Abstract
Gcn4, a yeast transcriptional activator that promotes the expression of amino acid and purine biosynthesis genes, is rapidly degraded in rich medium. Here we report that SCFCDC4, a recently characterized protein complex that acts in conjunction with the ubiquitin-conjugating enzyme Cdc34 to degrade cell cycle regulators, is also necessary for the degradation of the transcription factor Gcn4. Degradation of Gcn4 occurs throughout the cell cycle, whereas degradation of the known cell cycle substrates of Cdc34/SCFCDC4 is cell cycle regulated. Gcn4 ubiquitination and degradation are regulated by starvation for amino acids, whereas the degradation of the cell cycle substrates of Cdc34/SCFCDC4 is unaffected by starvation. We further show that unlike the cell cycle substrates of Cdc34/SCFCDC4, which require phosphorylation by the kinase Cdc28, Gcn4 degradation requires the kinase Pho85. We identify the critical target site of Pho85 on Gcn4; a mutation of this site stabilizes the protein. A specific Pho85-Pcl complex that is able to phosphorylate Gcn4 on that site is inactive under conditions under which Gcn4 is stable. Thus, Cdc34/SCFCDC4 activity is constitutive, and regulation of the stability of its various substrates occurs at the level of their phosphorylation.
INTRODUCTION
Regulation of cellular processes occurs in a large measure via modulation of steady-state levels of key regulatory proteins. This modulation can occur not only at the level of their synthesis but also at the level of their degradation. Gcn4, a yeast transcriptional activator involved in biosynthesis of amino acids and purines (Hinnebusch and Fink, 1983; Hope and Struhl, 1985), is under this dual control. Starvation for amino acids leads to an increase in Gcn4 translation (Hinnebusch, 1984), which in turn induces the transcription of amino acid biosynthetic genes, thereby providing the nutrients that were formerly limiting for growth. The levels of Gcn4 protein are controlled both at the level of synthesis and degradation. Gcn4 synthesis is increased by a translational control mechanism that involves phosphorylation of the general translation initiation factor eIF-2α by the kinase Gcn2 (Dever et al., 1992; for review, see Hinnebusch, 1997). In addition, Gcn4 is degraded extremely rapidly; however, under amino acid starvation conditions, Gcn4 is stabilized (Kornitzer et al., 1994).
The ubiquitin system is the major cytoplasmic pathway by which proteins are degraded (for a recent review, see Hershko and Ciechanover, 1998). Target proteins are first modified by the sequential addition of ubiquitin, a small, conserved protein. Ubiquitinated proteins are then degraded by a large multicatalytic protease, the 26S proteasome. Protein ubiquitination is catalyzed by a ubiquitin-conjugating enzyme, which accepts a ubiquitin “activated” as a thiolester by the ubiquitin-activating enzyme, and transfers it to the ε-amino group of an internal lysine on the target protein (or to the α-amino group of the polypeptide chain; Breitschopf et al., 1998). The transfer from the ubiquitin-conjugating enzyme to the target requires a third component, called ubiquitin ligase, which constitutes the main recognition component of the system and in some cases may form a second intermediate thiolester with ubiquitin.
Unlike the ubiquitin-conjugating enzymes, which carry a distinctive sequence signature, ubiquitin ligases fall in a number of groups with little sequence similarity. At least four types of ligases can be distinguished, two of which consist of large protein complexes (Hershko and Ciechanover, 1998). These are the anaphase-promoting complex (APC) or cyclosome (King et al., 1995; Sudakin et al., 1995) and the SCF complex (Feldman et al., 1997; Skowyra et al., 1997). The latter stands for Skp1-Cdc53 (or Cullin)-F box protein complex, in which the F-box protein constitutes a variable component that is thought to define the target specificity of the complex (for review, see Patton et al., 1998b). A fourth component of the SCF complex, Rbx1, recently was discovered (Kamura et al., 1999; Skowyra et al., 1999). The SCF ubiquitin ligase is found to interact with a single type of ubiquitin-conjugating enzyme, Cdc34 (Feldman et al., 1997; Skowyra et al., 1997; Mathias et al., 1998). Targets of the SCFCDC4 complex (the SCF complex containing Cdc4 as the F-box component) in yeast that have been identified so far include the cyclin-dependent kinase (CDK) inhibitors Sic1 (Feldman et al., 1997) and Far1 (Henchoz et al., 1997) and the DNA replication regulator Cdc6 (Drury et al., 1997; Elsasser et al., 1999; Sanchez et al., 1999). Other yeast SCF complexes were found to target other substrates, such as Cln1 and -2 and Gic2 (SCFGRR1; Skowyra et al., 1997; Jaquenoud et al., 1998; Patton et al., 1998a) and the inhibitory kinase Swe1 (SCFMET30; Kaiser et al., 1998). In mammalian cells, a specific SCF complex, SCFβ-TrCP, recently was found to target IκB for ubiquitination (Yaron et al., 1998; Spencer et al., 1999; Winston et al., 1999). Unlike APC substrates, which contain a consensus sequence required for degradation (the “destruction box”; Yamano et al., 1996), no such consensus has been found yet for the SCF substrates. However, common to many, and perhaps all, instances of ubiquitination by SCF complexes is the requirement for phosphorylation of the substrate (Hershko and Ciechanover, 1998). In yeast, ubiquitination of the CDK inhibitors Sic1 and Far1 and of the DNA replication regulator Cdc6 were shown to require activity of the major yeast cell cycle CDK, Cdc28 (Henchoz et al., 1997; Verma et al., 1997; Elsasser et al., 1999). A second yeast CDK, Pho85, which is not essential for cell cycle progression, recently was found to be also capable of phosphorylating Sic1 (Nishizawa et al., 1998).
Previous work has shown that Gcn4 is degraded via the ubiquitin system and stabilized in proteasome mutants, as well as in a mutant of the ubiquitin-conjugating enzyme Cdc34 (Kornitzer et al., 1994). However, neither the precise ubiquitination complex required for Gcn4 degradation nor the mechanism of regulation of Gcn4 degradation by starvation had been defined. Here we show that degradation of Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin–ligase complex. Our data indicate that Pho85 activity is repressed by starvation and suggest that the targeting of Gcn4 to the ubiquitin–ligase complex SCFCDC4 is determined by a specific Pho85-mediated phosphorylation event.
MATERIALS AND METHODS
Plasmids and Strains
The CUP1 vector plasmid KB354 was constructed by cloning the BamHI–EcoRI CUP1 promoter fragment from plasmid pYSK7 (Butt et al., 1984) to pRS314 (Sikorski and Hieter, 1989), followed by deletion of the SacII–BamHI sequence of the polylinker. PCR cloning (Ausubel et al., 1989) was used to fuse the coding sequence of GCN4 to the CUP1 promoter of KB354. The resulting plasmid was then digested with BamHI and HindIII to generate a fragment carrying part of the TRP1 sequence, the CUP1 promoter sequence, and the 5′ end of the GCN4 sequence. This fragment was cloned in KB64 digested with the same enzymes, to generate a CUP1p–GCN4–LacZ fusion (KB449). pGAL–CYC1–LacZ (KB496) was constructed by inserting a PCR-generated BglII fragment carrying the whole GCN4 coding sequence into the BamHI site of plasmid pLGSD5 (from L. Guarente, Massachusetts Institute of Technology). pGAL–CDC6–LacZ (KB448) was constructed by substituting the GCN4 coding region of plasmid KB64 (Kornitzer et al., 1994) with a PCR-generated BamHI–EcoRI fragment carrying the CDC6 coding sequence. The T165A mutation was introduced by site-directed mutagenesis into the ADE8-GCN4 construct KB105 (Kornitzer et al., 1994), to generate KB853. PCR cloning was used to subsequently transfer the mutation to the pGAL–GCN4–LacZ plasmid KB149, to generate KB854. The pCUP1–SIC1 plasmid (KB733) was generated by PCR cloning of an EcoRI–SalI fragment carrying SIC1 into the pCUP1 vector KB354 digested with the same enzymes, to generate KB733. pGAL1–CLN2(4T3S)HA expressing a stabilized version of Cln2 (Lanker et al., 1996) was obtained from Curt Wittenberg (Scripps Research Institute). The GCN4 deletion Δ(93–140) (KB 92) was described previously (Kornitzer et al., 1994). The GCN4 deletion Δ(151–167) fused to LacZ (KB295) was generated by digestion of the GCN4 coding sequence with KpnI and XbaI, filling-in, and religation. The plasmid expressing the myc-tagged Gcn4(62–202) fragment was constructed in two steps: first, PCR was used to generate an EcoRI site and an initiation codon in front of the Gcn4(62–202)–encoding fragment and an XhoI site preceded by a termination codon and a BamHI site at the other end of the fragment (forward primer: 5′ CCGAATTCATGTCGAACCTTGATTTTGATT; reverse primer: 5′ GGCCTCGAGTTAGGATCCCATATGATCCAGTCTCGATTCG). This fragment was then cloned in the EcoRI and XhoI sites of plasmid KB354 to generate KB 885 (wild type) and KB 887 (T165A). In a second step, a triple Myc-epitope–carrying BamHI fragment (Kornitzer and Kron, unpublished results) was cloned into the BamHI site of plasmids KB885 and KB887 to generate KB891 and KB893, respectively. The GST-Gcn4(62–202)–expressing plasmid was constructed by recloning the EcoRI-XhoI fragments of KB885 and KB887 into pGEX-4T-1 (Pharmacia Biotech, Piscataway, NJ) to generate KB927 and KB928, respectively. The plasmid expressing heptahistidine-tagged Gcn4 under the T7 promoter was constructed by cloning a Gcn4-carrying PCR fragment into pET11-his7 (obtained from S. Buratowski, Harvard Medical School) to generate KB363. Plasmids pGEX-PHO85, pGEX-pho85(E53A), pGEX-PCL1, pGEX-PHO80, and Yep-GAL10-HA-PCL1 are described in Nishizawa et al. (1998).
The cdc4-1 strain (KY301) was constructed by homologous recombination. The region extending from −270 to + 2575 relative to the CDC4 start site was cloned by PCR from a cdc4-1 strain (Fink laboratory collection) between the BamHI and EcoRI sites of plasmid YIp5, to yield KB527. Strain KY289 (an α strain isogenic to KY204) was transformed with KpnI-digested KB527, to yield a tandem mutant–wild-type structure in the chromosome, separated by the YIp5 sequence. Passage over 5-fluoroorotic acid to select for excision of the URA3 sequence of Yip5 by homologous recombination yielded a fraction of colonies in which the wild-type allele was excised and the mutant retained in the chromosome. The pho85Δ::hisG strain (DY4535) was constructed in several steps. First, the PHO85 gene in a diploid W303 strain was replaced with a pho85Δ::LEU2 allele using a disruption cassette provided by Brenda Andrews (University of Toronto). A pho85Δ::LEU2 haploid strain was derived from this diploid, and this was converted to a pho85Δ::hisG-URA3-hisG allele using plasmid pNKY85 (Alani et al., 1987). Finally, the strain with the pho85Δ::hisG allele was recovered after passage over 5-fluoro-orotic acid medium. The other yeast strains used and their origins are described in Table 1.
Table 1.
Yeast strains
| Name | Genotype | Source |
|---|---|---|
| KY26 | a ura3-52 leu2-Δ2 trp1-Δ1 bas1-2 bas2-2 gcn4-Δ1 | Kornitzer et al. (1994) |
| KY204 | KY26 ade8-GCN4 | Kornitzer et al. (1994) |
| KY205 | KY204 cdc34-2 | Kornitzer et al. (1994) |
| KY301 | KY204 cdc4-1 | This work |
| W303-1A | ura3-1 ade2-1 can1-100 leu2-3,112 trp1-1his3-11,15 | R. Rothstein |
| DY4535 | W303 pho85Δ∷hisG | This work |
| Y552 | W303 skp1-11 | Bai et al. (1996) |
| Y554 | W303 skp1-12 | Bai et al. (1996) |
| YPH1015 | ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CFIII(CEN3.L.YPH983)-HIS3-SUP11 | Connelly and Hieter (1996) |
| YPH1161 | YPH1015 LEU2∷skp1-4 | Connelly and Hieter (1996) |
| YPH1172 | YPH1015 LEU2∷skp1-3 | Connelly and Hieter (1996) |
| MTY668 | W303 cdc4-1 | M. Tyers |
| MTY670 | W303 cdc34-2 | M. Tyers |
| MTY740 | W303 cdc53-1 | Willems et al. (1996) |
Degradation Assays
The pulse–chase experiments were performed essentially as described previously (Kornitzer et al., 1994). Briefly, overnight cultures were diluted in 10 ml and grown to midlog phase, washed two times with medium lacking methionine, concentrated to 0.3 ml, and pulse-labeled for 5 min with 750–900 mCi [35S]methionine (“express,” NEN, Arlington, MA), pelleted again and chased in medium containing 10 mM methionine and 10 mM cysteine. For the experiments involving temperature-sensitive mutants, the cells were shifted from 30 to 37°C 20–40 min before labeling. Similarly, for the starvation studies, cells were harvested by centrifugation and grown in SD medium + adenine for 15–45 min before labeling and chased in the same medium + methionine and cysteine. At various times of the chase, an aliquot of the culture was removed and incubated 15 min on ice with 0.35 M NaOH and 1.5% 2-mercaptoethanol, followed by precipitation with 6% trichloroacetic acid. The protein precipitate was resuspended by boiling in 2.5% SDS and 5 mM EDTA, and equal amounts of trichloroacetic acid–precipitable radioactivity were immunoprecipitated in at least 10 volumes of buffer A (Hochstrasser and Varshavsky, 1990) containing protein A-Sepharose (Pharmacia). The immunoprecipitates were run on SDS-polyacrylamide gels, and the protein bands were quantitated using a Fujix Bas 2000 bioimage analyzer (Fuji, Tokyo, Japan). Antibodies used were either monoclonal anti-myc (9E10; Kolodziej and Young, 1991), anti-β-galactosidase (Cappel, Malvern, PA), or anti-Gcn4, which were generated in rabbits injected with recombinant GST-Gcn4 fusion protein. The proteins we tested were usually ectopically expressed either from the GAL1 promoter of from the CUP1 promoter. CUP1 is induced by copper ions; however, even in the absence of copper, enough protein is produced to permit detection in pulse-labeling assays. The half-life of Gcn4 expressed from the CUP1 promoter (Figures 1, 2A, 3, and 7A), 2–3 min, was somewhat shorter than that of the same protein expressed under the GAL1 promoter (3–5 min; see Kornitzer et al. [1994] and Figures 2D, 6, and 8). This difference may be due to the difference in the carbon source used (glucose vs. galactose). For the experiments performed in cell cycle-arrested cells, 3 μM α-factor or 0.2 M hydroxyurea were used to arrest cells in G1 and S, respectively. Cell cycle arrest was monitored microscopically; the labeling was initiated after >90% of the cells had arrested as unbudded or large-budded cells, respectively.
Figure 1.
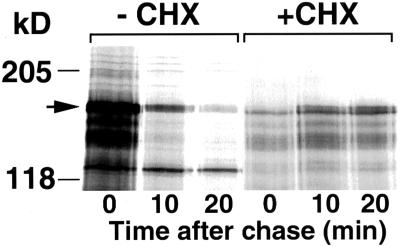
Inhibition of translation stabilizes Gcn4. Gcn4–LacZ was expressed from the CUP1 promoter of plasmid KB449. +CHX: cells were preincubated for 30 min in 0.5 μg/ml cycloheximide and then labeled and chased in the presence of the drug. The arrow on the left indicates Gcn4–LacZ.
Figure 2.
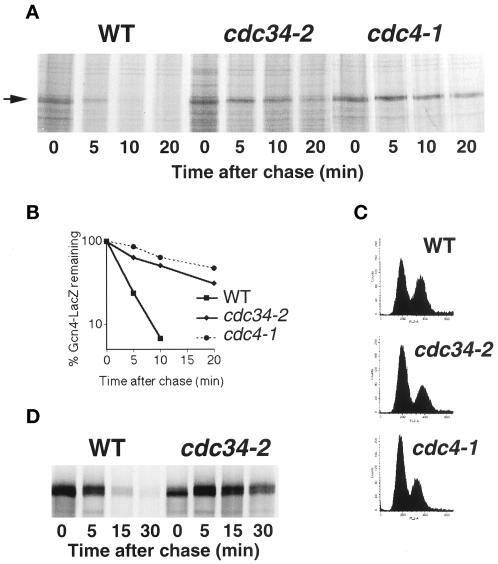
Gcn4 degradation is inhibited in cdc34 and cdc4 mutants. (A) Pulse-and-chase analysis of Gcn4–LacZ expressed from the CUP1 promoter of plasmid KB449 in strains KY204, KY205, and KY301. The cultures were shifted from 30 to 37°C, 20 min before the labeling. (B) Quantitation of the experiment shown in (A). (C) Fluorescence-activated cell sorter analysis of cells at the time of labeling. (D) Pulse–chase analysis of Gcn4–-LacZ expressed from the GAL1 promoter of plasmid KB64 in α-factor–arrested cells. Cells were arrested with α-factor at the permissive temperature and then shifted to 37° for 30 min before labeling.
Figure 3.
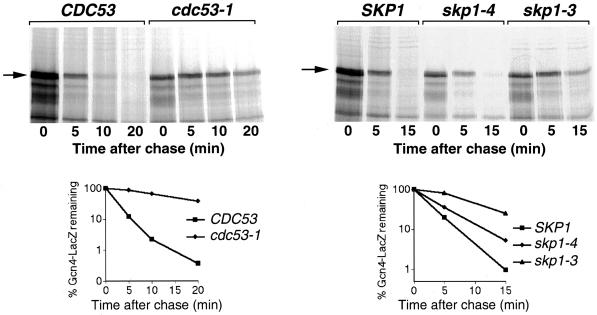
Gcn4 degradation is inhibited in cdc53 and skp1 mutants. Pulse–chase analysis of Gcn4–-LacZ expressed from the CUP1 promoter of plasmid KB449 in strains W303 and MTY740 (left) or YPH1015, YPH1161, and YPH1172 (right). The cultures were shifted from 30 to 37°C, 30 min before labeling.
Figure 7.
Degradation of Gcn4 in pho85Δ cells. (A) Pulse–chase analysis of Gcn4–LacZ expressed from the CUP1 promoter of plasmid KB449 in W303 and DY4535 cells. (B) Quantitation of the experiment shown in (A). (C) Pulse–chase analysis of native, endogenous Gcn4 in W303 and DY4535 cells. The lane marked “C” indicates an extract from a gcn4Δ strain. The anomalous migration of native Gcn4 (shown here) as well as of recombinant Gcn4 (Figures 10 and 11), a 33-kDa protein, has been noted before (Hope and Struhl, 1985). Note that a doublet of cross-reacting bands migrates slightly slower than Gcn4. (D) Expression of His4–LacZ from a HIS4 promoter derivative that is exclusively dependent on Gcn4 for activity (deletion number 203; Nagawa and Fink, 1985) in W303 and DY4535 cells. Overnight cultures were diluted and grown for 6 h to early log phase, and the β-galactosidase activity of the cultures was measured as described (Daignan-Fornier and Fink, 1992). (E) Degradation of the T105A mutant of Gcn4 in W303 and DY4535 cells. Gcn4(T105A)-LacZ was expressed from the GAL promoter of plasmid KB358 (Kornitzer et al., 1994).
Figure 6.
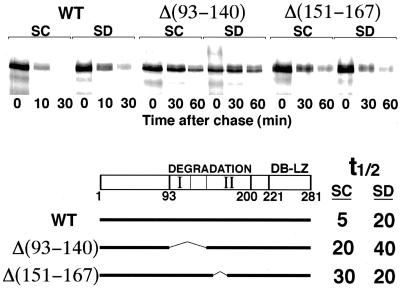
Effect of deletions in Gcn4 on the sensitivity to starvation. Full-length Gcn4 and two deletions fused to LacZ were subjected to pulse–chase analysis without (SC) or with (SD) prior mild starvation (15 min in medium lacking amino acids). Note the difference in time scale. t½ indicates the half-lives of the three proteins under both conditions as derived from quantitation of the data by phosphorimager. “DEGRADATION” denotes the minimal region of Gcn4 required for degradation, and “I” and “II” indicate the two domains of that region, as defined by deletion analysis (see text). “DB-LZ” denotes the DNA binding domain and the leucine zipper domain of Gcn4.
Figure 8.
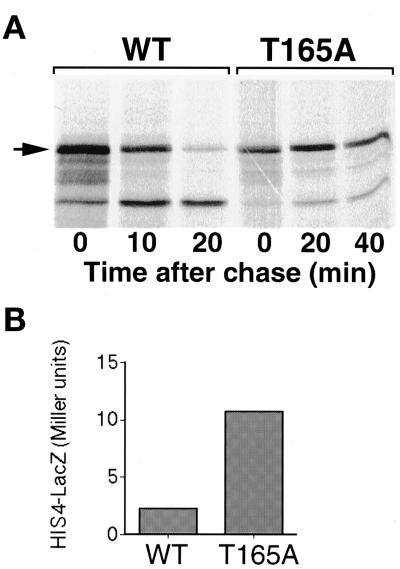
A mutation at residue Thr165 stabilizes Gcn4. (A) Pulse–chase analysis of wild-type (KB149) and mutant (KB854) Gcn4–LacZ expressed from the GAL promoter in W303 cells. Note the difference in time scale. (B) Expression of His4–LacZ from the full-length HIS4 promoter (plasmid pFN6; Nagawa and Fink, 1985). The strain used, bas1-2 bas2-2 gcn4Δ1 (KY26), is defective in both HIS4 transcription factors (Arndt et al., 1987). His4–LacZ expression is exclusively dependent on the plasmid-borne GCN4 alleles expressed from the ADE8 promoter of plasmids KB105 (WT) or KB853 (T165A). The β-galactosidase assay was performed as for Figure 7D.
In Vivo Phosphorylation Assay
For in vivo phosphate labeling, cells were grown overnight in yeast nitrogen base (YNB) with the required amino acids and a reduced KH2PO4 concentration (1 instead of 6 mM). The cells were then diluted in the same medium but with 0.1 mM KH2PO4, grown to optical density (OD)600 = 0.5, and treated with 1 μg/ml cycloheximide or transferred to YNB devoid of the required amino acids to induce starvation, as required. Cells were then washed twice with the same medium containing only 50 μM KH2PO4, and each sample (5 OD units of cells) was labeled for 15 min in 0.5 ml of the same medium containing 1 mCi 32PO4. Cell extract preparation and immunoprecipitation were performed as for the degradation assay.
Kinase Assays
For the in vitro kinase reactions with recombinant proteins, 10 ng GST-PHO85,10 ng GST-cyclin, and 0.5 μg substrate were incubated at 30°C for 30 min with 1 μCi [32P-γ]ATP and 0.1 mM cold ATP in 10 μl kinase assay buffer (50 mM Tris, 10 mM MgCl2, 2 mM EDTA, 1 mM dithiothreitol, pH 7.5). Recombinant proteins were expressed and purified according to standard protocols. An ultrafiltration centrifugal device was used for buffer exchange and concentration of the protein. For the immunoprecipitated kinase assays, extracts of 50 ml exponentially growing culture induced 4 h with galactose to express ha-Pcl1 and starved or cycloheximide-treated as indicated were made by breaking the cells for 5 min with glass beads in extract buffer (50 mM Tris, pH 7.5, 250 mM NaCl, 5 mM EDTA, 0.5% NP40, phosphatase inhibitors: 2 mM Na-pyrophosphate, 80 mM β-glycerolphosphate, 10 mM NaF, 0.3 mM NaVO3, protease inhibitors: 2 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, aprotinin 1:100, TPCK 50 μg/ml, TLCK 50 μg/ml, and 1:500 of an antiprotease cocktail containing leupeptin, pepstatin, and chymostatin, each 10 mg/ml in dimethyl sulfoxide). Five to 20 μl of 0.2 mg protein extract were incubated with the 12CA5 monoclonal antibody for 30 min on ice and then 20 μl of a 50% slurry of protein A-agarose beads (Pharmacia) and 200 μl extract buffer were added, and the tubes were incubating while tumbling for 1 h at 4°C. The agarose beads were then washed three times with extract buffer lacking aprotinin, TPCK, TLCK, and the 500× antiprotease cocktail, and two times with kinase buffer. For each phosphorylation reaction, the agarose bead pellet was incubated 20 min at 30°C with 0.2 μg substrate in 10 μl kinase buffer with 1 μCi [32P-γ]ATP and 20 μM cold ATP. All the kinase reactions were terminated by the addition of 10 μl protein-loading buffer and electrophoresed on an SDS-PAGE gel.
RESULTS
Inhibition of Translation Leads to Gcn4 Stabilization
Degradation of Gcn4 occurs through the ubiquitin pathway and is inhibited under amino acid starvation conditions or in an amino acyl-tRNA synthetase mutant (Kornitzer et al., 1994). To define further the signal that inhibits Gcn4 degradation, we examined the role of translation on Gcn4 degradation. In previous pulse–chase experiments designed to measure the rate of Gcn4 decay, we noticed that the addition of the translational inhibitor cycloheximide at the beginning of the chase typically reduced the rate of Gcn4 decay starting 10 min into the chase (see Kornitzer et al., 1994). To test the possibility that inhibition of protein synthesis can reduce Gcn4 turnover, Gcn4-expressing cells were preincubated with 0.5 μg/ml cycloheximide, so as to reduce protein synthesis by 80–90%. This pretreatment with the translation inhibitor led to a complete stabilization of the protein (Figure 1). In a separate experiment, cycloheximide was added to 0.1 μg/ml or the translational inhibitor paromomycin was added to 10 mg/ml; under these conditions, translation was inhibited twofold at most, and the half-life of Gcn4 was increased twofold (our unpublished results).
Several SCF Components Are Implicated in Gcn4 Degradation
To clarify the regulatory mechanism of Gcn4 degradation, we first attempted to identify the ubiquitin ligase required for Gcn4 degradation. It was previously shown that Gcn4 is stabilized in a cdc34 mutant (Kornitzer et al., 1994). We asked whether mutants of the SCF complex would affect Gcn4 degradation. A Gcn4–LacZ fusion, previously shown to be degraded with the same kinetics as native Gcn4 (Kornitzer et al., 1994), was placed under the control of the CUP1 promoter (see MATERIALS AND METHODS). The cdc4-1 mutant displays a cell cycle arrest phenotype similar to that of cdc34-2: both arrest at the G1 to S transition, with a multibudded phenotype (Yochem and Byers, 1987). We tested degradation of Gcn4 in cdc34-2 and cdc4-1 cells 20 min after they were shifted up to the nonpermissive temperature of 37°C. As can be seen in Figure 2, the half-life of Gcn4 is extended from 2.5 min to 12 and 20 min, respectively. We also tested whether the F-box protein Grr1, which is required for the degradation of G1 cyclins (Barral et al., 1995; Skowyra et al., 1997), is involved in Gcn4 degradation. No effect was seen on the half-life of Gcn4 in strains carrying a null allele of GRR1 (our unpublished results).
One concern is that the stabilization of Gcn4 in the cdc34 and cdc4 mutants may be an indirect consequence of their cell cycle arrest rather than a direct consequence of their function in the ubiquitin pathway. However, if Gcn4 degradation were cell cycle dependent, then it should be biphasic in a population of cycling cells. Because we find that in cycling cells, Gcn4 degradation is exponential over two orders of magnitude (e.g., see Figures 2 and 3), if there is a cell cycle window where Gcn4 is stable, it has to be extremely narrow. It is still possible in theory that the cdc34 and cdc4 mutants arrest in that window. To address this concern, we analyzed the cell cycle distribution of the various strains at the time of labeling. Although the mutant strains do display a higher proportion of G1 cells than the wild-type strains (see Figure 2C), fully one-third of the mutant cells were still in G2/M at the time of labeling. Thus, the fact that cells in different stages of the cell cycle show stabilization of Gcn4 argues against the stabilization being an indirect effect of cell cycle arrest. This possibility was further addressed in an alternative way: wild-type and cdc34 mutant cells expressing Gcn4–LacZ under the GAL1,10 promoter were arrested in G1 with α-factor, the cells were shifted to the nonpermissive temperature, and Gcn4 degradation was assayed. As shown in Figure 2D, the pattern of degradation was identical to that in cycling cells (rapid in the wild type, very slow in the cdc34 mutant), demonstrating that the stabilization of Gcn4 is unrelated to the cell cycle phenotype of cdc34.
Two other SCF components, Cdc53 and Skp1, were tested for effects on Gcn4 degradation. The cdc53-1 mutant arrests with the same cell cycle phenotype as cdc4 and cdc34 (Mathias et al., 1996). Pulse–chase analysis showed that Gcn4 is strongly stabilized in cdc53-1, with an increase in the half-life of the protein from 2 to 15 min (Figure 3). SKP1 was isolated as a suppressor of cdc4-1 (Bai et al., 1996), and various skp1 alleles arrest in either G1 or G2/M of the cell cycle (Bai et al., 1996; Connelly and Hieter, 1996). We found that, although the G1-arresting alleles skp1-3 (Figure 3) and skp1-11 (our unpublished results) strongly stabilized Gcn4, the effect of the G2-arresting alleles skp1-4 (Figure 3) and skp1-12 (our unpublished results) was more moderate.
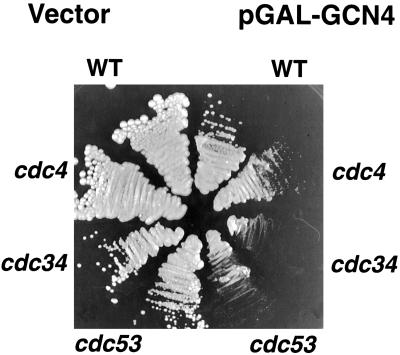
We next tested the effects of overexpression of Gcn4 in wild-type versus mutant cells. Overexpression of Gcn4 leads to slower growth of wild-type cells but does not involve an obvious cell cycle delay (Kornitzer and Fink, unpublished observations). We reasoned that, if Gcn4 is a direct substrate of the Cdc34/SCFCDC4 complex, in mutant cells, where Cdc34/SCFCDC4 activity is limiting, Gcn4 overexpression may compete with the cell cycle substrates and lead to cell cycle arrest even at the normally permissive temperature. To test this, a Gcn4 construct under the strong galactose-inducible GAL1–CYC1 hybrid promoter was expressed in wild-type and mutant cells. As can be seen in Figure 4, at the normally permissive temperature of 30°C, the presence of the Gcn4-overexpressing plasmid led to complete growth arrest of the cdc34-2 and cdc53-1 mutants on galactose medium and to strong growth repression of the cdc4-1 mutant. Microscopic observation of these cells showed the characteristic multibudded phenotype of this class of cell division cycle mutants (our unpublished observations). The fact that overexpression of Gcn4 in these mutant backgrounds results in a mutant phenotype at 30° that mimics the phenotype at 37° supports the assumption that Gcn4 is a direct substrate of Cdc34/SCFCDC4 and that its overexpression competitively inhibits the complex.
Figure 4.
Effect of Gcn4 overexpression on the growth of various SCF mutants. Plasmid KB496, expressing Gcn4 under the strong GAL1–CYC1 hybrid promoter, or the vector plasmid pLGSD5, were transformed into the indicated strains. The strains were streaked on selective galactose plates and incubated for 3 d at 30°C.
Starvation Does Not Affect Degradation of Other SCF Substrates
Is Gcn4 ubiquitination regulated by modulation of Cdc34/SCFCDC4 activity or by modification of the substrate, Gcn4? If the Cdc34/SCFCDC4 complex were regulated, one would predict that amino acid starvation would stabilize other substrates of this complex. To test this, two additional substrates of the Cdc34/SCFCDC4 complex, the CDK inhibitor Sic1 and Cdc6, a protein involved in initiation of DNA replication, were analyzed for their stability. Sic1 degradation usually occurs at the end of G1 and is dependent upon phosphorylation by Cln1 or -2/Cdc28 (Schneider et al., 1996; Tyers, 1996; Verma et al., 1997). To dissociate the effects of starvation on Sic1 degradation from effects on the cell cycle progression, degradation was tested in S-phase–arrested cells in which a stable version of Cln2 (Lanker et al., 1996) was ectopically expressed from the GAL1 promoter. Sic1 was constitutively expressed from the CUP1 promoter. Under these conditions, in rich medium, 90% of Sic1 was degraded in the first 10 min of the chase. The residual fraction appeared to be stable (Figure 5A) and could represent the Sic1 protein from a subpopulation of cells having lost the toxic GAL–CLN2 plasmid. Cells starved for amino acids display the same kinetics of Sic1 degradation, indicating that starvation does not affect the ubiquitination of this protein by the complex.
Figure 5.
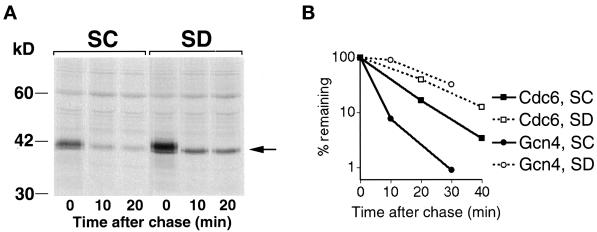
Starvation does not affect degradation of two other SCFCDC4 substrates. (A) Degradation of Sic1. Cells carrying plasmids KB733 (CUP1-SIC1) and pGAL1-CLN2(4T3S)HA (Lanker et al., 1996) were arrested in S-phase with hydroxyurea, induced with galactose to produce Cln2, and subjected to pulse–chase analysis without (SC) or with (SD) prior amino acid starvation. The Sic1 protein was visualized by immunoprecipitation with a specific antibody. The arrow on the right indicates Sic1. (B) Degradation of Cdc6–LacZ versus Gcn4–LacZ. Cells carrying plasmid KB448 (pGAL1–CDC6–LacZ) or KB64 (pGAL1–GCN4–LacZ) were grown in galactose, arrested in G1 with α-factor, and subjected to pulse–chase analysis without (SC) or with (SD) prior amino acid starvation (30 min in medium lacking amino acids). The lacZ fusion proteins were visualized by immunoprecipitation with a specific LacZ antibody. The graph depicts the quantitation of the fusion protein band by phosphorimager.
Cdc6 degradation is also cell cycle regulated and peaks in late G1/early S (Drury et al., 1997). To measure Cdc6 degradation, a Cdc6–LacZ fusion was placed under the control of the GAL promoter, and degradation of the fusion protein was assayed in cells arrested in G1 with α-factor. The half-life of the Cdc6–lacZ fusion protein was 8 min in standard synthetic medium; in cells shifted to starvation medium 30 min before the chase, the half-life increased approximately twofold, to 15 min (Figure 5B). By comparison, in a parallel experiment, using the same starvation regimen, the half-life of Gcn4 increased from 3 to 21 min, i.e., sevenfold (Figure 5B). Thus, these results suggest that SCFCDC4 activity is not regulated by amino acid starvation.
Identification of a Sequence in Gcn4 Responsive to the Starvation Signal
If SCFCDC4 activity is constitutive, then the regulation of Gcn4 degradation must occur at the level of substrate modification. To identify the sequence in Gcn4 that is the target of this regulation, we tested a series of GCN4 deletions for degradation under starvation versus nonstarvation conditions. Previous deletion analysis identified an extended region of Gcn4, including at least residues 93–200, that is required for degradation (Kornitzer et al., 1994). Finer deletion mapping allowed us to subdivide that region in at least two domains, extending from 93 to 118 (“I”) and from 140 to 200 (“II”). This subdivision is based on the observation that a deletion of residues 118–140 barely affects stability of the protein, whereas deletions N-terminal or C-terminal to these residues (e.g., Δ151–167) significantly stabilize the protein (Kornitzer et al., 1994; our unpublished results; Figure 6). Within domain I, point mutations at positions S101, T105, and P106 were sufficient to partially stabilize the protein (Kornitzer et al., 1994). If this degradation signal were the only target modified in starved versus unstarved cells, then mutations in that signal would render the degradation unresponsive to starvation. However, we found that mutation T105A was still stabilized further by starvation (our unpublished results). Likewise, a protein lacking residues 93–140 was partially stabilized when compared with the full-length protein but still stabilized further under starvation conditions (Figure 6). In contrast, a deletion of residues 151–167 led also to partial stabilization of the protein in rich medium, but it was not stabilized further upon starvation; in fact, its half-life was somewhat reduced under starvation conditions (Figure 6). Degradation of Δ(151–167) was tested in two additional experiments and consistently found to be slightly faster under starvation conditions (unpublished results). Thus, deletion Δ(151–167) may define a sequence that is differentially modified in sated versus starved cells.
Gcn4 Degradation Requires Pho85 Activity
It was reasonable to assume that Gcn4 might be modified by phosphorylation because other known SCF substrates require phosphorylation by the main yeast cell cycle CDK, Cdc28, before ubiquitination. Although Gcn4 degradation is unaffected in cdc28-1 mutant cells even at the nonpermissive temperature (Kornitzer and Fink, unpublished results), the yeast genome contains four additional CDK homologues, including Pho85, Kin28, and Srb10. An SRB10 null mutant, srb10-3 (Liao et al., 1995), and a KIN28 temperature-sensitive mutant, kin28-ts3 (Valay et al., 1993), were not defective in Gcn4 degradation (unpublished results). We found, however, that PHO85 is required for Gcn4 degradation. Initially, two observations suggested a genetic interaction between PHO85 and GCN4. First, we found it impossible to transform a pho85Δ strain with a plasmid carrying a constitutive GCN4 mutation (Hinnebusch, 1984). In addition, the pho85Δ mutant was highly sensitive to overexpression of GCN4 under the GAL promoter (unpublished results).
These genetic interactions led us to test Gcn4 degradation in a pho85Δ mutant. As shown in Figure 7A, Gcn4 was markedly, but not completely, stabilized in the pho85Δ strain, with a half-life of 10 min instead of 2.5 min. In this assay, we used a Gcn4–LacZ fusion overexpressed under a heterologous promoter. To confirm that this stabilization was not due to overexpression, we used antibodies against Gcn4 to follow the degradation of the native, endogenous protein. Although the expression of the endogenous protein is weak, we were able to see a rapid degradation in the PHO85 strain, and a strong stabilization in the pho85Δ strain (Figure 7C). We also tested transcriptional activity of endogenous Gcn4 in the pho85Δ mutant, using as reporter an HIS4–LacZ fusion exclusively dependent on Gcn4 for expression (Nagawa and Fink, 1985). HIS4 expression was increased twofold in the mutant versus the wild-type strain, indicating that the stabilization of Gcn4 was accompanied by an increase in transcriptional activity (Figure 7D).
Identification of the Critical Pho85 Target Site on Gcn4
CDK target sites are characterized by a serine or threonine residue followed by a proline. Gcn4 contains five such sites, at positions 17, 61, 105, 165, and 218. Previous deletion analysis excluded positions 17, 61, and 218 from being required for Gcn4 degradation. Of the two remaining sites, Thr105 had already been shown to be required for degradation (Kornitzer et al., 1994). However, the T105A mutant was still stabilized further in PHO85 versus pho85Δ cells (Figure 7E), suggesting that Pho85 may target another site on Gcn4. The last potential site, Thr165, is located in domain II, shown by deletion analysis to be required for degradation, and for the starvation response (see above, Figure 6). Strikingly, Thr165 is embedded within a sequence, TPVL, which conforms to a proposed Pho85 target consensus sequence, S/TPXI/L (O'Neill et al., 1996). To test the requirement of Thr165 for Gcn4 degradation, the residue was subjected to site-directed mutagenesis. Substitution of an alanine for Thr165 resulted in strong stabilization of the protein (Figure 8A), with an increase in half-life from 5 to >30 min. In addition, this mutant displayed a significant increase in Gcn4-dependent transcriptional activity, as measured by HIS4–LacZ activity (Figure 8B). The high initial stability of this mutant precluded us from testing for further stabilization upon starvation.
Phosphorylation of Thr165 in Vivo
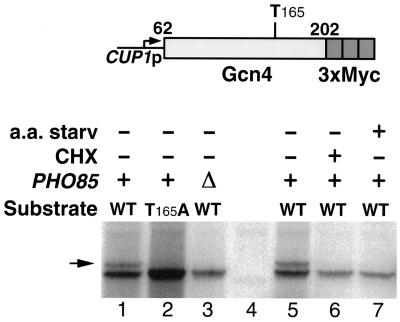
To directly show Pho85-dependent phosphorylation of Thr165 in vivo, we fused a fragment of Gcn4 extending from residue 62 to residue 202 to a triple Myc epitope. This fragment, although more stable than full-length Gcn4, is still rapidly degraded (our unpublished results) and contains fewer potential kinase target sites than the full-length protein. The epitope-tagged fragment was then expressed in cells subjected to phosphate labeling and immunoprecipitated. As shown in Figure 9, (lanes 1 and 5) the immunoprecipitated phosphate-labeled protein migrated as two distinct bands, indicating that it still contained at least two phosphorylation sites. However, the protein fragment carrying the T165A mutation migrated as the single faster-migrating band only (lane 2), indicating that the slower-migrating band represents phosphorylation at Thr165 (the stronger signal of the mutant versus the wild-type protein fragment reflects the fact that the mutant protein is more stable). We then tested the phosphorylation pattern of Gcn4 in pho85Δ cells. Strikingly, the slower-migrating band was absent in these cells (lane 3), indicating that one site, presumably Thr165, is underphosphorylated in the pho85Δ mutant. We further tested phosphorylation of the 62–202 fragment under conditions that stabilize Gcn4. As shown in lanes 6 and 7, cycloheximide treatment or amino acid starvation also resulted in the disappearance of the slower-migrating band, suggesting loss of phosphorylation at Thr165 under these conditions.
Figure 9.
In vivo phosphorylation of a fragment of Gcn4 encompassing residues 62–202 fused to a triple Myc epitope and expressed from the CUP1 promoter. The upper phosphorylation band (indicated by an arrow) is 30–40% as intense as the lower band in lanes 1 and 5 and is undetectable in the other lanes, even upon prolonged exposure (we estimate the limit of detection at <2% of the main band). Lane 4 is a no-tag control. CHX, cycloheximide treatment (1 μg/ml, 30 min); a.a. starv, 30-min incubation in YNB medium lacking all amino acids.
Pho85 Phosphorylates Gcn4 in Vitro
The data shown above indicate that Gcn4 phosphorylation on Thr165 depends on Pho85 activity in vivo. However, the possibility still existed that this is an indirect effect and that another kinase is directly responsible for Gcn4 phosphorylation. To show that Pho85 is able to directly phosphorylate Gcn4, we first attempted to reconstitute the phosphorylation reaction in vitro with pure recombinant proteins. Pho85, like all CDKs, requires a regulatory cyclin subunit for its activity. Pcl1 is one of the 10 different Pho85 cyclins (Pcls) identified to date (Andrews and Measday, 1998). We used recombinant GST-Pho85 together with GST-Pcl1 (Nishizawa et al., 1998) to phosphorylate either full-length heptahistidine-tagged Gcn4 or the same 62–202 fragment of Gcn4 used for the in vivo phosphate labeling, fused to GST. As shown in Figure 10, efficient phosphorylation of full-length Gcn4 depended both on an active Pho85 kinase and on Pcl1. Furthermore, phosphorylation of the wild-type 62–202 Gcn4 fragment fused to GST yielded two bands, but with the mutant T165A construct only the lower band was evident, indicating that Pho85 is able to phosphorylate Thr165 directly.
Figure 10.
In vitro phosphorylation of Gcn4 by Pho85-Pcl1. Different substrates (indicated at the bottom) were incubated with wild-type or catalytically inactive (E53A) GST-Pho85, with or without GST-Pcl1. Asterisk indicates background phosphorylation on bacterial proteins copurifying with the recombinant kinase and cyclin. The two arrowheads indicate the two phosphorylation bands of GST-Gcn462–202. The two weak signals (<10% intensity) visible over the main band in the T165A mutant presumably represent phosphorylation at alternative sites. The right panel shows that GST alone is not phosphorylated by Pho85-Pcl1 (the GST protein band would be expected to migrate slightly faster than the 30-kDa marker).
Finally, we asked whether cellular Pho85 and Pcl1 activity toward Gcn4 was dependent on physiological conditions favoring Gcn4 degradation. ha-tagged Pcl1 (Nishizawa et al., 1998) was immunoprecipitated from untreated exponentially growing cells, or from amino acid–starved or cycloheximide-treated cells. As shown in Figure 11, kinase activity toward both the full-length Gcn4 protein and the 62–202 fragment, was strongly reduced in the starved or cycloheximide-treated cells. Thus, Pcl1-associated Gcn4 kinase activity correlated with the conditions under which Gcn4 is rapidly degraded.
Figure 11.
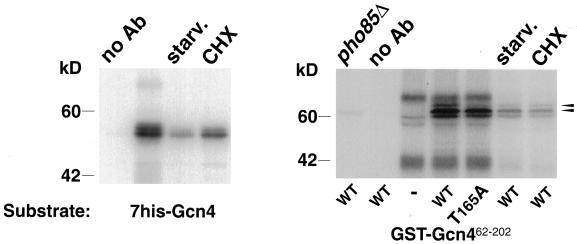
In vitro phosphorylation of Gcn4 by ha-Pcl1–associated kinase activity. Different substrates (indicated at the bottom) were incubated with Protein A-agarose beads carrying Pcl1-associated kinase activity immunoprecipitated from various cell extracts. pho85Δ indicates that the extract was made from a pho85 mutant strain; all other extracts were from wild-type cells. starv. and CHX refer to the same treatments as in Figure 9. no Ab refers to a blank immunoprecipitation in the absence of antibody. The two arrowheads at the right indicate the two phosphorylation bands of the GST-Gcn462–202 substrate.
DISCUSSION
The findings presented here begin to unravel the mechanism by which Gcn4 degradation is regulated: a) inhibition of protein synthesis, even in the absence of starvation, is sufficient to stabilize Gcn4; b) the SCFCDC4 ubiquitination complex is required for Gcn4 degradation, but its activity is not affected by starvation; c) Gcn4 degradation requires the activity of the cyclin-dependent kinase Pho85; d) a specific domain of Gcn4 is required for its starvation-sensitive degradation; e) a specific site within that region, Thr165, is required for Gcn4 degradation; f) phosphorylation of this site depends on Pho85 activity and correlates with the conditions that allow rapid degradation of Gcn4; g) Pho85 is able to phosphorylate Thr165; and h) Pho85 activity toward Gcn4 is reduced in starved cells. The simple model emerging from these results is that Gcn4 phosphorylation by Pho85 on Thr165 transforms it into a substrate of the SCFCDC4 ubiquitination complex (Figure 12). The differential degradation of Gcn4 versus other SCFCDC4 substrates is explained by the different kinases required to transform the proteins into effective substrates of the ubiquitination complex. The regulation of Pho85 activity by starvation explains the stabilization of Gcn4 under these conditions.
Figure 12.
A model of the proposed pathway of regulation of Gcn4 degradation by starvation. See text for details.
Recognition sites of the SCF complexes are ill defined, beyond the general requirement for phosphorylation. Deletion analysis allowed to define two separate domains, I and II, within the region of Gcn4 required for its rapid degradation (Figure 7). We previously described a number of mutations in and around position 105 that stabilize Gcn4 (Kornitzer et al., 1994). Thr105 is located in domain I. Thr165, which we characterize in the present study, is located in domain II. It is possible that these two residues constitute each the core of partially redundant SCFCDC4 recognition sites. Phosphorylation of the 62–202 fragment of Gcn4 (Figure 9), which carries both sites, suggests that in vivo, Pho85 activity is required for Thr165 phosphorylation but that phosphorylation of (an)other site(s) in this fragment, possibly Thr105, does not require Pho85. Conversely, our in vitro phosphorylation data (Figures 10 and 11) indicate that this same fragment can be phosphorylated by Pho85 at Thr165 as well as at another site, possibly Thr105. Thus, Pho85 activity is necessary and sufficient only for phosphorylation of Thr165.
Interestingly, contrary to Nishizawa et al. (1998), who found that recombinant Pho85-Pcl1 will phosphorylate Sic1 only after being “activated” with a yeast cell extract, we found that phosphorylation of Gcn4 does not require such an activation. Our results are in agreement with the recent findings of Wilson et al. (1999), that Gsy2 is efficiently phosphorylated in vitro by recombinant Pho85-Pcl10. Thus, it is possible that in the case of Sic1 phosphorylation, the cell extract confers a specificity factor required for Sic1 recognition, rather than an activation factor.
The Thr165 mutation was not isolated in our previous screen, probably because that screen depended on the enhanced transcriptional activity of Gcn4—the Thr105 mutation yielded a 15-fold increase in transcriptional activity (Kornitzer et al., 1994), whereas the Thr165 mutation only increased transcription 5-fold (Figure 8). Interestingly, Thr105 lies within the region defined by deletion analysis as the Gcn4 activation domain (Hope and Struhl, 1986; Hope et al., 1988). More recent mutational mapping showed that the activation domain can be subdivided into two redundant subdomains, the N-terminal and central acidic activation domains (Drysdale et al., 1995; Jackson et al., 1996). The Thr105 region lies between these two subdomains. Thus, it is possible that the mutations that were isolated in that region, in addition to stabilizing the protein, increase its specific transcriptional activity by modification of the activation region. In line with the recent findings of Komeili and O'Shea (1999), showing that phosphorylation of the transcription factor Pho4 by Pho85 affects its transcriptional activity, it is conceivable that the role of phosphorylation at Thr105 is to modulate the transcriptional activity of Gcn4 more than its stability.
The main difference between the requirements of Gcn4 degradation compared with that of the cell cycle substrates of SCF is the specific kinase involved. Gcn4 degradation requires Pho85 activity, whereas the other known SCF substrates require Cdc28 activity. The known functions of Pho85 in association with specific cyclins relate to metabolic regulation, e.g., phosphate assimilation (cyclin: Pho80; Kaffman et al., 1994), or glycogen synthesis (cyclins: Pcl8 and -10; Huang et al., 1998). In this respect, a function in Gcn4 degradation and therefore, indirectly, in amino acid metabolism, fits this pattern. The fact that the degradation of the cell cycle substrates of the SCFCDC4 complex depends on the phase of the cell cycle, whereas Gcn4 degradation is constitutive during the cell cycle, can be accounted for by difference in the kinase involved. Indeed, the nine Cdc28-associated cyclins, which confer its substrate specificity, are each present only during a limited window of each cell cycle. In contrast, most, but not all, of the 10 Pho85 cyclins are present throughout the cell cycle (Andrews and Measday, 1998). Strikingly however, Pcl1 is one of the Pho85 cyclins that is strongly cell cycle regulated, at least at the level of transcription (Measday et al., 1997). This apparent contradiction would be resolved if different Pcls were able to promote Gcn4 phosphorylation. Two observations indicate that this is in fact the case: a) if Pcl1 were the only Pcl able to promote Gcn4 phosphorylation, then Gcn4 should be stabilized in a pcl1Δ mutant. However, a number of single and multiple combinations of pcl mutants that we tested, including pcl1Δ, are unaffected in Gcn4 degradation (our unpublished results); and b) preliminary data indicate that at least one other Pcl, Pho80, is able to promote Gcn4 phosphorylation on Thr165 by Pho85 in vitro (our unpublished results). These two findings indicate that there may be a high redundancy in the Pcls able to activate Pho85 for Gcn4 phosphorylation.
How is Gcn4 phosphorylation by Pho85 regulated by starvation? Starvation could induce an inhibitor of Pho85 activity. Previous studies demonstrated that inhibition of an amino acyl-tRNA–charging enzyme also leads to Gcn4 stabilization (Kornitzer et al., 1994), suggesting that uncharged tRNA constitutes the signal for Gcn4 stabilization. An elevated concentration of uncharged tRNA is also thought to constitute the primary signal for the translational control of Gcn4 (Wek et al., 1995). In the current study, we found that inhibition of translation by cycloheximide is sufficient to stabilize Gcn4. Under these conditions, charged rather than uncharged tRNA accumulates in the cell. It should be noted that this does not contradict the earlier results obtained with the aminoacyl-tRNA–charging enzyme mutant; indeed, inhibition of one of the 20 aminoacyl tRNA synthetases, although resulting in an increase in the concentration of the uncharged form of its cognate tRNAs, would also, via inhibition of translation, increase the concentration of the charged form of all the other tRNAs. However, there is no direct evidence that Pho85 activity is regulated by the charged tRNA concentration or by any other signal generated by the stalled cellular biosynthetic machinery. An alternative possibility is that continued protein synthesis per se is required for maintaining Pho85 activity; therefore, starvation, or direct inhibition of protein synthesis, would result in reduced kinase activity.
Limitation of protein synthesis is known to arrest the cell cycle in G1 (Unger and Hartwell, 1976; Shilo et al., 1978; Pardee, 1989). The fact that the SCFCDC4 complex is required for the G1/S transition and for Gcn4 turnover raised the possibility that amino acid starvation coordinately inhibits cell cycle progression and Gcn4 degradation via inhibition of the SCFCDC4 complex (Kornitzer et al., 1994). However, our data do not support this hypothesis. Cdc6 and Sic1, two other substrates of the SCFCDC4 complex, are not stabilized upon starvation. Rather, our data indicate that Gcn4 degradation is regulated via modulation of Pho85 activity. Might modulation of Pho85 activity coordinately regulate Gcn4 turnover and cell cycle progression? Recent reports indicate a role for the G1 cyclin Cln3 in the cell cycle arrest in face of reduced protein synthesis (Polymenis and Schmidt, 1997; Danaie et al., 1999). However, it cannot be excluded that modulation of Pho85 activity also participates in this regulation. Pho85, although not essential for cell cycle progression, displays cell cycle–related phenotypes. For example, a deletion of the Cdc28 cyclins CLN1 and CLN2 is synthetically lethal with a deletion of the Pho85 cyclins PCL1 and PCL2 or of PHO85 itself (Espinoza et al., 1994; Measday et al., 1994). Thus, it is possible that Pho85 is one of the transducers of physiological signals, such as amino acid starvation, that need to be integrated by the cell cycle machinery.
ACKNOWLEDGMENTS
We thank Masafumi Nishizawa, Mike Tyers, Steve Elledge, Wade Harper, Steve Buratowski, Angelika Amon, Phil Hieter, Brenda Andrews, Helen Causton, Gerard Faye, and Curt Wittenberg for strains and plasmids; Mike Tyers for the Sic1 antibody; and Aaron Ciechanover and Sara Selig for critical reading of the manuscript. This work was supported by grants from the Israel Science Foundation and the Israel Cancer Research Fund to D.K., National Institutes of Health (NIH) grants GM35010 to G.R.F. and GM48624 and GM39067 to D.J.S., and NIH training grant GM07464 to H.J.M.
REFERENCES
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- Arndt KT, Styles CA, Fink GR. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1989. [Google Scholar]
- Bai C, Sen P, Hoffman K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt TR, Sternberg EJ, Gorman JA, Clark P, Hamer D, Rosenberg M, Crooke ST. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc Natl Acad Sci USA. 1984;81:3332–3336. doi: 10.1073/pnas.81.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B, Fink GR. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaie P, Altmann M, Hall MN, Trachsel H, Helliwell SB. CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E. Biochem J. 1999;340:135–141. [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale CM, Duenas E, Jackson BM, Reusser U, Braus GH, Hinnebusch AG. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of cdc6p destruction: a biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza FH, Ogas J, Herskowitz I, Morgan DO. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies RJ, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin- dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Fink GR. Positive regulation in the general control of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M, Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990;61:697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- Hope IA, Mahadevan S, Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Hope IA, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hope IA, Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985;43:177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Huang D, Moffat J, Wilson WA, Moore L, Cheng C, Roach PJ, Andrews B. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BM, Drysdale CM, Natarajan K, Hinnebusch AG. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M, Gulli MP, Peter K, Peter M. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 1998;17:5360–5373. doi: 10.1093/emboj/17.18.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Herskowitz I, Tjian R, O'Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Sia RA, Bardes EG, Lew DJ, Reed SI. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kolodziej PA, Young RA. Epitope tagging and protein surveillance. In: Guthrie C, Fink G R, editors. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. pp. 508–520. [DOI] [PubMed] [Google Scholar]
- Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- Kornitzer D, Raboy B, Kulka RG, Fink GR. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanker S, Valdivieso MH, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, van Vuuren HJ, Young RA. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- Mathias N, Johnson SL, Winey M, Adams AE, Goetsch L, Pringle JR, Byers B, Goebl MG. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias N, Steussy CN, Goebl MG. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J Biol Chem. 1998;273:4040–4045. doi: 10.1074/jbc.273.7.4040. [DOI] [PubMed] [Google Scholar]
- Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman AM, Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin- dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa F, Fink GR. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa M, Kawasumi M, Fujino M, Toh-e A. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol Biol Cell. 1998;9:2393–2405. doi: 10.1091/mbc.9.9.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Patton E, Willems A, Sa D, Kuras L, Thomas D, Craig K, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998a;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998b;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Calzada A, Bueno A. The Cdc6 protein is ubiquitinated in vivo for proteolysis in Saccharomyces cerevisiae. J Biol Chem. 1999;274:9092–9097. doi: 10.1074/jbc.274.13.9092. [DOI] [PubMed] [Google Scholar]
- Schneider B, Yang Q, Futcher A. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- Shilo B, Simchen G, Pardee AB. Regulation of cell-cycle initiation in yeast by nutrients and protein synthesis. J Cell Physiol. 1978;97:177–187. doi: 10.1002/jcp.1040970207. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger MV, Hartwell LH. Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc Natl Acad Sci USA. 1976;73:1664–1668. doi: 10.1073/pnas.73.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valay JG, Simon M, Faye G. The kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J Mol Biol. 1993;234:307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]
- Verma R, Annan R, Huddleston M, Carr S, Reynard G, Deshaies R. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Wilson WA, Mahrenholz AM, Roach PJ. Substrate targeting of the yeast cyclin-dependent kinase Pho85p by the cyclin Pcl10p. Mol Cell Biol. 1999;19:7020–7030. doi: 10.1128/mcb.19.10.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano H, Gannon J, Hunt T. The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J. 1996;15:5268–5279. [PMC free article] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Yochem J, Byers B. Structural comparison of the yeast cell division cycle gene CDC4 and a related pseudogene. J Mol Biol. 1987;195:233–245. doi: 10.1016/0022-2836(87)90646-2. [DOI] [PubMed] [Google Scholar]