Abstract
A nested polymerase chain reaction (nPCR)-based assay, was developed and evaluated for rapid detection of Trypanosoma evansi in experimentally infected mice and naturally infected camels (Camelus dromedarius). Four oligonucleotide primers (TE1, TE2, TE3 and TE4), selected from nuclear repetitive gene of T. evansi, were designed and used for PCR amplifications. The first amplification, using a pair of outer primers TE1 and TE2, produced a 821-bp primary PCR product from T. evansi DNA. The second amplification, using nested (internal) pair of primers TE3 and TE4, produced a 270-bp PCR product. T. evansi DNAs extracted from blood samples of experimentally infected mice and naturally infected Sudanese breed of dromedary camels were detected by this nested PCR-based assay. The nested primers TE3 and TE4 increased the sensitivity of the PCR assay and as little as 10 fg of T. evansi DNA (equivalent to a single copy of the putative gene of the parasite) was amplified and visualized onto ethidium bromide-stained agarose gels.
Amplification products were not detected when the PCR-based assay was applied to DNA from other blood parasites including Thieleria annulata, Babesia bigemina or nucleic acid free samples. Application of this nPCR-based assay to clinical samples resulted in direct detection of T. evansi from a variety of tissue samples collected from experimentally infected mice and blood from naturally infected camels. The described nPCR-based assay provides a valuable tool to study the epidemiology of T. evansi infection in camels and other susceptible animal populations.
Background
Trypanosoma evansi (T. evansi), the cause of trypanosomiasis (Surra), constitutes one of the major veterinary problems worldwide. The disease causes significant morbidity and mortality in camels in the Sudan, which has a population of over 3 million camels [1]. trypanosomiasis in camels occurs both in chronic and acute forms [2]. The chronic form of the disease is most common and is likely to be associated with secondary infections due to immunosuppression [3]. Clinical signs and pathological lesions caused by T. evansi in camels are unreliable for definitive diagnosis [4]. In addition, detection of parasites in the blood is difficult because parasitaemia is intermittent [5]. Serological tests have been developed and evaluated for diagnosis of trypanosomiasis in camels. They include card agglutination test and enzyme-linked immunosorbent assay (Ab-ELISA) [6-8]. In general serological techniques are useful for detection of a past infection but not for detection of an active infection with T. evansi. To address these problems, nucleic acid hybridization probes have been developed and evaluated for detection of T. evansi. However, the concentration of T. evansi DNA in suspected samples may be below the detection limit of the hybridization assay and hence positive cases are likely to be missed [9]. It is, therefore, becoming increasingly obvious that the development of molecular diagnostic techniques for rapid, sensitive and specific detection of T. evansi would be advantageous in a variety of circumstances including clinical disease investigation and epidemiologic surveys for the disease as well as in control programs. Recently, the polymerase chain reaction (PCR) has been widely used as highly sensitive and specific assay for detection of trypanosomes. Primers targeting subgroup Trypanozoon were previously described [10,11]. In addition, nuclear repetitive T. evansi specific PCR assays have also been developed [10-12]. The PCR has been used in surveys to determine the prevalence of T. evansi in camels from different regions in Kenya [3] and in buffaloes in Vietnam (Holland et al., 2001) and T. equiperdum in Mongolian horses [13]. However, PCR has not been systematically used for the detection of T. evansi in camels in Sudan. In this study, we described a simple, rapid, sensitive and specific assay for detection of T. evansi in Sudanese breed of dromedary camels (Camelus dromedarius) using nPCR amplification.
Results
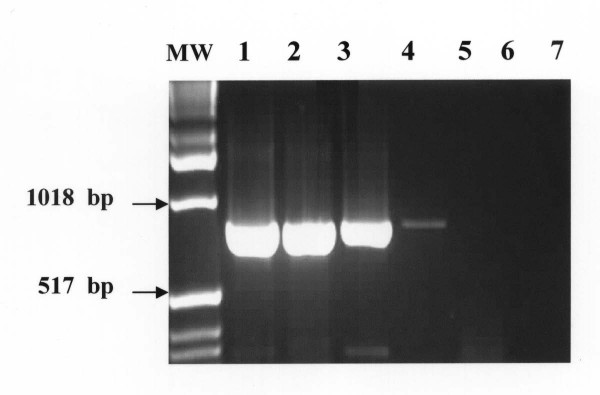
The nPCR-based assay afforded sensitive and specific detection of all T. evansi strains obtained from experimentally infected mice and naturally infected camels. The outer pair of primers TE1 and TE2 produced a primary 810-bp PCR product from T. evansi DNA. The primary 810-bp PCR product was visualized on ethidium bromide-stained gel from ≥ 10 pg of T evansi DNA. (Figure 1). Using the nested primers (T3 and T4), the PCR assay resulted in amplification of a 270 bp PCR product from the primary PCR product. The nested 270 bp PCR product was detected from as little as as 10 fg of T. evansi DNA (equivalent to a single copy of nuclear repetitive gene of T. evansi). The nested amplification increased the sensitivity of the PCR assay and confirmed the identity of the nucleotide sequences of the primary amplified PCR product (Figure 2).
Figure 1.
Visualization of the primary 810-bp PCR product onto ethidium-bromide-stained agarose gel from 10 pg of DNA of T. evansi. Lane MW: molecular weight marker (1 Kb ladder); Lane 1–7: T. evansi DNA at a concentration of 10 ng, 1.0 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, respectively.
Figure 2.
Amplifcation of the nested 270-bp specific-PCR product on ethidium-bromide-stained agarose gel from 10 fg of T. evansi DNA. Lane MW: molecular weight marker (1 Kb ladder); Lane 1–7: T. evansi DNA at a concentration of 10 ng, 1.0 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, respectively.
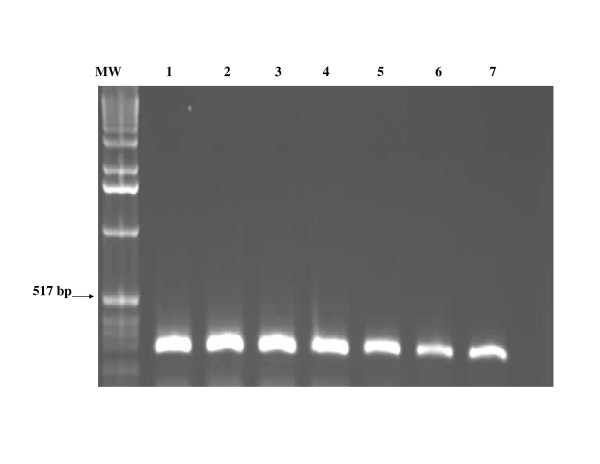
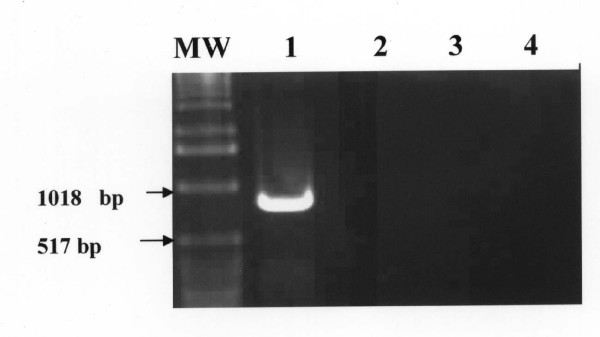
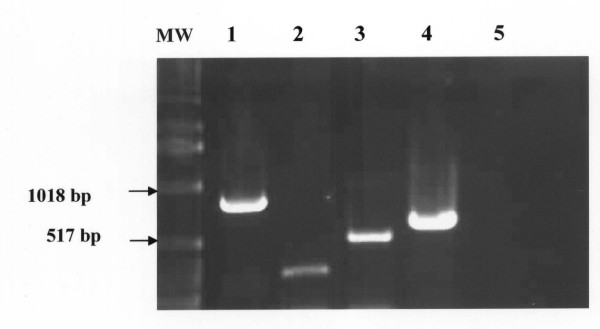
The amount of 1.0 ng DNA from other blood parasites including Thieleria annulata, Babesia bigemina; nucleic acid-free samples, blood and tissue samples from uninfected mice and camels failed to produce the primary or the nested amplification products (Figure 3). The specific PCR products were detected directly from a variety of infected tissue samples including, unfractionated lysed blood, spleen, lung and liver homogenates from experimentally infected mice (Figure 4) and from blood of naturally infected camels (Figure 5). Using the pair of primers semi-nested 1 (TE1 and TE4), the PCR assay resulted in amplification of a 490-bp PCR product. Likewise, the use of pair of primers semi-nested 2 (TE3 and TE2) produced a 590-bp PCR product (figure 6).
Figure 3.
Specificity of the polymerase chain reaction for T. evansi. The 810-bp PCR amplification product was not detected from 1.0 ng of DNA extracted from Babesia and Theileria spp. Lane MW: molecular weight marker (1 Kb ladder); Lane 1: 1.0 ng of T. evansi DN (positive control), Lane 2: 1.0 pg DNA of Theileria anulata, Lane 3: 1.0 pg DNA of Babesia bigemina, Lane 4: nucleic acid free sample (negative control).
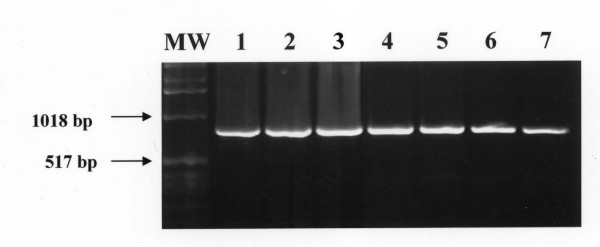
Figure 4.
Detection of the 810-bp T. evansi PCR product from tissue samples collected from experimentally infected mice. Lane MW: molecular weight marker (1 Kb ladder); Lane 1: blood samples, Lane 2: spleen; Lane 3: liver; Lane: blood from non infected mouse, Lane 5: blood from non infected camel (negative controls).
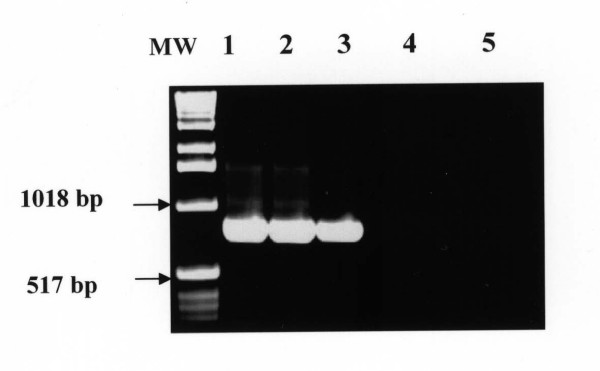
Figure 5.
Detection of the 810-bp T. evansi PCR product from blood samples collected from naturally infected camels. Lane MW: molecular weight marker (1 Kb ladder); Lane 1–7: blood samples from naturally infected camels.
Figure 6.
Detection of T. evansi DNA by PCR using four different primers. Lane MW: molecular weight marker (1 Kb ladder); Lane 1: amplification of the primary 810 bp-PCR product using the outer pair of primers (TE1 and TE2); Lane 2: amplification of the nested 270-bp PCR product using the internal pair of primers (TE3 and TE4); Lane 3: amplification of the semi-nested 490-bp PCR product using the pair of primers semi-nested 1 (TE1 and TE4); Lane 4: amplification of the semi-nested 590-bp PCR product using the pair of primers semi-nested 2 (TE3 and TE2). Lane 5: primers-free PCR amplification (negative control).
Discussion
The economic importance of T. evansi infection is mainly attributed to clinical disease in camels in many parts of the world [14-18]. PCR has been used successfully in detecting infection with T. evansi in buffaloes [12,19], horses [13] and in camels [11]. So far, there is no comprehensive data on the use of PCR for detecting infection in Sudanese breed of dromedary camels (Camelus dromedarius). In the absence of other diagnostic techniques, the camel owners and ethno-veterinarians rely on the smell of urine for diagnosis of T. evansi infections. The characteristic smell of urine is due to ketones produced after breakdown of amino acids by the parasite in infected animals [20].
Conventional parasitological techniques will remain important for understanding the biology, ecology and molecular epidemiology of different strains of the parasites. The described nPCR assay provides simple, rapid, sensitive and specific method for detection of T. evansi in naturally infected camels (Camelus dromedarius) and can be recommended for inclusion in survey and control programmes. The nPCR-based assay using primers derived from nuclear repetitive gene of T. evansi reproducibly and specifically detected all strains of the parasite used in this study. Selection of these universal primers was based on the observation that the nuclear repetitive gene has highly conserved nucleotide sequences [11]. The nPCR assay was a simple procedure that efficiently detected all T. evansi strains under the stringency condition used in this study. The laboratory detection limit indicated that the nPCR protocol was capable of detecting the amount of 10 fg of total T. evansi genomic DNA.
The specificity studies indicated that the primary specific 821-bp and the nested 270-bp PCR products were not amplified even from concentrations of 10 ng of DNA from other blood parasite including Thieleria annulata, Babesia bigemina and blood cells from non infected mice and camels under the same stringency condition described in this study. Based on these sensitivity and specificity results of this study, the described nPCR should be considered as the most highly sensitive and specific assay compared to the previously described PCR-based detection assays [10,12,13,19]. The second amplification step using the nested primers TE3 and TE4 is necessary to confirm the specificity of the primary amplified product and to increase the sensitivity of the PCR-based assay by at least 1000 times particularly, when the concentration of the T. evansi DNA in the sample is less than 10 pg. The use of nested amplification for confirmatory diagnosis of T. evansi infection renders this PCR assay a rapid and an inexpensive assay. The nested PCR does not required hybridization assay and this removes the hazardous and cumbersome radioactive laboratory procedures of working with 32P or 33P in hybridization assays [23]. Successful amplification was also obtained from tissue samples from whole blood of experimentally infected mice, unlike other PCR assays where fractionation of blood leukocyte was necessary for detection of the parasite DNA [10]. Sample preparation and DNA extraction using QIAamp extraction kit was a simple procedure, which takes one hour. The thermal cycling profiles for production of the primary and the nPCR products were consistently 4 hours. Running of the agarose gel and electrophoresis usually takes one hour. Thus, confirmatory diagnosis of T. evansi infection using the described nPCR assay could be made within the same working day.
Cross contamination in nPCR amplification reactions is also a problem. The problem of contamination could be minimized by using nucleic acid-free aerosol resistant tips and separate rooms for DNA extraction and amplification. Negative and positive controls should be included in each PCR assay to maintain the higher limit of sensitivity and the lower limit of specificity. The rapidity, sensitivity and specificity of the nPCR assay would greatly facilitate rapid detection of early or chronic infection with T. evansi in camels and other susceptible animals. Application of this nested PCR on a practical scale to determine the prevalence of T. evansi in naturally infected camels in different states of Sudan is in progress.
Conclusion
The described nPCR assay, using well characterized T. evansi-specific primers, provides a simple, rapid, sensitive and specific detection of T. evansi in naturally infected camels (Camelus dromedarius) and can be used as a valuable tool during epidemiological surveys and control programmes.
Methods
Collection of blood samples
Blood samples were collected in clean sterile vacutainers, containing ethylene diamine tetra acetic acid (EDTA), from experimentally infected mice and from naturally infected camels. The blood samples were used for extraction of total nucleic acids for use as target DNA for PCR amplification.
Parasitological examination and mouse inoculation
Two capillary tubes were prepared from each heparinised blood sample and centrifuged in a microhaematocrit centrifuge for 5 min at 12,000 g. The capillary tubes were examined for the presence of motile trypanosomes using the microhaematocrit centrifugation technique. Four hundred microlitres of blood from infected camels were inoculated into Swiss-bred mice, which were screened for infection with T. evansi by wet smear examination of tail-tip blood every day for detection of parasitaemia.
Extraction of DNA from blood samples
Whole blood was used for extraction of total genomic DNA using commercially available QIAamp blood kit (QIAGEN Inc. Chatsworth, CA, USA) according to the manufacturer's instructions. Briefly, 200 μl of whole blood, 20 μl of proteinase K stock solution, and 200 μl of lysing buffer were pipetted into 1.5 ml eppendorf tube and the mixture was incubated at 37°C for 30 minutes and then at 70°C for 10 minutes. 200 μl of absolute ethanol was added to the sample and the mixture was mixed by vortexing and spinning. The mixture was transferred to the QIAspin column, and placed in a clean 2 ml collection tube and centrifuged at 8000 g for 1 minute. The QIAspin column was washed twice using 500 μl of washing buffers W1 and W2, respectively. The QIAamp spin column was then placed in a clean 1.5 ml eppendorf tube and the DNA was eluted with 200 μl double distilled water preheated at 70°C. Maximum DNA yield was obtained by spinning at 12,000 g for 1 min. The DNA concentration was determined by spectrophotometer at 260 nm wave length. Five μl of the extracted DNA was used in the PCR amplification.
Extraction of DNA from tissue samples
Five ml distilled water was added to 1 gm of each tissue sample collected from lungs, Liver and spleen of experimentally infected mice. The tissue sample was then mixed by homogenization. The homogenate was treated by freezing and thawing and finally centrifuge at 3000 RPM for 10 minutes. 200 μl of the supernatant was used for DNA extraction following the same procedure used for extraction of DNA from blood as described above.
Primers selection
Primers (TE1 and TE2) were selected from a highly conserved region of the published sequence of nuclear repetitive gene of Indonesian strain of T. evansi [24]. This pair of primers was used for the synthesis of the primary PCR amplification product. TE1 included bases 91–110 of the positive sense strand of T. evansi putative gene (5)-AGG ACG CAG AAA TAG CAG TA-(3). TE2 included bases 881–900 of the complementary strand: (5)-ATT TAA TTG AGT GGC GTG AG-(3). The PCR using primer TE1 and TE2 will result in a 810-bp PCR product. For the nested amplification step, oligonucleotide primers (TE3 and TE4) were selected from the same published sequence cited above. TE3 consisted of bases 311–330 of the positive strand (5)-CTT TTA TAC GAG GAG AGG GA-(3). TE4 was designed from the complementary strand between bases 561–580 (5)-TAT GGG CGT GCA GAT TTC AC-(3). PCR amplification using TE3 and TE4 will result in a 270 bp PCR product, internal to the annealing sites of TE1 and TE2. Using the semi-nested pair of primers (TE1 and TE4), the PCR assay will result in amplification of a 490-bp semi-nested PCR product. The pair of semi-nested primers (TE3 and TE2) will be expected to produce a 590-bp semi-nested PCR product. Details for primers design including position of nucleotides, nucleotide sequences, and expected PCR products are shown in (Table 1).
Table 1.
| Primer | Position | Nucleic acid sequence | PCR product |
| External (outer primers) | |||
| TE1 | 91–110 | (5)-AGGACGCAGAAATAGCAGTA (3) | 810-bp |
| TE2 | 881–900 | (5)-ATTTAATTG AGTGGCGTGAG (3) | |
| Internal (nested Primers) | |||
| TE3 | 311–330 | 5)-CTTTTATACGAGGAG GGGA (3) | 270-bp |
| TE4 | 561–580 | 5)-TATGGGCGTGCAGATTTC AC (3) | |
| Semi-nested 1 | |||
| TE1 | 91–110 | 5)-AGGACGCAGAAATAGCAGTA (3) | 490-bp |
| TE4 | 561–580 | 5)-TATGGGCGTGCAGATTTC AC (3) | |
| Semi-nested 2 | |||
| TE3 | 311–330 | 5)-CTTTTATACGAGGAG GGGA (3) | 590-bp |
| TE2 | 881–900 | 5)-ATTTAATTG AGTGGCGTGAG (3) | |
All primers were synthesized on a DNA synthesizer (Milliigen/Biosearch, a division of Millipore Burlington, MA) and purified using oligo-pak oligonucleotide purification columns (Glen Research Corporation, Sterling, VA.) as per manufacturer's instructions.
Polymerase chain reaction (PCR)
A stock buffered solution containing 250 μl 10× PCR buffer, 100 μl of magnesium chloride, 12.5 μl of each dATP, dTTP, dGTP and dCTP was prepared in 1.5 ml eppendorf tube. The primers were used at a concentration of 20 pg/μl, and double distilled water was added to bring the volume of the stock buffer solution to 1.5 ml. Two μl of primers, 5.0 μl of the target DNA and 42 μl of the stock solution were added onto 0.5 ml PCR tubes and mixed by vortexing. One μl of Taq DNA polymerase (Perkin Elmer) was used at a concentration of 5.0 U/μl. All PCR amplification reactions were carried out in a final volume of 50 μl. The thermal cycling profiles were as follows: a 2 min initial incubation at 95°C, followed by 40 cycles of 95°C for 1 min, 55°C for 30 sec and 72°C for 45 sec, and a final incubation at 72° for 10 min. Thermal profiles were performed on a Techne PHC-2 thermal cycler (Techne, Princeton, NJ.). Following amplification, 15 μl from each PCR containing amplified product were loaded onto gels of 1.0% SeaKem agarose (FMC Bioproduct, Rockland ME) and electrophoresed. The gels were stained with ethidium bromide and T. evansi primary PCR products were easily identified following visualization under UV light.
Nested Polymerase Chain Reaction (nPCR)
For the nested PCR amplification, 2.0 μl of the primary amplified product produced by TE1 and TE2 were transferred to 0.5 ml PCR tube containing (2 μl of nested primers and; 42 μl of stock PCR buffer and Taq DNA polymerase was used at a concentration of 5.0 U/μl. The nested pair of primers (TE3 and TE4) was expected to amplify a 270 bp PCR product, internal to the annealing sites of primers PSL1 and PSR2. All PCR amplifications were carried out in a final volume of 50 μl. The thermal cycling profiles were as follows: a 2 min incubation at 95°C, followed by 30 cycles of 94°C for 1 min, 55°C for 30 sec and 72°C for 45 sec, and a final incubation at 72°C for 10 min. Thermal profiles were performed on a Techne PHC-2 thermal cycler (Techne, Princeton, NJ.). Following amplification, 15 μl from each PCR containing amplified products were loaded onto gels of 1.5% SeaKem agarose (FMC Bioproduct, Rockland ME) and electrophoresed. The gels were stained with ethidium bromide and the nested PCR products were easily identified following visualization under UV light.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
AAM designed the study and prepared the manuscript. IEA performed the experimental work and optimized the nested polymerase chain reaction-based detection assay.
Acknowledgments
Acknowledgements
The authors gratefully acknowledge the financial support from the Ministry of Science and Technology (MOST) and the Sudanese Standard and Meterology Organization (SSMO). We also thank Mr. Abdalla M. Fadl Elmoula for technical assistance.
Contributor Information
Imadeldin E Aradaib, Email: aradaib@yahoo.com.
Ali A Majid, Email: ali.majid@gmail.com.
References
- Omer RA, Elamin SMM, El Nahas AE, Aradaib IE. PCR for detection of Echinococus granulosus hydatid cysts collected from camels (Camelus dromerarius) The Sudan J of Vet Sci & Anim Husbandry. 2004;43:139–143. [Google Scholar]
- Gutierrez G, Juste MC, Corbera JA, Magnus E, Verloo D, Montoya JA. Camel Trypanosomiasis in the Canary Islands: assessment of seroprevalence and infection rates using the card agglutination test (CATT/T. evansi) and parasite tests. Vet Parasitol. 2000;90:155–159. doi: 10.1016/S0304-4017(00)00225-9. [DOI] [PubMed] [Google Scholar]
- Njiru ZK, Constantine CC, Ndung'u JM, Robertsonb I, Okayec S, Thompsona RCA, Reida SA. Detection of Trypanosoma evansi in camels using PCR and CATT/T. evansi tests in Kenya. Vet Parasitolo. 2004;124:187–19. doi: 10.1016/j.vetpar.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Chaudhary ZI, Iqbal MJ. Incidence, biochemical and haematological alterations induced by natural trypanosomosis in racing dromedary camels. Acta Trop. 2000;77:209–213. doi: 10.1016/S0001-706X(00)00142-X. [DOI] [PubMed] [Google Scholar]
- Mahmound MM, Gray AR. Trypanosomosis due to T. evansi (Istell, 1885; Balbiani, 1885). A review of recent research. Trop Anim Health Prod. 1980;12:35–47. doi: 10.1007/BF02242629. [DOI] [PubMed] [Google Scholar]
- Davison HC, Thrusfield MV, Muharsini S, Hussein A, Partoutomo S, Rae PF, Masake R, Luckins AG. Evaluation of antigen and antibody detection tests for Trypanosoma evansi infection of buffaloes in Indonesia. Epidemiol Infect. 1999;123:149–155. doi: 10.1017/S0950268899002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SA, Copeman DB. The development and validation of an antibody-ELISA to detect Trypanosoma evansi infection in cattle in Australia and Papua New Guinea. Prev Vet Med. 2003;61:195–208. doi: 10.1016/j.prevetmed.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Nantulya VM. Suratex: a simple latex agglutination antigen test for diagnosis of Trypanosoma evansi Suratex: infections (Surra) Trop Med Parasitol. 1994;45:9–12. [PubMed] [Google Scholar]
- Viseshakul N, Panyim S. Specific DNA probe for the sensitivity detection of Trypanosoma evansi, Southeast Asian. J Trop Med Public Health. 1990;21:21–27. [PubMed] [Google Scholar]
- Moser DR, Cook GA, Ochs DA, Bailey CP, McKane MR, Donelson JE. Detection of Trypanosoma congolense and T. brucei ssp by DNA amplification using the polymerase chain reaction. Parasitolo. 1980;99:57–66. doi: 10.1017/s0031182000061023. [DOI] [PubMed] [Google Scholar]
- Masiga RC, Nyang'ao JM. Identification of trypanosome species from camel using polymerase chain reaction and procyclic transformation test. Camel Pract Res. 2001;8:17–22. [Google Scholar]
- Holland WG, Claes F, My LN, Thanh NJ, Tam PT, Verloo D, Buscher P, Goddeeris B, Vercruysse J. A comparative evaluation of parasitological tests and a PCR for Trypanosoma evansi diagnosis in experimentally infected water buffaloes. Vet Parasitol. 2000;97:23–33. doi: 10.1016/S0304-4017(01)00381-8. [DOI] [PubMed] [Google Scholar]
- Clausen PH, Chuluun S, Sodnomdarjaa R, Greiner M, Noeckler K, Staak C, Zessin KH, Schein E. A field study to estimate the prevalence of Trypanosoma equiperdum in Mongolian horses. Vet Parasitol. 2003;115:9–18. doi: 10.1016/S0304-4017(03)00160-2. [DOI] [PubMed] [Google Scholar]
- Dia ML, Diop C, Aminetou M, Jacquiet P, Thiam A. Some factors affecting the prevalence of Trypanosoma evansi in camels in Mauritania. Vet Parasitol. 1997;72:111–120. doi: 10.1016/S0304-4017(97)00054-X. [DOI] [PubMed] [Google Scholar]
- Diall O, Bocoum Z, Diarra B, Sanogo Y, Coulibaly Z, Waigalo Y. Epidemiology of trypanosomosis caused by T. evansi in camels in Mali: results of parasitological and clinical survey. Rev Elev Med Vet Pays Trop. 1993;46:455–461. [PubMed] [Google Scholar]
- Elsaid HM, Nantulya VM, Hilali M. Diagnosis of Trypanosoma evansi infection among Sudanese camels imported to Egypt using card agglutination test (CATT) and antigen detection latex agglutination test (Suratex) J Protozool Res. 1998;8:194–200. [Google Scholar]
- Gibson WC, Wilson AL, Moloo SK. Characterization of Trypanosoma (Trypanozoon) T. evansi from camels in Kenya using isoenzyme electrophoresis. Res Vet Sci. 1983;34:114–118. [PubMed] [Google Scholar]
- Atarhouch T, Rami M, Bendahman MN, Dakkak A. Camel trypanosomosis in Morocco 1: results of a first epidemiological survey. Vet Parasitol. 2003;111:277–286. doi: 10.1016/S0304-4017(02)00382-5. [DOI] [PubMed] [Google Scholar]
- Omanwar S, Rao JR, Basagoudanavar SH, Singh RK, Butchaiah G. Direct and sensitive detection of Trypanosoma evansi by polymerase chain reaction. Acta Vet Hung. 1999;47:351–359. doi: 10.1556/AVet.47.1999.3.9. [DOI] [PubMed] [Google Scholar]
- Hunter AG. Urine odour in a camel suffering from Surra T. evansi infection. Trop Anim Health Prod. 1986;18:146–148. doi: 10.1007/BF02359524. [DOI] [PubMed] [Google Scholar]
- Borst P, Fowler SF, Gibson WC. Kinetoplast DNA of Trypanosoma evansi. Mol Biochem Parasitol. 1987;23:31–38. doi: 10.1016/0166-6851(87)90184-8. [DOI] [PubMed] [Google Scholar]
- Wuyts N, Chokesajjawatee N, Panyim S. A simplified and highly sensitive detection of Trypanosoma evansi by DNA amplification. S E Asian J Trop Med Publ Health. 1994;25:266–271. [PubMed] [Google Scholar]
- Aradaib IE, Abbas AM, Abbas B, Elsanousi SM. Evaluation of conventional methods and nested PCR for detection of paratunerculosis in goats. Vet Res Commun. 2005;29:381–383. doi: 10.1007/s11259-005-1433-y. [DOI] [PubMed] [Google Scholar]
- Artama WT, Agey MW, Danelson JE. DNA comparison of Trypanosoma evansi (Indonesia) and Trypanosoma brucei spp. Parasitology. 1992;104:67–74. doi: 10.1017/s0031182000060819. [DOI] [PubMed] [Google Scholar]