Abstract
Gene amplification in eukaryotes plays an important role in drug resistance, tumorigenesis, and evolution. The Schizosaccharomyces pombe sod2 gene provides a useful model system to analyze this process. sod2 is near the telomere of chromosome I and encodes a plasma membrane Na+(Li+)/H+ antiporter. When sod2 is amplified, S. pombe survives otherwise lethal concentrations of LiCl, and >90% of the amplified sod2 genes are found in 180- and 225-kilobase (kb) linear amplicons. The sequence of the novel joint of the 180-kb amplicon indicates that it is formed by recombination between homologous regions near the telomeres of the long arm of chromosome I and the short arm of chromosome II. The 225-kb amplicon, isolated three times more frequently than the 180-kb amplicon, is a palindrome derived from a region near the telomere of chromosome I. The center of symmetry of this palindrome contains an inverted repeat consisting of two identical 134-base pair sequences separated by a 290-base pair spacer. LiCl-resistant mutants arise 200–600 times more frequently in strains deficient for topoisomerases or DNA ligase activity than in wild-type strains, but the mutant cells contain the same amplicons. These data suggest that amplicon formation may begin with DNA lesions such as breaks. In the case of the 225-kb amplicon, the breaks may lead to a hairpin structure, which is then replicated to form a double-stranded linear amplicon, or to a cruciform structure, which is then resolved to yield the same amplicon.
INTRODUCTION
Genomic alterations, often deleterious, can also confer unique advantages to a cell. Gene amplification, defined as a relative increase in copy number of a part of the genome smaller than a chromosome, provides a means to increase the amounts of certain proteins without changing the DNA sequence near the relevant genes (Wu and Black, 1987). Amplification plays important roles in drug resistance (Stark et al., 1989), adaptive mutability (alterations in the specificity or rate of mutation during stress), and divergent evolution. Amplification also provides a selective advantage to many tumors, which often contain amplified oncogenes (Bishop, 1987). Amplification of a gene provides additional targets for mutation (Andersson et al., 1998) and thus can be the first step in the evolution of a new gene, because the extra copies are free to provide new functions (Kimura and Ota, 1974).
Two main classes of amplified structures (amplicons) have been characterized. Tandem head-to-tail arrays predominate in prokaryotes. In examples from both Escherichia coli (Whoriskey et al., 1987) and phage T4 (Wu et al., 1991), short (<20 base pairs [bp]), imperfect, direct repeats flank the duplicated region, suggesting that homologous recombination is a major mechanism. In the eukaryote Leishmania, homologous recombination between longer (541 bp) direct repeats flanking the P-glycoprotein gene produces amplicons harboring tandem duplications. Palindromic head-to-head amplification, which is widespread in many eukaryotes, including Leishmania, Tetrahymena, yeast, and mammals, has been observed as both intrachromosomal and extrachromosomal (linear and circular) amplified structures (Ford et al., 1985; Yao et al., 1985; Walton et al., 1986; Hightower et al., 1988; Ouellette et al., 1991).
Determining amplification mechanisms has been difficult in mammalian cells, in large part because of limitations in analyzing the novel joints formed. Novel joints are created when genomic sequences from distinct locations in the wild-type genome become juxtaposed. The work of several laboratories has made it clear that dicentric chromosomes are often present early in amplification events in mammalian cells and that they can be generated by either telomere–telomere fusions (Smith et al., 1992) or double-strand breaks, perhaps at fragile sites (Windle et al., 1991; Coquelle et al., 1997). The precise location of these breakage events is required to determine the mechanism responsible for forming the amplification structures.
The fission yeast Schizosaccharomyces pombe provides an excellent model system for studying gene amplification. Its simple genome comprises three chromosomes, ranging in size from 3.5 to 5.7 megabases. Fission yeast is permissive for gene amplification, and spontaneous amplification has been observed at a frequency of ∼10−6 per cell per generation (Patterson et al., 1999). The sod2 gene of S. pombe encodes a Na+/H+ antiporter that is essential for Na+ export (Jia et al., 1992). Amplification of sod2 is the major mechanism by which cells become resistant to high concentrations of LiCl. Preliminary characterization of the predominant amplicons in LiCl-resistant cells revealed that they are extrachromosomal and, strikingly, of only two sizes, 225 or 180 kilobases (kb). The amplicons are linear, contain telomeric sequences, and are stable only if LiCl selection is maintained (Jia et al., 1992; Patterson et al., 1999). Genetic studies have indicated that mutant strains defective in the DNA-damage checkpoint, but not in the S-phase-completion checkpoint, have an increased frequency of sod2 amplification (Patterson et al., 1999). We now present a detailed structural characterization of the two predominant sod2 amplicons and show that mutations likely to produce DNA lesions also enhance amplicon formation. The new data suggest specific models for amplicon formation.
MATERIALS AND METHODS
Selection of Strains Resistant to LiCl
The three temperature-sensitive mutant strains were h−, top2-191, leu1-32; h−, top1::swi3, top2-110, leu1-32; the strain defective in ligase function was h+, cdc17-K42. All were obtained from the laboratory of Paul Nurse. Supplemented yeast extract medium and Edinburgh minimal medium were described previously (Moreno et al., 1991; Jia et al., 1992). Selections were performed as described previously (Patterson et al., 1999). All the mutant strains but one had the same sensitivity to LiCl as the wild-type strain, but the ligase-deficient strain, cdc17, was more sensitive. Cells were grown in liquid minimal medium prewarmed to a semirestrictive temperature (32 or 34°C) overnight and plated in 40 mM LiCl (16 mM for cdc17) in minimal medium agar, pH 5. Plates were incubated at the permissive temperature of 25°C for 5–7 d.
Isolation of Chromosomal DNA and Pulsed-Field Gel Electrophoresis
Agarose-embedded chromosomal DNA was isolated essentially as described by Alfa et al. (1993). Cells from a 20-ml portion of a dense culture (A595 ∼ 4) were pelleted, washed in H2O, and resuspended in ∼0.3 ml of buffer with lysing enzymes plus 0.4 ml of 1% agarose (Nusieve GTG agarose, FMC Bioproducts, Rockland, ME). The mixture was placed into plug molds (CHEF disposable plug molds, Bio-Rad, Richmond, CA). Before restriction enzyme digestion, the agarose plugs were rinsed twice in TE (10 mM Tris-HCl, pH 7.5, 1 mM EDTA) and equilibrated with restriction enzyme digestion buffer (20–50 ml in 50-ml conical tubes). The plugs were loaded onto an agarose gel (ultrapure DNA-grade agarose, Bio-Rad), and electrophoresis was performed with a CHEF Mapper XA system (Bio-Rad). The electrophoresis conditions used in Figure 1 were 1% agarose, 0.5× standard Tris-borate electrophoresis buffer, 120° included angle, 6 V/cm, 14°C. The switch time was 60 s for the first 15 h and 90 s for the next 9 h.
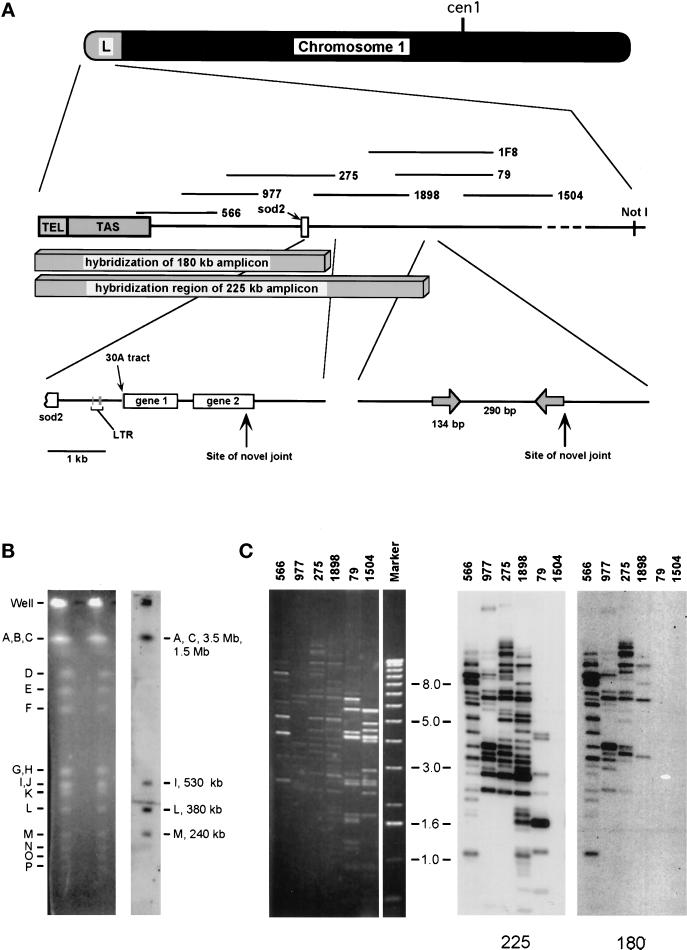
Figure 1.
Hybridization of the 225- and 180-kb amplicons with cosmids representing the telomere-proximal third of NotI fragment L (∼120 kb). (A) The subregion of NotI fragment L that hybridizes with the amplicons and characteristics of the amplified DNAs are shown. NotI fragment L is 380 kb long, and approximately one-third of it hybridizes to the amplicons. TEL, telomere; TAS, telomere-associated sequences; LTR, long terminal repeat. The cosmids, which are numbered, are drawn at their approximate locations. The sod2 gene is depicted as a rectangle. The centromere-proximal 5 kb of hybridization between wild-type genomic DNA and the 180-kb amplicon contains sod2, a long terminal repeat sequence, a stretch of 30 adenosine residues, and two ORFs. The centromere-proximal 1 kb of hybridization between wild-type genomic DNA and the 225-kb amplicon contains a 134-bp inverted repeat and a 290-bp spacer. (B) Wild-type chromosomal DNA digested with NotI. The digest was separated by pulsed-field gel electrophoresis. Left, ethidium bromide–stained gel. Right, hybridization with labeled 225-kb amplicon. NotI fragment L contains sod2 and a telomere. NotI fragments C, I, and M all contain telomeres. A indicates intact chromosome III. NotI fragments C and A do not separate well under the running conditions used in this experiment. (C) Cosmids from the Mizukami library (Mizukami et al., 1993) digested with HindIII, with the most telomere-proximal cosmid on the left. Left, ethidium bromide staining. Right, duplicate transfers hybridized with the 225- or 180-kb amplicon. All of NotI fragment L was tested, and only cosmids in the region shown hybridized. DNA Marker X (Boehringer Mannheim) was used for size markers.
Southern Hybridization
Probes were prepared with a random-primer-labeling kit (Megaprime DNA-labeling system, Amersham, Arlington Heights, IL). To obtain amplicon DNAs, appropriate bands were cut from a pulsed-field gel, run as described above. The agarose was melted and diluted with sterile water to ∼1 ng/l, based on the amount of DNA (10–25 ng) estimated with ethidium bromide. Hybridizations were carried out in Church buffer (Church and Gilbert, 1984) at 65°C and washed with Church wash No. 2. Plasmids obtained from Neal Sugawara (laboratory of J. Szostak, Department of Molecular Biology, Massachusetts General Hospital, Boston, MA) contained S. pombe telomeres or telomere-adjacent regions (Sugawara, 1988). The 800-bp TaqI fragment of plasmid pSPT16 was used as a probe to determine whether telomeric sequences were present in cosmids 566 and 977 (Mizukami et al., 1993). A probe for the telomere-adjacent region was made from the entire plasmid pNSU64.
Cloning and Sequencing
Transformations of competent E. coli (DH5) were performed with CaCl2. The 1.7- and 3.2-kb EcoRI–BamHI restriction fragments from cosmid 1898 were ligated into pBluescript II KS (+/−) and transformed by electroporation into Sure supercompetent E. coli (Stratagene, La Jolla, CA). Transformation of these plasmids, especially the 3.2-kb fragment, into DH5 cells produced no viable plasmid-containing clones. These regions were difficult to clone, possibly because of the presence of repeat sequences and secondary structures. However, we were able to recover clones of the 3.2-kb fragment when the plasmid was transformed into Sure cells. The sequence of the 1.7-kb fragment was obtained by cutting it into the following nonoverlapping fragments, which were subcloned into pBluescript and sequenced: BamHI–XbaI, XbaI–KpnI, KpnI–EcoRV, EcoRV–HindIII (pBluescript was cut with HindIII–HincII), and HindIII–EcoRI. These plasmids were transformed into DH5 and sequenced from both ends with T7 and M13 reverse primers with the use of an ABI model 377 version 3.0 sequencer.
Cloning of Amplicon Novel Joints
PCR was performed on DNA extracted from strains containing the 225-kb amplicon. The primers (5′ to 3′) were For 5 (GCTTTGCTATCATCGCCTAGC) and For 13 (ATACCTATACTTAGTTGCTAC), and the conditions were 59°C (or 63°C) annealing, 45-s extension, Taq DNA polymerase (Boehringer Mannheim, Indianapolis, IN). The PCR products were cloned into pCR2.1 (Invitrogen, Carlsbad, CA). The first 180-kb amplicon novel joint was cloned with the use of a PCR-linker method. DNA from a strain carrying the 180-kb amplicon was digested with EcoRI, and the enzyme was inactivated by heating to 65°C for 15 min. The ∼270-bp PvuII–EcoRI fragment from Bluescript II KS (+/−), purified from agarose with Qiaex II (Qiagen, Chatsworth, CA), was used as the linker. The ligation reaction included the following: 0.25 ng/l linker, 5 ng/l digested genomic DNA, and ∼1 Weiss unit of T4 DNA ligase (Boehringer Mannheim). Properly ligated novel joints were recovered by PCR with the use of the pBluescript KS primer (CGAGGTCGACGGTATCG), located within the linker and oriented toward the EcoRI site, and with For 3.2A (AGTTCCGTTTCCTATCTGCG), present in the 6.5-kb novel EcoRI fragment, also oriented toward the EcoRI site. After transformation into E. coli, two plasmids were isolated and sequenced. To obtain the two additional 180-kb amplicon novel joints, the primer R-180 (TTATTTAATGCTGAATAAACCTTCT), specific for DNA on the centromere-proximal side of the novel joint, was used with the primer For 3.2A in a standard PCR reaction (63°C annealing temperature) with the use of genomic DNA from two additional independent strains carrying the 180-kb amplicon. (The products of the PCR reaction with primers R-180 and For 3.2A were sequenced.) R-180 has only four mismatches compared with the genomic sequence of the 3.2-kb region. To confirm the specificity of this primer pair for the novel joint, rather than for the unrearranged genomic sequence, the primer R-Gen (TTATTTAATGCTGAACAGATCTTCA) was designed from the homologous sequence within the 3.2-kb EcoRI–BamHI fragment of the sod2 locus. When used with primer For 3.2A, primer R-180 amplified a 1.1-kb fragment from plasmids containing the novel joint, and primer R-Gen amplified a 1.1-kb fragment from plasmids containing the 3.2-kb fragment. No PCR product was obtained when the templates were placed in the opposite primer mix.
Sequences have been submitted to GenBank. The 180-kb amplicon-break-region genomic sequence has the accession number AF192974. The 180-kb amplicon novel joints (see Figure 6) have accession numbers AF207957, AF207958, and AF207959 for A, B, and C, respectively. The 225-kb amplicon novel joint (see Figure 4) has the accession number AF207956.
Figure 6.
A novel restriction fragment in the 180-kb amplicon. (A) Sheared genomic DNA from the wild-type (Wt) strain and from two strains carrying 180-kb amplicons (Amp) was digested with EcoRI, XbaI, or SnaBI, and the fragments were separated by gel electrophoresis. The transfers were hybridized with the 3.2-kb BamHI–EcoRI fragment of cosmid 1898. The numbers indicate sizes in kilobases. Boxed numbers indicate novel fragments detected only in strains containing the amplicon. (B) Restriction map of the novel fragment. The partial map of chromosome I is derived from our sequence and restriction data (our unpublished results). The 4.9-kb genomic fragment sequence has been submitted to GenBank (No. AF192974). (C) Sequence of the 1.1-kb novel joint PCR products from the 180-kb amplicon. The novel joint sequences have been submitted to GenBank (Nos. AF207957, AF207958, and AF207959 for A, B, and C, respectively). The region to the left (telomere-proximal) of the site of recombination is identical to the 3.2-kb BamHI–EcoRI fragment from cosmid 1898. Numbers correlate to the sequence of cosmid 1898 beginning at the telomere-proximal end. The 150 bp of sequence to the right (centromere-proximal) of the novel joints abuts the novel EcoRI site. Genomic indicates wild-type genomic DNA represented by cosmid 1898 sequence. A, B, and C represent three independent sequences. The recombination has clearly occurred in all three amplicons by bp 3534. Differences from the genomic sequence before this are believed to be Taq-induced errors. Only portions of PCR product C outside the stars were sequenced with fidelity. The underlined ATG represents the putative start site of the L-asp homologue, the longest ORF. Also underlined is the binding site of primer R-180 and the novel EcoRI site.
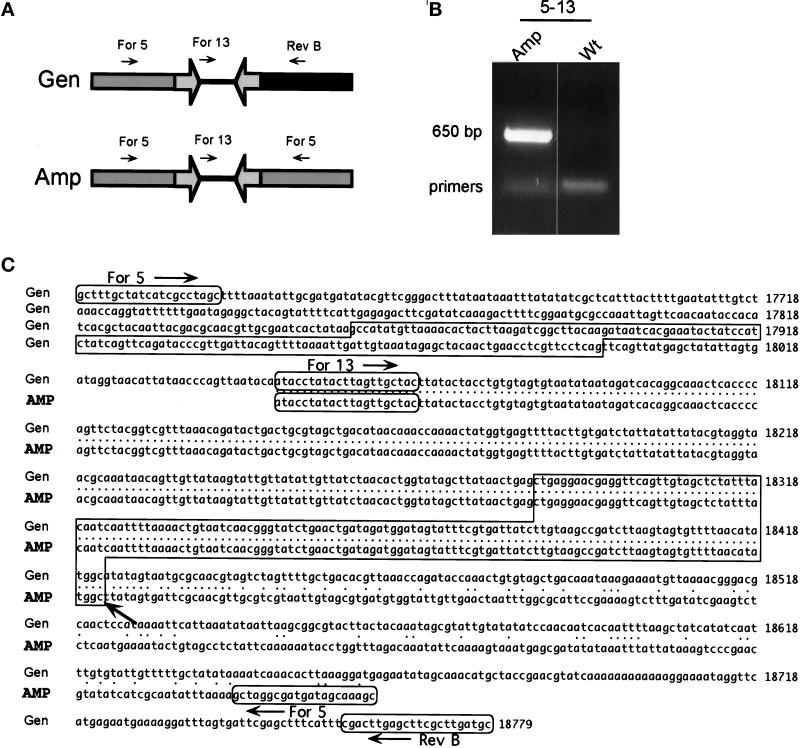
Figure 4.
Sequence of the novel joint of the 225-kb amplicon. (A) The PCR strategy used to clone the novel joint. The proposed structure of the amplicon is shown, together with the PCR primers For 5, For 13, and Rev B. Open arrows represent the inverted repeat. The primer pair For 5–For 13 together can produce a PCR product only if an inversion of one of the primers has occurred. The primer pairs For 5–Rev B and For 13–Rev B can produce products only from wild-type DNA. (B) The PCR products obtained with primers For 5 and For 13. Total DNA isolated from the wild-type (Wt) strain and from strains containing the 225-kb amplicon were used as templates for PCR. The 650-bp PCR product For 5–For 13 was observed only with template DNA from strains carrying the 225-kb amplicon. The other PCR products were observed with all templates tested, as expected (our unpublished results). (C) The sequence of the novel joint. Gen, sequence of wild-type chromosome I. Amp, sequence of the 650-bp PCR product shown in B. The inverted repeats are boxed, and the novel joint lies precisely at the end of the centromere-proximal repeat (arrow). Four independent 225-kb novel joints were sequenced, and all were identical (GenBank No. AF207956).
RESULTS
Linear Extrachromosomal Amplicons Are the Predominant sod2 Amplification Product
Selection with LiCl of wild-type S. pombe cells yielded 20 independent resistant strains. Three different amplification structures were observed by pulsed-field gel electrophoresis. As found previously (Patterson et al., 1999), the frequency of resistance was ∼10−6 per cell per generation. Extrachromosomal amplicons were predominant, but an intrachromosomal amplicon was observed in one case. Linear extrachromosomal amplicons of 225 or 180 kb have been reported (Patterson et al., 1999). All 20 strains contained either the 225-kb amplicon (15 of 20) or the 180-kb amplicon (5 of 20). No strain contained both the 225- and 180-kb amplicons. Three strains contained megabase-long extrachromosomal amplicons in addition to a 225- or 180-kb linear extrachromosomal amplicon. One of these amplicons migrated differently relative to size markers depending on the switch time during pulsed-field gel electrophoresis (our unpublished results), suggesting a circular structure. One strain appeared to have sod2 amplified intrachromosomally as well as extrachromosomally. In this strain, NotI fragment L (380 kb), which contains the endogenous sod2 gene, was absent and was replaced by a new band of 610 kb (our unpublished results). We have studied in detail the two most frequently observed extrachromosomal linear amplicons.
The 225- and 180-kb Amplicons Both Contain Genomic DNA from the Telomere-proximal Third of NotI Fragment L
Figure 1A shows an overview of the region of chromosome I contained in the linear amplicons and of cosmids covering the region. To determine the origin of the amplified DNA, a Southern blot of NotI-digested wild-type genomic DNA was hybridized with gel-purified, radiolabeled amplicon DNA. Probes from both amplicons hybridized with the highest intensity to fragment L, but they also hybridized to the other telomeric fragments (A, C, I, and M) (data for the 225-kb amplicon are shown in Figure 1B; data for the 180-kb amplicon are not shown). NotI fragment L (380 kb) represents the end of the long arm of chromosome I and contains both the telomere and sod2 (Fan et al., 1989; Patterson et al., 1999). The amplicons contain telomeric DNA (Patterson et al., 1999), and both of the amplicon probes hybridized with the five telomeric NotI fragments (chromosome III is not cut by NotI) and the telomeric SfiI fragments. We noted that both the amplicons were smaller than NotI fragment L.
Cosmids from the Hoheisel library (Hoheisel et al., 1993) representing NotI fragment L plus cosmids from the Mizukami library (Mizukami et al., 1993) near the telomere were used. The Mizukami library represents more of the telomeric end of NotI fragment L. Cosmids from the latter library are shown in Figure 1C. Southern hybridization of cosmids covering NotI fragment L indicated that the 225-kb amplicon hybridizes with the region covered by cosmids 566 (the cosmid closest to the telomere), 977, 275, 1898, and 79, altogether ∼120 kb of fragment L. The 180-kb amplicon hybridized to cosmids 566, 977, 275, and 1898, a region of ∼80 kb. These estimations are based on cosmid insert sizes plus ∼10 kb of telomere-adjacent sequences. Cosmid 566 does hybridize with telomere-adjacent sequences but does not hybridize with true telomeric sequences (our unpublished results). Thus, the insert of cosmid 566 extends into the telomere-adjacent region, but the distance between the end of the insert and the end of the chromosome is not known. The telomere-adjacent DNA has been estimated to be >19 kb long (Sugawara, 1988). We estimate that 100–130 kb of chromosome I DNA is represented in the 225-kb amplicon and that 70–90 kb of chromosome I DNA is represented in the 180-kb amplicon. These sizes are consistent with amplicons that each contain two copies of the region.
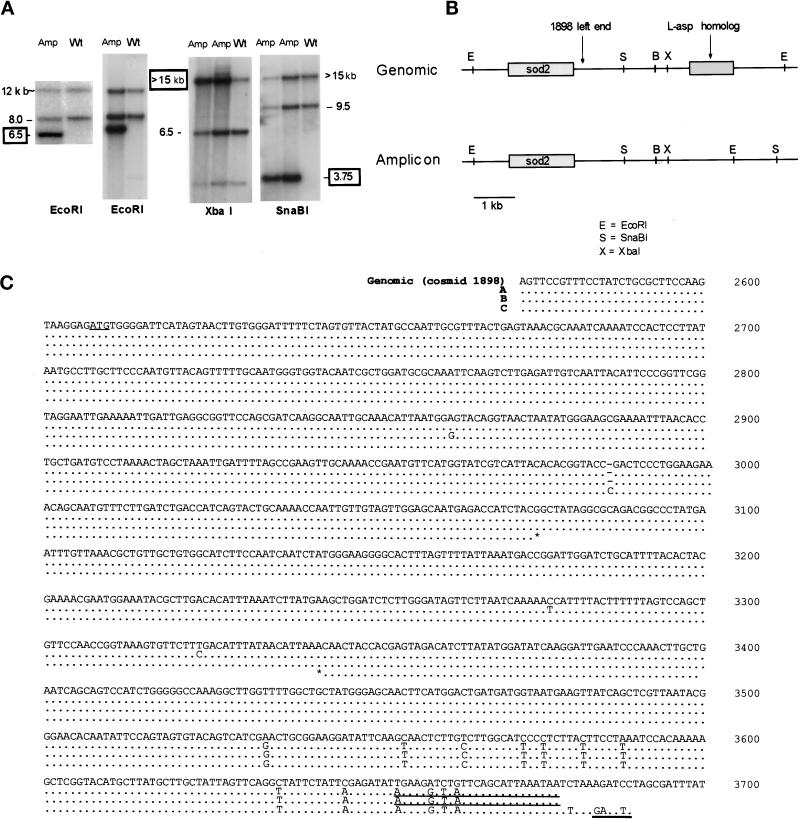
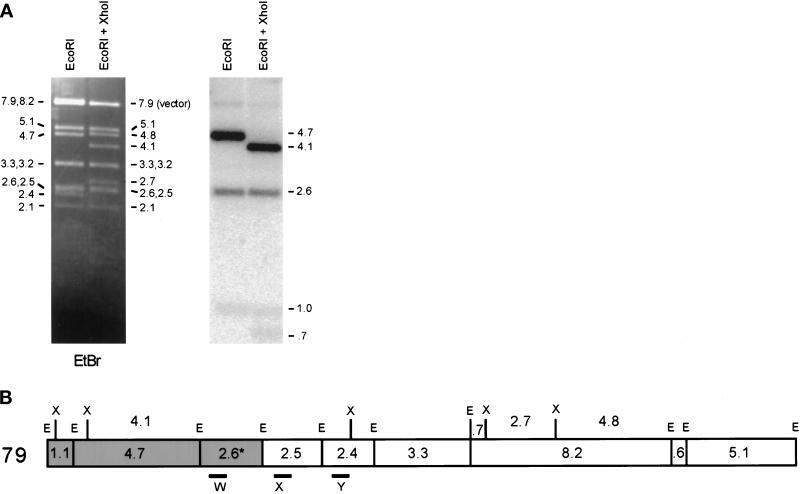
To locate the end point of the genomic region contained in the 225-kb amplicon, we performed Southern hybridization experiments with restriction digests of cosmid 79 (Mizukami et al., 1993). Only the telomere-proximal 8.4 kb (the 1.1-, 4.7-, and 2.6-kb EcoRI fragments) of the cosmid was present (Figure 2). Two fragments comigrated at 2.6 kb, designated 2.5 and 2.6 in Figure 2A. The sequence of cosmid 1F8 (Hoheisel et al., 1993; GenBank No. Z81312) (Figure 1A) predicts a 3.0-kb EcoRI fragment. Inspection of the sequence reveals a 500-bp direct repeat within this 3.0-kb fragment. We believe that this duplication is an artifact of sequencing and is not present in either the cosmid or the genomic sequence. A restriction map based on our data is shown in Figure 2B, with a 2.6-kb fragment in place of the 3.0-kb fragment. Use of PCR probes from each of the EcoRI fragments in the area (Figure 2B, W, X, and Y) showed that, as expected, sequences in the 2.5- and 2.4-kb fragments were not in the 225-kb amplicon (our unpublished results) and that sequences from the 2.6-kb fragment were present in the amplicon. Therefore, the 2.6-kb EcoRI fragment contains the boundary of the genomic DNA duplicated in the 225-kb amplicon.
Figure 2.
Hybridization of the 225-kb amplicon ends within a 2.6-kb restriction fragment of cosmid 79 that lies near an inverted repeat. (A) Cosmid 79 (Mizukami et al., 1993) was digested with EcoRI (E) or EcoRI plus XhoI (X), followed by gel electrophoresis. Left, ethidium bromide (EtBr) staining. Right, hybridization with labeled 225-kb amplicon. The vector bands exhibit weak hybridization with this probe. (B) Restriction map of cosmid 79. The fragments that hybridize with the 225-kb amplicon are shaded. The end point of the amplified DNA is within the 2.6-kb EcoRI fragment. PCR products W, X, and Y were used as probes to check the restriction map (our unpublished results).
The Novel Joint of the 225-kb Amplicon Contains an Inverted Repeat
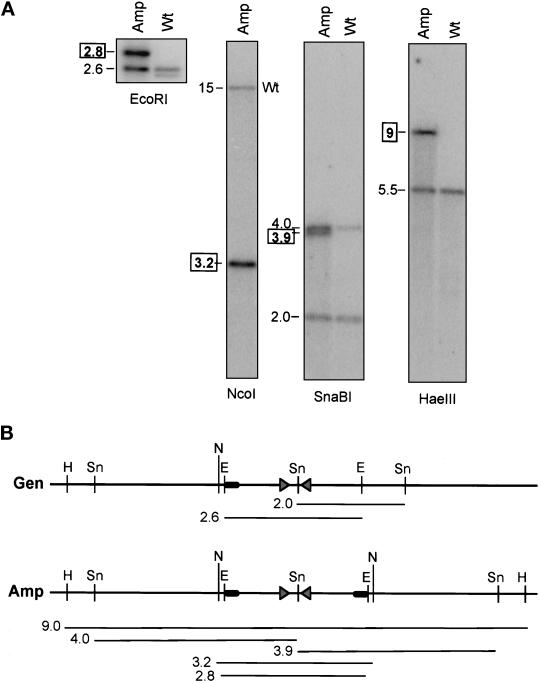
The 2.6-kb EcoRI fragment of cosmid 1F8 hybridizes with novel restriction fragments of the 225-kb amplicon (Figure 3). We conclude that the 225-kb amplicon extends into and stops within the sequence of this 2.6-kb EcoRI fragment at a novel joint where DNA from one region of the genome joins DNA from another region. The 2.6-kb fragment includes two exact copies of a 134-bp sequence in an inverted orientation, separated by a 290-bp spacer. Beyond the ends of the 134-bp elements, their similarity declines rapidly. The inverted repeat is homologous to the long terminal repeat (LTR) of Tf2 and Tf1-107 retrotransposons (GenBank Nos. L10324 and M38526) but contains only a portion of the LTR sequence and is not an intact retrotransposon. We used the genomic 2.6-kb EcoRI fragment to identify novel fragments from the 225-kb amplicon. The sizes of these novel fragments (Figure 3A) support the interpretation that the palindrome extends for at least 4 kb on either side of the original inverted repeat. The presence of two amplified SnaBI fragments (3.9 and 4.0 kb) of approximately equal intensity indicates that the 290-bp spacer, which contains a SnaBI site, has been conserved in the amplicon. The wild-type restriction fragments appear to be conserved.
Figure 3.
Novel restriction fragments detected in the 225-kb amplicon by the 2.6-kb EcoRI fragment. (A) Sheared genomic DNA from the wild-type (Wt) strain and from a strain carrying a 225-kb amplicon (Amp) was digested with EcoRI, SnaBI, HaeIII, or NcoI, and the fragments were separated by gel electrophoresis. The transfers were hybridized with labeled PCR products from within the 2.6-kb EcoRI fragment of cosmid 79. Numbers indicate the sizes of the fragments in kilobases. Boxed numbers indicate novel fragments, which were detected only in strains containing the 225-kb amplicon. (B) Restriction map of the novel fragment. Gen, map of the genomic region. Amp, map of the amplicon, deduced from the sizes of the novel fragments. The map of chromosome I is deduced from sequence data (the S. pombe genome project) and restriction data (our unpublished results). The sizes of the novel fragments are consistent with a palindromic amplicon with a novel joint immediately after the inverted repeat. The triangles represent the inverted repeats. The black ovals represent unique sequences found only telomere-proximal to the inverted repeats in wild-type DNA. E, EcoRI; Sn, SnaBI; H, HaeIII; N, NcoI
Sequence of the Novel Joint of the 225-kb Amplicon
PCR was used to clone the novel joint (Figure 4A). Primer For 5 binds on the telomeric side of the inverted repeats and is oriented to prime DNA synthesis toward them. Primer For 13 lies within the 290-bp spacer, which is oriented in the same direction as primer For 5. These primers cannot amplify a fragment from wild-type DNA, but they can amplify a fragment from a sequence that is palindromic around the inverted repeats. As expected, no PCR product was obtained when these two primers were used with wild-type DNA, but a fragment of ∼650 bp was produced when the DNA from a strain containing a 225-kb amplicon was used as a template (Figure 4B). The 650-bp For 5/For 13 PCR products from four independent amplicons were identical and contained the spacer, one repeat element, and DNA upstream of the inverted repeat in an inverted orientation, as expected for a palindromic novel sequence (Figure 4C; GenBank No. AF207956).
The Novel Joint of the 180-kb Amplicon Is Not Formed at an Inverted Repeat
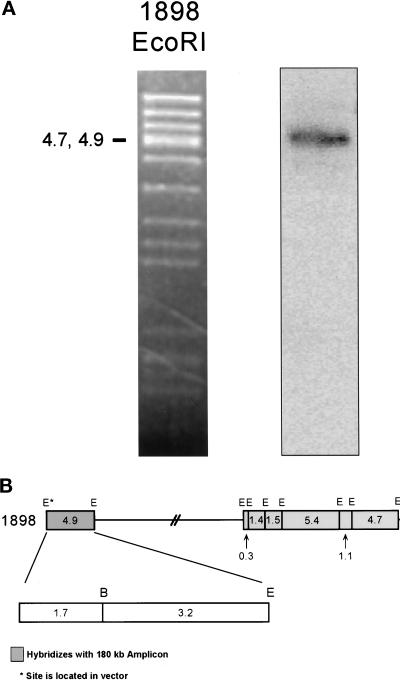
To locate the end of the genomic region in the 180-kb amplicon, we performed Southern hybridization experiments with restriction digests of cosmid 1898 (Mizukami et al., 1993). Only the telomere-proximal 4.9 kb of this cosmid is present in the 180-kb amplicon (Figure 5). The 4.9-kb genomic sequence was submitted to GenBank (No. AF192974). Note that the 4.9- and 4.7-kb fragments are not resolved in this figure, but the 4.7-kb fragment is a part of cosmid 79, which does not hybridize to the 180-kb amplicon (Figure 1C). Additional experiments (our unpublished results) narrow the end point region to within the 3.2-kb BamHI–EcoRI fragment, which was therefore used to probe a Southern transfer of genomic DNA from amplicon-containing strains. One amplified novel fragment and two wild-type fragments were observed per digest (Figure 6A). Notably, the size of the EcoRI novel fragment is ∼6.5 kb, whereas the wild-type fragments are 8 and 12–15 kb long. The native 8-kb fragment is from chromosome I, whereas the 12- to 15-kb fragment appears to be from chromosome II (our unpublished results). The restriction map of the amplicon is derived from the sizes of novel fragments compared with those of the chromosomal fragments (Figure 6B). We conclude that the 180-kb amplicon is not a palindrome.
Figure 5.
Hybridization of the 180-kb amplicon ends within the 3.2-kb BamHI–EcoRI restriction fragment of cosmid 1898. (A) Cosmid 1898 (Mizukami et al., 1993) was digested with EcoRI, and the fragments were separated by gel electrophoresis. Left, ethidium bromide–stained gel. Right, hybridization with labeled 180-kb amplicon. Only the 4.9-kb EcoRI restriction fragment hybridized. (B) Partial restriction map of cosmid 1898. Hybridization data from A are shown in relationship to the restriction map. The end point of the amplified DNA lies within the 4.9-kb EcoRI fragment. Additional data (our unpublished results) localize the end point to within the 3.2-kb BamHI–EcoRI fragment. E, EcoRI; B, BamHI.
The 1.7- and 3.2-kb EcoRI–BamHI fragments in the region of the novel joint were subcloned from cosmid 1898 into pBluescript II KS and sequenced. The first 375 bp of cosmid 1898 are identical to the 3′-untranslated region of a cDNA plasmid containing the sod2 gene, the precise genome location of which was previously unknown. The sequence of cosmid 1898 begins ∼284 bp downstream of the stop codon of sod2. A run of 30 contiguous adenosine residues, an unusual sequence in S. pombe DNA, begins at position 1361 of cosmid 1898. Retrotransposon sequences were found within this region of the 1.7-kb fragment; interestingly, this is the same portion of the LTR sequence that constituted the inverted repeat associated with the novel joint of the 225-kb amplicon. There are several large ORFs within the 4.9-kb sequence represented in Figure 1A. The first has high similarity to l-asparaginase (l-asp) (GenBank No. Y11944), which is near the telomere of the short arm of chromosome I (Bonthron, personal communication). Both ORFs are present in the expressed sequence tag database, indicating that they are expressed genes (GenBank Nos. AU013226 and AU009847).
The 180-kb Amplicon Is Formed as a Result of an Interchromosomal Event
The sequences of novel joints from three independent 180-kb amplicons were very similar. They have been submitted to GenBank (Nos. AF207957, AF207958, and AF207959). The sequence centromere-proximal to the novel joint shares approximately the same degree of similarity with the l-asp sequence as does the 4.9-kb EcoRI sequence from cosmid 1898. The sequence centromere-proximal to the novel joint is not present in the expressed sequence tag database. Alignment of the novel joints with the 4.9-kb EcoRI fragment of cosmid 1898 (Figure 6C) reveals a pattern of nucleotide changes consistent with a mechanism involving homologous recombination between the two sequences homologous to l-asp. Examination of these sequences reveals that the novel joints have occurred by bp 3534. The sequence of C between the stars (Figure 6C) was not obtained, but the lack of this sequence does not detract from the above conclusions. The observations are consistent with recombination occurring at one site within this region of cosmid 1898.
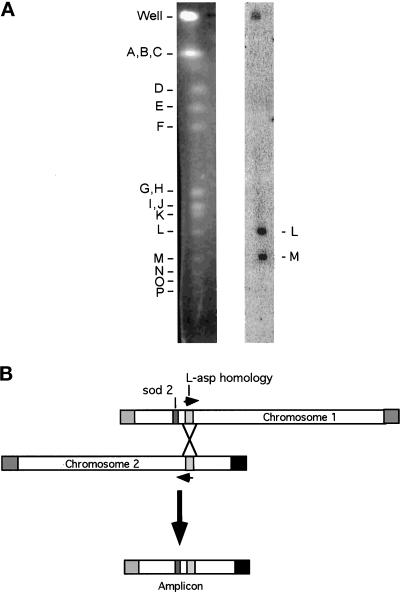
To determine the chromosomal origin of the sequence on the centromeric side of the novel joint, the 320-bp EcoRV fragment from plasmid 1.1D (corresponding to the terminal 320 bp of the 1.1-kb novel joint sequence) was used to probe a pulsed-field gel electrophoresis transfer containing NotI restriction fragments from wild-type S. pombe DNA (Figure 7A). Fragments L (the sod2-containing telomeric fragment from chromosome I) and M (the telomeric fragment from the short arm of chromosome II) hybridized with the probe. The result was confirmed with a smaller probe from the centromere-proximal sequence of the novel joint, which hybridized only to NotI fragment M (our unpublished results). We conclude that the 180-kb amplicon is formed by homologous mitotic recombination between the transcribed l-asp homologue located ∼5 kb away from the sod2 gene on the centromeric side (and ∼70 kb from the telomere) and another l-asp homologue located on the short arm of chromosome II, ∼110 kb from the telomere. The l-asp homologue on chromosome II apparently is oriented with the direction of transcription toward the telomere.
Figure 7.
Hybridization of the 180-kb novel joint region with NotI fragments L and M. (A) Wild-type S. pombe DNA, digested with NotI, was separated by pulsed-field gel electrophoresis. Left, ethidium bromide–stained gel. Right, hybridization with novel joint region fragment. The 320-bp novel joint region fragment was an EcoRV fragment of plasmid 1.1D. The experiment was repeated with a smaller probe from the centromere-proximal sequence of the novel joint, which hybridized only to NotI fragment M (our unpublished results). (B) A homologous recombination model for the 180-kb amplicon. The L-asp homologue on chromosome I is oriented with the direction of transcription away from the telomere (our unpublished results). sod2 is represented as a dark gray rectangle. The L-asp homology region is represented as a light gray rectangle. The arrows denote the direction of transcription of the L-asp homologues. The squares represent the telomeres: the chromosome I long arm telomere is gray and the chromosome II short arm telomere is black. The horizontally striped box is the telomere of the short arm of chromosome I, and the vertically striped box is the telomere of the long arm of chromosome II.
Topoisomerase and Ligase Mutations Increase the Frequency of sod2 Amplification
Mutations increasing the probability of DNA breakage in a cell may affect the frequency of DNA amplification. Taking mutations in DNA ligase and topoisomerase as examples, we determined sod2 amplification frequencies in haploid strains bearing temperature-sensitive alleles of these genes at a semirestrictive temperature (Nasmyth, 1979) (Table 1). In S. pombe, both top1 and top2 have type I topoisomerase activity (Uemura and Yanagida, 1984). Consequently, a substantial decrease in topoisomerase I activity is observed only in the top1 top2 double mutant. A top1 top2 strain had a 200-fold increase in amplification frequency, and a top2(mut) strain with a normal top1 gene had only a 3-fold increase. A strain with a temperature-sensitive mutation in DNA ligase (cdc17) exhibited a 600-fold increase in amplification frequency at the semirestrictive temperature compared with the wild-type strain. Both size classes of amplicon were observed in the mutant strains at about the same ratio as in wild-type cells (our unpublished results). In 10 ligase-deficient sod2-amplified strains, nine 225-kb amplicons and one 180-kb amplicon were observed. In 12 top1 top2–deficient strains amplified for sod2, nine 225-kb amplicons and three 180-kb amplicons were observed.
Table 1.
Increases in sod2 amplification in S. pombe strains with temperature-sensitive mutations in the ligase and topoisomerase genes
| Strain | Temperature (°C) | Hours at temperature shown | Viability (%) | Fold increase in amplification frequencya |
|---|---|---|---|---|
| cdc-17 (ligase) | 25b | 16 | 100 | 6 |
| cdc-17 (ligase) | 34c | 16 | 10 | 600 |
| top1 top2 | 25b | 16 | 100 | 3 |
| top1 top2 | 34c | 16 | 20 | 200 |
| top2 | 25b | 16 | 100 | 1 |
| top2 | 32c | 7 | 5 | 3 |
Cells grown for the times and at the temperatures shown were pelleted, resuspended, and plated at 5 × 107 cells per selection plate. The plates were incubated at the permissive temperature of 25°C for 5–7 d before scoring of LiCl-resistant colonies.
Compared with wild type and corrected for viability.
Permissive temperature.
Semirestrictive temperature.
DISCUSSION
Identification of the novel joint sequences for each class of amplicon characterized was essential for a molecular characterization of the amplicons. The novel joints and structure were different between the two amplicon classes. Each LiCl-resistant strain studied contained a sod2 linear extrachromosomal amplicon of 180 kb (25%) or 225 kb (75%). The 225-kb amplicon is a palindrome of a region bounded by an inverted repeat and a telomere. The 180-kb linear amplicon results from the fusion of two chromosome ends through a region of homology.
The novel joint of the 180-kb amplicon occurs within homologues of L-asp located near the telomeres of the long arm of chromosome I and the short arm of chromosome II, indicating recombination between these two homologous regions (Figure 7B). Because the amplified strains studied have intact chromosomes I and II and are haploid, we conclude that this recombination event must occur after DNA replication. The event must be mitotic because the cells do not enter meiosis during selection. Additional cell cycles can generate multiple copies of the amplicon. Similar recombination-based mechanisms of gene amplification that use repeats of homologous regions have been proposed (Edlund and Normark, 1981; Whoriskey et al., 1987; Wu et al., 1991; Grondin et al., 1993). In E. coli and phage T4, head-to-tail arrays of 50–100 copies are generated when the amplified DNA is flanked by short (<20 bp) direct repeats (Whoriskey et al., 1987; Wu and Black, 1987). In Leishmania, amplicons of the P-glycoprotein gene are extrachromosomal circles containing two head-to-tail copies of the region and are formed with the participation of 541-bp direct repeats (Grondin et al., 1993, 1996). In the amplification of sod2 in S. pombe, a long run of homology is involved, similar to the situation in Leishmania. When the full genomic sequence for this region of S. pombe becomes available, the length of the homologous region will be revealed.
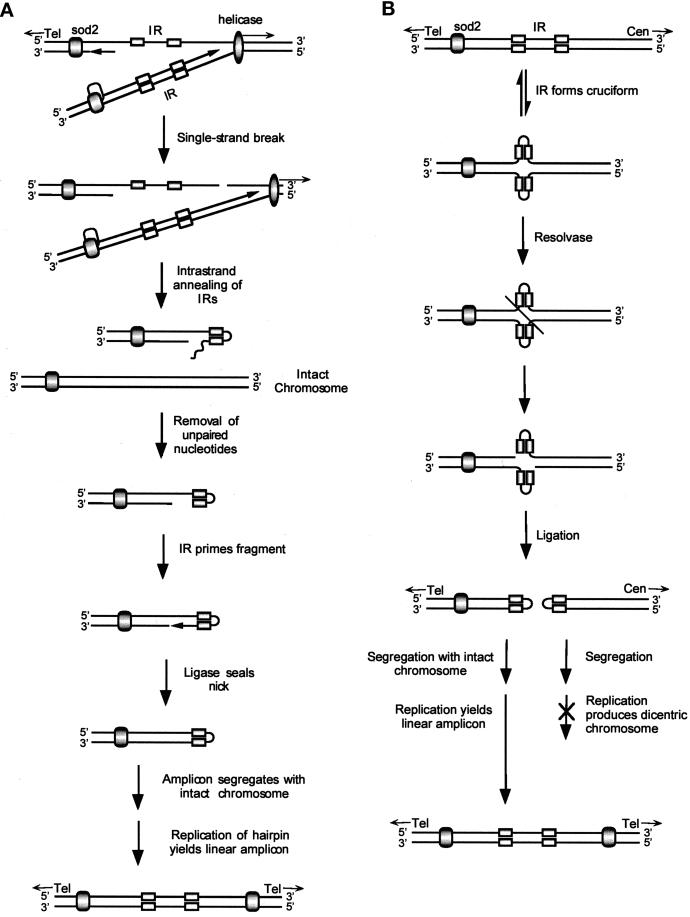
The novel joint of the 225-kb amplicon occurs precisely at the end of an exact inverted repeat, arguing for an essential role of the repeat in amplicon formation. An aberrant replication model (Figure 8A) invokes a single-strand break in the DNA downstream of both the inverted repeat and the replication fork, allowing the inverted repeat to form a hairpin. After the unpaired nucleotides are removed, the inverted repeat primes replication of an additional DNA synthesis fragment, and DNA ligase seals the nick. A large hairpin duplication of the left end of the chromosome results, and a telomere-deleted chromosome is a by-product. The hairpin segregates together with an intact chromosome I to generate a viable cell with a small selective advantage. An additional cell cycle is required to form the mature double-stranded amplicon. Additional cycles yield cells with multiple copies of the amplicon and a correspondingly increased growth advantage in selective medium.
Figure 8.
Replication and recombination models for the 225-kb amplicon. (A) Replication model. sod2 is flanked by a telomere (Tel) on the left and an inverted repeat (IR) on the right. sod2 is shown as a shaded oval, the inverted repeats are shown as rectangles, and the helicase/replication complex is shown as an elongated oval. A single-strand DNA break on the lagging strand, to the right of the inverted repeat and ahead of the replication fork, allows the inverted repeat to form a hairpin. Replication of the other DNA strand is unaffected. Removal of unpaired nucleotides allows the hairpin to prime DNA synthesis. After ligase seals the nick, a larger hairpin, consisting of a duplication of the region from the inverted repeat to the telomere, is formed. The large hairpin and intact chromosome I must segregate together to yield a viable cell resistant to LiCl. The mature palindrome is formed in the next cell cycle. (B) Recombination model. The inverted repeat to the right of sod2 forms a cruciform analogous to a Holliday structure. If resolvase cuts across the axis of symmetry and ligase joins the free ends of the different DNA strands, two hairpins will be produced that include sod2 and the centromere (Cen), respectively. The sod2 hairpin must segregate with intact chromosome I to form a viable cell resistant to LiCl. The next round of replication produces the mature palindrome. Because the fission yeast S. pombe is haploid during selection with LiCl, this mitotic recombination event must occur after replication of the DNA.
Similar models have been presented for both Tetrahymena and Leishmania (Ford et al., 1985; Yasuda and Yao, 1991; Butler et al., 1996; Grondin et al., 1996). When the Leishmania P-glycoprotein locus is flanked by inverted repeats, it forms an inverted duplication upon amplification (Grondin et al., 1996). White et al. (1988) suggested that annealing of these repeats during a block in replication could serve as a primer for DNA polymerase. The newly synthesized strand could then serve as a template to generate a large inverted duplication. Others (Ouellette et al., 1991; Cohen et al., 1994) have suggested that stalling of the replication fork would allow the inverted repeats to anneal, enabling strand switching, or “U-turn” replication. Our model differs in that formation of the hairpin does not require a single strand to invade duplex DNA. For our model to produce both a palindromic amplicon and an intact chromosome, a single-strand DNA break is required between the inverted repeat and the centromere. A double-strand break, causing stalling of replication and switching of both strands, is incompatible with maintaining an intact chromosome.
An alternative recombination-resolvase model (Figure 8B) uses a recombination event that leads to a large hairpin precursor of the amplicon. This model, which is very similar to those proposed previously for Tetrahymena thermophila (Yasuda and Yao, 1991; Butler et al., 1996) and S. pombe (Patterson et al., 1999), invokes the formation of a cruciform structure analogous to a Holliday structure (Holliday, 1964; Leach, 1994). After resolvase cuts across the cruciform in a precise manner (Figure 9), a hairpin can form after DNA ligase seals the gap between the 3′ and 5′ ends of the different DNA strands. A dicentric chromosome is also formed. If the large hairpin segregates with an intact chromosome I, a cell resistant to LiCl results. As in the model shown in Figure 8A, one more round of replication is required to form the mature double-stranded amplicon and additional rounds of replication can generate multiple copies of the amplicon.
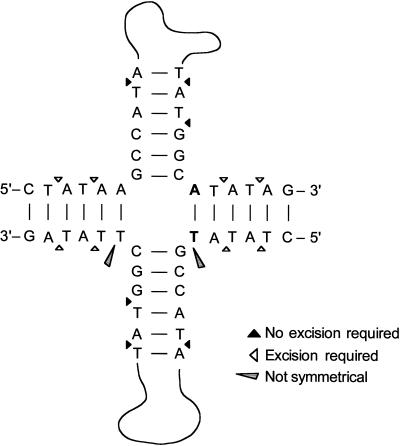
Figure 9.
Possible sites for resolvase action on the cruciform structure. Only the base of the cruciform is shown. Triangles indicate predicted sites of resolvase action. Black triangles, sites of resolvase action that would require subsequent mismatch excision repair to yield observed amplicon. White triangles, sites that would not require such excision. Carrot shape, sites lacking symmetry.
The sequence of the novel joint reveals that the junction formed (Figure 9) is consistent with the expected products of S. pombe resolvase, which cuts Holliday junctions across the axis of symmetry and prefers phosphodiester bonds 3′ to thymidine residues, cutting 3′ to the cruciform structure (Whitby and Dixon, 1997; Oram et al., 1998). Resolvase cuts within 6 bp of the junction in most cases. The locations within the cruciform structure at which subsequent excision is required or not required for our model are indicated in Figure 9. The areas lacking symmetry are also indicated; the lack of symmetry prevents this cruciform from being a true Holliday structure. The sequence of the novel joint of the 225-kb amplicon suggests that the cut may have occurred either 2 or 4 bp away from the cruciform if mismatch repair has occurred. Additional experiments to study the effects of base pair changes in this region should reveal whether resolvase plays a key role in forming the amplicon. For example, the removal of thymidine residues from this region should prevent amplification if resolvase is required.
Previous work has shown that mutations in the DNA-checkpoint pathway increased the frequency of sod2 amplification (Patterson et al., 1999). We now find that strains defective for ligase or for both topoisomerase I and topoisomerase II activity also have greatly increased frequencies of amplification. Both types of mutation increase the likelihood of DNA breakage. Topoisomerase is necessary to untangle chromosomes, removing supercoiling and helping to maintain chromatin organization (Uemura et al., 1987). DNA ligase is needed to seal single-strand DNA breaks. Together, these results make it likely that DNA breaks facilitate gene amplification in S. pombe. DNA breaks are also known to increase the frequency of mitotic recombination (Sipiczki et al., 1990), and a single-strand break has been invoked in our replication model. The genetic data are consistent with both models for generating the 225-kb amplicon and the recombination model for the 180-kb amplicon. The aberrant replication model (Figure 8A) requires a single-strand break, and the recombination-resolvase model (Figure 8B) does not require DNA breakage at all. However, mitotic recombination is increased by DNA breakage and also by mutations in DNA ligase (Sipiczki et al., 1990). Analysis of amplification frequencies in strains deficient in mitotic recombination may help to rule out this class of mechanisms for generating the 225-kb amplicon (see Gysler-Junker et al., 1991).
We have presented the novel joint sequences for two different sod2 amplicon structures. In the first case, the sequence of the novel joint indicates that homologous recombination is responsible for generating the 180-kb amplicon. In the second case, the sequence of the novel joint indicates that inverted repeats play a key role in generating the 225-kb amplicon. Two different mechanisms, one involving aberrant replication and the other involving resolvase action on a cruciform structure, were presented as models of the mechanism of amplification by means of inverted repeats. All three models presented for generating the amplicons are expected to be stimulated by the presence of DNA breaks, as observed in our genetic data. Further studies, both molecular and genetic, should elucidate the mechanistic details of how the 225-kb palindromic sod2 amplicon is formed. We have made observations consistent with circular amplicons being formed in this system. In both wild-type and mutant strains, megabase-size extrachromosomal amplicons have been observed. In one case, the amplicon was observed to migrate differently when different pulse times were used, consistent with it being circular. These observations have not been investigated further. However, they add to the argument that amplification in S. pombe may parallel amplification in higher organisms. The conservation observed between amplification in S. pombe and other organisms suggests that detailed studies in fission yeast will be relevant to the analysis of amplification mechanisms in higher eukaryotes.
ACKNOWLEDGMENTS
We thank members of the Stark laboratory for helpful discussions, Kurt Runge, Helen Salz, and David Setzer for critical reading of the manuscript, the S. pombe sequencing center, the Sanger Center, David Botstein, and Tomo Matsumoto for cosmids, Paul Nurse for S. pombe strains, Neal Sugawara for plasmids containing S. pombe telomere and telomere-adjacent sequences, and David Bonthron for sharing unpublished information on the L-asp gene in S. pombe. This work was supported by National Institutes of Health grant GM 49345 to G.R.S.
REFERENCES
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- Andersson DI, Slechta ES, Roth JR. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- Bishop JM. The molecular genetics of cancer. Science. 1987;235:305–311. doi: 10.1126/science.3541204. [DOI] [PubMed] [Google Scholar]
- Butler DK, Yasuda LE, Yao MC. Induction of large DNA palindrome formation in yeast: implications for gene amplification and genome stability in eukaryotes. Cell. 1996;87:1115–1122. doi: 10.1016/s0092-8674(00)81805-x. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Hassin D, Karby S, Lavi S. Hairpin structures are the primary amplification products: a novel mechanism for generation of inverted repeats during gene amplification. Mol Cell Biol. 1994;14:7782–7791. doi: 10.1128/mcb.14.12.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Edlund T, Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981;292:269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- Fan JB, Chikashige Y, Smith CL, Niwa O, Yanagida M, Cantor CR. Construction of a NotI restriction map of the fission yeast Schizosaccharomyces pombe genome. Nucleic Acids Res. 1989;17:2801–2818. doi: 10.1093/nar/17.7.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M, Davies B, Griffiths M, Wilson J, Fried M. Isolation of a gene enhancer within an amplified inverted duplication after “expression selection.”. Proc Natl Acad Sci USA. 1985;82:3370–3374. doi: 10.1073/pnas.82.10.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin K, Papadopoulou B, Ouellette M. Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania. Nucleic Acids Res. 1993;21:1895–1901. doi: 10.1093/nar/21.8.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin K, Roy G, Ouellette M. Formation of extrachromosomal circular amplicons with direct or inverted duplications in drug-resistant Leishmania tarentolae. Mol Cell Biol. 1996;16:3587–3595. doi: 10.1128/mcb.16.7.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gysler-Junker A, Bodi Z, Kohli J. Isolation and characterization of Schizosaccharomyces pombe mutants affected in mitotic recombination. Genetics. 1991;128:495–504. doi: 10.1093/genetics/128.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hightower RC, Ruiz-Perez LM, Wong ML, Santi DV. Extrachromosomal elements in the lower eukaryote Leishmania. J Biol Chem. 1988;263:16970–16976. [PubMed] [Google Scholar]
- Hoheisel JD, Maier E, Mott R, McCarthy L, Grigoriev AV, Schalkwyk LC, Nizetic D, Francis F, Lehrach H. High resolution cosmid and P1 maps spanning the 14 Mb genome of the fission yeast S. pombe. Cell. 1993;73:109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Jia Z-P, McCullough N, Martel R, Hemmingsen S, Young PG. Gene amplification at a locus encoding a putative Na+/H+ antiporter confers sodium and lithium tolerance in fission yeast. EMBO J. 1992;11:1631–1640. doi: 10.1002/j.1460-2075.1992.tb05209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ota T. On some principles governing molecular evolution. Proc Natl Acad Sci USA. 1974;71:2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- Mizukami T, et al. A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell. 1993;73:121–132. doi: 10.1016/0092-8674(93)90165-m. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nasmyth KA. Genetic and enzymatic characterization of conditional lethal mutants of the yeast Schizosaccharomyces pombe with a temperature-sensitive DNA ligase. J Mol Biol. 1979;130:273–284. doi: 10.1016/0022-2836(79)90541-2. [DOI] [PubMed] [Google Scholar]
- Oram M, Keeley A, Tsaneva I. Holliday junction resolvase in Schizosaccharomyces pombe has identical endonuclease activity to the CCE1 homologue YDC2. Nucleic Acids Res. 1998;26:594–601. doi: 10.1093/nar/26.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M, Hettema E, Wust D, Fase-Fowler F, Borst P. Direct and inverted DNA repeats associated with P-glycoprotein gene amplification in drug resistant Leishmania. EMBO J. 1991;10:1009–1016. doi: 10.1002/j.1460-2075.1991.tb08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson TE, Albrecht EB, Nurse P, Sazer S, Stark GR. Effects of genome position and the DNA damage checkpoint on the structure and frequency of sod2 gene amplification in fission yeast. Mol Biol Cell. 1999;10:2199–2208. doi: 10.1091/mbc.10.7.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M, Grossenbacher-Grunder AM, Bodi Z. Recombination and mating type switching in a ligase defective mutant of Schizosaccharomyces pombe. Mol Gen Genet. 1990;220:307–313. [Google Scholar]
- Smith KA, Stark MB, Gorman PA, Stark GR. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc Natl Acad Sci USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Debatisse M, Giulotto E, Wahl GM. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell. 1989;57:901–908. doi: 10.1016/0092-8674(89)90328-0. [DOI] [PubMed] [Google Scholar]
- Sugawara NF. DNA Sequences at Telomeres of the Fission Yeast Schizosaccharomyces pombe. Ph.D. Thesis. Cambridge, MA: Harvard University; 1988. [Google Scholar]
- Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JD, Paquin CE, Kaneko K, Williamson VM. Resistance to antimycin A in yeast by amplification of ADH4 on a linear, 42 kb palindromic plasmid. Cell. 1986;46:857–863. doi: 10.1016/0092-8674(86)90067-x. [DOI] [PubMed] [Google Scholar]
- Whitby MC, Dixon J. A new Holliday junction resolving enzyme from Schizosaccharomyces pombe that is homologous to CCE1 from Saccharomyces cerevisiae. J Mol Biol. 1997;272:509–522. doi: 10.1006/jmbi.1997.1286. [DOI] [PubMed] [Google Scholar]
- White TC, Fase-Fowler F, van Luenen H, Calafat J, Borst P. The H circles of Leishmania tarentolae are a unique amplifiable system of oligomeric DNAs associated with drug resistance. J Biol Chem. 1988;263:16977–16983. [PubMed] [Google Scholar]
- Whoriskey SK, Nghiem VH, Leong PM, Masson JM, Miller JH. Genetic rearrangements and gene amplification in Escherichia coli: DNA sequences at the junctures of amplified gene fusions. Genes Dev. 1987;1:227–237. doi: 10.1101/gad.1.3.227. [DOI] [PubMed] [Google Scholar]
- Windle B, Draper BW, Yin YX, O'Gorman S, Wahl GM. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes Dev. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- Wu DG, Wu CH, Black LW. Reiterated gene amplifications at specific short homology sequences in phage T4 produce Hp17 mutants. J Mol Biol. 1991;218:705–721. doi: 10.1016/0022-2836(91)90260-d. [DOI] [PubMed] [Google Scholar]
- Wu DG, Black LW. Gene amplification mechanism for the hyperproduction of T4 bacteriophage gene 17 and 18 proteins. J Mol Biol. 1987;195:769–783. doi: 10.1016/0022-2836(87)90483-9. [DOI] [PubMed] [Google Scholar]
- Yao MC, Zhu SG, Yao CH. Gene amplification in Tetrahymena thermophila: formation of extrachromosomal palindromic genes coding for rRNA. Mol Cell Biol. 1985;5:1260–1267. doi: 10.1128/mcb.5.6.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda LF, Yao MC. Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell. 1991;67:505–516. doi: 10.1016/0092-8674(91)90525-4. [DOI] [PubMed] [Google Scholar]