [DOI] [PubMed] [Google Scholar]
- Fremaux I, Mazeres S, Brisson-Lougarre A, Arnaud M, Ladurantie C, Fournier D. Improvement of Drosophila acetylcholinesterase stability by elimination of a free cysteine. BMC Biochem. 2002;3:21. doi: 10.1186/1471-2091-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub C, Alies C, Lougarre A, Ladurantie C, Czaplicki J, Fournier D. Mutation of exposed hydrophobic amino acids to arginine to increase protein stability. BMC Biochem. 2004;5:9. doi: 10.1186/1471-2091-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB, Scheraga HA. Experimental and theoretical aspects of protein folding. Adv Protein Chem. 1975;29:205–300. doi: 10.1016/s0065-3233(08)60413-1. [DOI] [PubMed] [Google Scholar]
- Plaza del Pino IM, Ibarra-Molero B, Sanchez-Ruiz JM. Lower kinetic limit to protein thermal stability: a proposal regarding protein stability in vivo and its relation with misfolding diseases. Proteins. 2000;40:58–70. doi: 10.1002/(SICI)1097-0134(20000701)40:1<58::AID-PROT80>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Clarke J, Fersht AR. Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- Eijsink VG, Bjork A, Gaseidnes S, Sirevag R, Synstad B, van den Burg B, Vriend G. Rational engineering of enzyme stability. J Biotechnol. 2004;113:105–120. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Hall LM, Spierer P. The Ace locus of Drosophila melanogaster: structural gene for acetylcholinesterase with an unusual 5' leader. Embo J. 1986;5:2949–2954. doi: 10.1002/j.1460-2075.1986.tb04591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutero A, Fournier D. Post-translational modifications of Drosophila acetylcholinesterase. In vitro mutagenesis and expression in Xenopus oocytes. J Biol Chem. 1992;267:1695–1700. [PubMed] [Google Scholar]
- Harel M, Kryger G, Rosenberry TL, Mallender WD, Lewis T, Fletcher RJ, Guss JM, Silman I, Sussman JL. Three-dimensional structures of Drosophila melanogaster acetylcholinesterase and of its complexes with two potent inhibitors. Protein Sci. 2000;9:1063–1072. doi: 10.1110/ps.9.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://bioweb.ensam.inra.fr/ESTHER/general?what=index
- http://www.ncbs.res.in/~faculty/mini/dsdbase/
- Dani VS, Ramakrishnan C, Varadarajan R. MODIP revisited: re-evaluation and refinement of an automated procedure for modeling of disulfide bonds in proteins. Protein Eng. 2003;16:187–193. doi: 10.1093/proeng/gzg024. [DOI] [PubMed] [Google Scholar]
- Ogasahara K, Tsunasawa S, Soda Y, Yutani K, Sugino Y. Effect of single amino acid substitutions on the protease susceptibility of tryptophan synthase alpha subunit. Eur J Biochem. 1985;150:17–21. doi: 10.1111/j.1432-1033.1985.tb08979.x. [DOI] [PubMed] [Google Scholar]
- Braxton S, Wells JA. Incorporation of a stabilizing Ca(2+)-binding loop into subtilisin BPN'. Biochemistry. 1992;31:7796–7801. doi: 10.1021/bi00149a008. [DOI] [PubMed] [Google Scholar]
- Villafranca JE, Howell EE, Voet DH, Strobel MS, Ogden RC, Abelson JN, Kraut J. Directed mutagenesis of dihydrofolate reductase. Science. 1983;222:782–788. doi: 10.1126/science.6356360. [DOI] [PubMed] [Google Scholar]
- Perry LJ, Wetzel R. Disulfide bond engineered into T4 lysozyme: stabilization of the protein toward thermal inactivation. Science. 1984;226:555–557. doi: 10.1126/science.6387910. [DOI] [PubMed] [Google Scholar]
- Wakarchuk WW, Sung WL, Campbell RL, Cunningham A, Watson DC, Yaguchi M. Thermostabilization of the Bacillus circulans xylanase by the introduction of disulfide bonds. Protein Eng. 1994;7:1379–1386. doi: 10.1093/protein/7.11.1379. [DOI] [PubMed] [Google Scholar]
- Villafranca JE, Howell EE, Oatley SJ, Xuong NH, Kraut J. An engineered disulfide bond in dihydrofolate reductase. Biochemistry. 1987;26:2182–2189. doi: 10.1021/bi00382a017. [DOI] [PubMed] [Google Scholar]
- Matsumura M, Signor G, Matthews BW. Substantial increase of protein stability by multiple disulphide bonds. Nature. 1989;342:291–293. doi: 10.1038/342291a0. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Katsuda C, Kimura S, Nakai T, Kitakuni E, Nakamura H, Katayanagi K, Morikawa K, Ikehara M. Stabilization of Escherichia coli ribonuclease H by introduction of an artificial disulfide bond. J Biol Chem. 1991;266:6038–6044. [PubMed] [Google Scholar]
- Mitchinson C, Wells JA. Protein engineering of disulfide bonds in subtilisin BPN'. Biochemistry. 1989;28:4807–4815. doi: 10.1021/bi00437a043. [DOI] [PubMed] [Google Scholar]
- Eder J, Wilmanns M. Protein engineering of a disulfide bond in a beta/alpha-barrel protein. Biochemistry. 1992;31:4437–4444. doi: 10.1021/bi00133a008. [DOI] [PubMed] [Google Scholar]
- Mansfeld J, Vriend G, Dijkstra BW, Veltman OR, Van den Burg B, Venema G, Ulbrich-Hofmann R, Eijsink VG. Extreme stabilization of a thermolysin-like protease by an engineered disulfide bond. J Biol Chem. 1997;272:11152–11156. doi: 10.1074/jbc.272.17.11152. [DOI] [PubMed] [Google Scholar]
- Ivens A, Mayans O, Szadkowski H, Jurgens C, Wilmanns M, Kirschner K. Stabilization of a (betaalpha)8-barrel protein by an engineered disulfide bridge. Eur J Biochem. 2002;269:1145–1153. doi: 10.1046/j.1432-1033.2002.02745.x. [DOI] [PubMed] [Google Scholar]
- Clarke J, Henrick K, Fersht AR. Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography. J Mol Biol. 1995;253:493–504. doi: 10.1006/jmbi.1995.0568. [DOI] [PubMed] [Google Scholar]
- van den Akker F, Feil IK, Roach C, Platas AA, Merritt EA, Hol WG. Crystal structure of heat-labile enterotoxin from Escherichia coli with increased thermostability introduced by an engineered disulfide bond in the A subunit. Protein Sci. 1997;6:2644–2649. doi: 10.1002/pro.5560061219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA, Powers DB. In vivo formation and stability of engineered disulfide bonds in subtilisin. J Biol Chem. 1986;261:6564–6570. [PubMed] [Google Scholar]
- Betz SF. Disulfide bonds and the stability of globular proteins. Protein Sci. 1993;2:1551–1558. doi: 10.1002/pro.5560021002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz BA, Kossiakoff A. The crystallographically determined structures of atypical strained disulfides engineered into subtilisin. J Biol Chem. 1986;261:15480–15485. [PubMed] [Google Scholar]
- Betz SF, Marmorino JL, Saunders AJ, Doyle DF, Young GB, Pielak GJ. Unusual effects of an engineered disulfide on global and local protein stability. Biochemistry. 1996;35:7422–7428. doi: 10.1021/bi9528558. [DOI] [PubMed] [Google Scholar]
- Mozhaev VV. Mechanism-based strategies for protein thermostabilization. Trends Biotechnol. 1993;11:88–95. doi: 10.1016/0167-7799(93)90057-G. [DOI] [PubMed] [Google Scholar]
- Volkin DB, Klibanov AM. Thermal destruction processes in proteins involving cystine residues. J Biol Chem. 1987;262:2945–2950. [PubMed] [Google Scholar]
- Morel N, Bon S, Greenblatt HM, Van Belle D, Wodak SJ, Sussman JL, Massoulie J, Silman I. Effect of mutations within the peripheral anionic site on the stability of acetylcholinesterase. Mol Pharmacol. 1999;55:982–992. doi: 10.1124/mol.55.6.982. [DOI] [PubMed] [Google Scholar]
- Ulbrich-Hofmann R, Arnold U, Mansfeld J. The concept of the unfolding region of approaching the mechanism of enzyme stabilization. J Mol Catalysis B: Enzymatic. 1999;7:125–131. doi: 10.1016/S1381-1177(99)00026-0. [DOI] [Google Scholar]
- Chaabihi H, Fournier D, Fedon Y, Bossy JP, Ravallec M, Devauchelle G, Cerutti M. Biochemical characterization of Drosophila melanogaster acetylcholinesterase expressed by recombinant baculoviruses. Biochem Biophys Res Commun. 1994;203:734–742. doi: 10.1006/bbrc.1994.2243. [DOI] [PubMed] [Google Scholar]
- Ellman GL, Courtney KD, Andres V, Jr, Feather-Stone RM. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Charpentier A, Menozzi P, Marcel V, Villatte F, Fournier D. A method to estimate acetylcholinesterase-active sites and turnover in insects. Anal Biochem. 2000;285:76–81. doi: 10.1006/abio.2000.4738. [DOI] [PubMed] [Google Scholar]
- Betz SF, Pielak GJ. Introduction of a disulfide bond into cytochrome c stabilizes a compact denatured state. Biochemistry. 1992;31:12337–12344. doi: 10.1021/bi00164a007. [DOI] [PubMed] [Google Scholar]