Abstract
The cytoplasm-to-vacuole targeting (Cvt) pathway and macroautophagy are dynamic events involving the rearrangement of membrane to form a sequestering vesicle in the cytosol, which subsequently delivers its cargo to the vacuole. This process requires the concerted action of various proteins, including Apg5p. Recently, it was shown that another protein required for the import of aminopeptidase I (API) and autophagy, Apg12p, is covalently attached to Apg5p through the action of an E1-like enzyme, Apg7p. We have undertaken an analysis of Apg5p function to gain a better understanding of the role of this novel nonubiquitin conjugation reaction in these import pathways. We have generated the first temperature-sensitive mutant in the Cvt pathway, designated apg5ts. Biochemical analysis of API import in the apg5ts strain confirmed that Apg5p is directly required for the import of API via the Cvt pathway. By analyzing the stage of API import that is blocked in the apg5ts mutant, we have determined that Apg5p is involved in the sequestration step and is required for vesicle formation and/or completion.
INTRODUCTION
To survive in their natural environments, many organisms must endure significant periods of nutrient starvation. Accordingly, eukaryotic cells have evolved mechanisms for delivering intracellular proteins, membranes, and organelles to specialized recycling compartments. In yeast, protein degradation in the cytosol occurs through a proteasome-dependent process. Because of physical constraints, the proteasome is not suited for the recycling of larger structures, including membrane-bound organelles. In addition, cytosolic localization would be problematic for degradative machinery with the capacity to break down organelles. In contrast, the vacuole contains a wide variety of hydrolytic enzymes and provides the cell a membrane-bound degradative environment, which accounts for the majority of cellular protein turnover. Compartmentalization, however, necessitates a specialized mechanism for delivering substrates across the vacuole membrane and into the lumen.
Macroautophagy is the process that cells use to package and transport bulk cytoplasmic material to the vacuole. During starvation, cytosol is nonspecifically sequestered within a double-membrane vesicle termed an autophagosome (Baba et al., 1994). Subsequent fusion with the vacuole releases the inner vesicle into the lumen, where it is broken down, allowing access to the cargo. Genetic analyses have identified mutants, including apg and aut, blocked in macroautophagic protein uptake (Tsukada and Ohsumi, 1993; Thumm et al., 1994). Many of these mutants were shown to be allelic to cvt (cytoplasm-to-vacuole targeting) mutants that were isolated based on defects in the localization of aminopeptidase I (API), a resident vacuolar hydrolase (Harding et al., 1995, 1996; Scott et al., 1996). Biochemical and cytological studies have supported the overlap between macroautophagy and the Cvt pathway. As in macroautophagy, API import occurs through a vesicle-mediated process (Baba et al., 1997; Scott et al., 1997).
Most macroautophagy mutants are blocked in API import, and the majority of the cvt mutants are similarly defective in macroautophagic protein degradation (Harding et al., 1996; Scott et al., 1996). These observations suggest that the pathways share a common set of machinery. Despite the overlap between these pathways, there are significant physiological and morphological differences. API import is a biosynthetic process that is rapid, selective, and occurs constitutively. Precursor API (prAPI) oligomerizes into homododecamers, which subsequently assemble into higher-order cytosolic complexes (Kim et al., 1997). The Cvt vesicles that surround these prAPI complexes are 140–160 nm in diameter and appear to exclude bulk cytosol (Baba et al., 1997). In contrast, macroautophagy is a nonspecific degradative process with much slower kinetics. Uptake of bulk cytosol by macroautophagy plateaus at ∼30% and is induced by nutrient starvation (Noda et al., 1995; Scott et al., 1996). Autophagosomes are 300–900 nm in diameter and, befitting their role in protein turnover, contain bulk cytosol along with prAPI (Baba et al., 1997). Because of these fundamental differences between the two pathways, it is possible that the dual defects in macroautophagy and prAPI import observed in the apg and aut mutants are the result of indirect effects. All of the characterized apg, aut, and cvt strains are null mutants, making it impossible to determine which phenotypes are direct consequences of the mutations.
Recently, the APG5 gene was cloned (Kametaka et al., 1996), and Apg5p was shown to be involved in a unique protein-modification event involving at least three other proteins, which are also required for prAPI import and autophagy (Mizushima et al., 1998a). Apg7p, a member of the E1-like family of proteins, which includes the well-characterized ubiquitin-activating enzymes, forms a thioester linkage with Apg12p. The Apg12 protein subsequently forms a thioester intermediate with Apg10p, a protein that functionally corresponds to the E2 ubiquitin-conjugating enzyme (Shintani et al., 1999). Apg12p is finally conjugated at its C-terminal glycine to lysine 149 of Apg5p. To gain a better understanding of the molecular basis of protein transport into the vacuole from the cytoplasm, we have undertaken an analysis of the APG5 gene product. We have isolated a temperature-sensitive allele of APG5 and used it to determine the site of Apg5p function. prAPI is in a protease-sensitive, membrane-associated form in the apg5ts mutant, indicating that Apg5p is involved in the membrane-sequestration event, acting at the stage of Cvt vesicle/autophagosome formation and/or completion.
MATERIALS AND METHODS
Materials
Restriction endonucleases, T4 DNA ligase, and calf intestinal phosphatase were from New England Biolabs (Beverly, MA). Antiserum to API was as described previously (Klionsky et al., 1992). mAbs to the hemagglutinin epitope were from Covance Research Products (Richmond, CA), and mAbs to Dpm1p and alkaline phosphatase (ALP) were from Molecular Probes (Eugene, OR). Antiserum to Vma2p was prepared as described previously (Tomashek et al., 1996). Antisera to Pep12p and phosphoglycerate kinase (PGK) were generously provided by Dr. Scott Emr (University of California, San Diego) and Dr. Jeremy Thorner (University of California, Berkeley), respectively. EXPRE35S35S protein-labeling mix was from Dupont–New England Nuclear Research Products (Boston, MA). Complete EDTA-free protease inhibitor cocktail was from Roche Molecular Biochemicals (Indianapolis, IN). Vectors for engineering and expressing green fluorescent protein (GFP) fusions and hemagglutinin fusion proteins in yeast were a kind gift from Dr. Jodi Nunnari (University of California, Davis).
Strains and Media
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. The following media were used: SMD (synthetic minimal medium containing ammonium sulfate, essential amino acids and vitamins, and 2% glucose); SD(−N) (synthetic minimal medium without ammonium sulfate and amino acids but containing 2% glucose); and YPD (rich medium containing 1% yeast extract, 2% peptone, and 2% glucose).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| MGY101 | MATα apg5Δ∷LEU2 ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | This study |
| MT37-4-2 | MATα apg5-1 ura3-52 | Kametaka et al. (1996) |
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | Robinson et al. (1988) |
| TID7-1 | MATa ura3 leu2 trp1 apg7Δ∷LEU2 | This study |
| YNM101 | MATα ura3 leu2 trp1 his3 apg12Δ∷HIS3 | Mizushima et al. (1998a) |
| YNM124 | MATα ura3 leu2 trp1 his3 apg16Δ∷LEU2 | Mizushima et al. (1999) |
| KA311B | MATα ura3 leu2 trp1 his3 | Irie et al. (1993) |
| MT28-4-4 | MATa apg8-1 ura3-52 | Tsukada and Ohsumi (1993) |
| MT54-4-2 | MATa apg14-1 ura3-52 | Tsukada and Ohsumi (1993) |
| SSY101 | MATα ura3-52 trp1-Δ901 apg5Δ∷LEU2 pep40∷HIS3 | This study |
Construction of Hemagglutinin Epitope and GFP Fusions with APG5
The APG5 gene was amplified by PCR with the use of oligonucleotides that incorporated ApaI and NcoI restriction sites into the 5′ and 3′ ends of the gene, respectively (5′-CATGGGCCCAGCGTGAAGG-3′, 5′-GCTCCATGGAAGCTTTATCG-3′). Plasmid pSF19 contains the hemagglutinin (HA) epitope followed by a stop codon and actin termination sequence. The restriction enzyme–digested PCR product was cloned into pSF19 vector that had been restriction enzyme digested with ApaI and NcoI. The resulting plasmid, pCAPG5-HA, is a pRS314 vector in which the sequence encoding the HA epitope is fused in frame to the C terminus of APG5.
pCAPG5-GFP is a pRS414-based plasmid expressing modified GFP (S65T) from the jellyfish Aequorea victoria fused to the C terminus of APG5. Briefly, APG5 was PCR amplified with the use of oligonucleotides that incorporated BamHI sites into both the 5′ and 3′ ends (5′-TGGGGGGGATCCTACAGCG-3′, 5′-GAGCTCAGAGGATCCTTTATC-3′). The plasmid pRS305Mip1-GFP contains GFP followed by an actin termination sequence. The BamHI-digested PCR fragment was cloned into the BamHI site of pRS305Mip1p. The resulting plasmid, pRS305APG5-GFP, was digested with PstI and EagI to release the entire APG5-GFP cassette, which was then cloned into PstI–EagI-digested pRS414 and pRS424 to generate CEN and 2μ APG5-GFP plasmids, respectively.
Pulse-Chase Analysis
Yeast strains were grown to 1.0 OD600 unit/ml in SMD and pulse labeled for the indicated times (Figures 1 and 4). Labeled cells were then suspended at 1.0 OD600 unit/ml in SMD containing cold cysteine and methionine at concentrations of 20 and 10 μM, respectively, and chased for the indicated times and temperatures. Cells were precipitated immediately at the indicated time by the addition of trichloroacetic acid to 10% and washed twice with acetone. The dried pellet was resuspended in 100 μl of MES-urea resuspension buffer [50 mM sodium phosphate, 25 mM 2-(N-morpholino)ethanesulfonic acid, pH 7.0, 1% SDS, 0.5% 2-mercaptoethanol, 1 mM sodium azide] and lysed with glass-bead mixing for 4 min at full speed on a multi-mixer (model MT-360, Tomy, Palo Alto, CA). The resulting supernatants were then immunoprecipitated with the indicated antiserum, as described previously (Klionsky et al., 1992). Immunoprecipitated proteins were resolved by SDS-PAGE and detected and quantified with the use of a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA), as indicated. The data on the kinetics of API import are expressed as the ratio of mature API to total prAPI plus mature API.
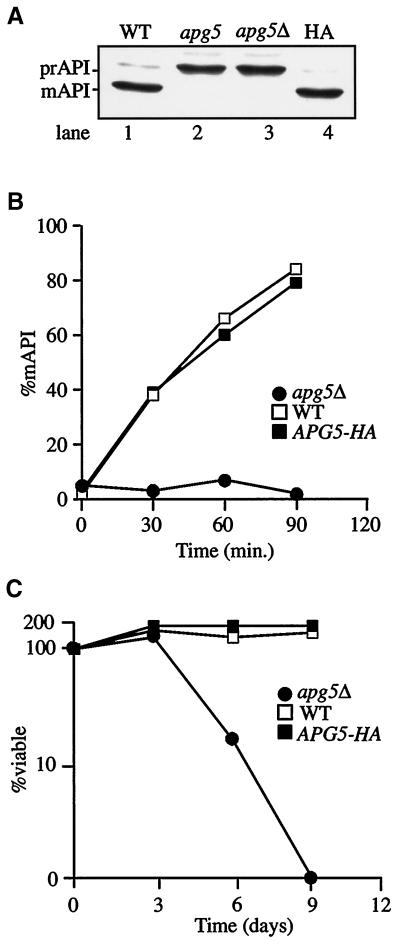
Figure 1.
Epitope-tagged Apg5p complements defects in prAPI import and macroautophagy in an apg5Δ strain. (A) API maturation in steady-state cultures. Cells from strains SEY6210 (WT, lane 1), apg5 (lane 2), apg5Δ (lane 3), and apg5Δ expressing Apg5HAp from a CEN plasmid (HA, lane 4) were grown to log phase, and protein extracts were prepared and immunoblotted with antiserum to API, as described in MATERIALS AND METHODS. mAPI, mature API. (B) Kinetic analysis of prAPI import. Cells from strains SEY6210 (WT), apg5Δ, and apg5Δ expressing Apg5HAp from a CEN plasmid (APG5-HA) were pulse labeled for 10 min at 30°C and subjected to a nonradioactive chase for the indicated times. API was recovered by immunoprecipitation and detected with a STORM PhosphorImager. The percentage of mature API (%mAPI) was calculated by dividing mature API by mature API plus prAPI at each time point. (C) Viability during nitrogen starvation. Cells from SEY6210 (WT), apg5Δ, and apg5Δ expressing Apg5HAp from a CEN plasmid (APG5-HA) were analyzed for sensitivity to starvation, as described in MATERIALS AND METHODS.
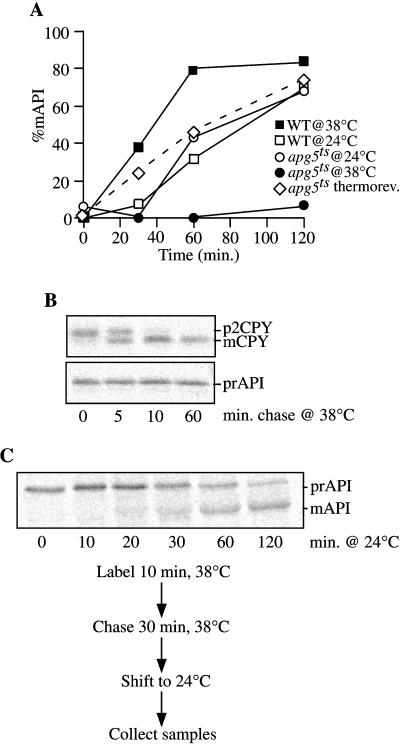
Figure 4.
The apg5ts mutant displays rapid inactivation kinetics for prAPI import. (A) Strains apg5Δ expressing apg5ts from a centromeric plasmid (apg5ts) and SEY6210 (WT) were preincubated for 5 min at 24 and 38°C, labeled for 10 min, and subjected to a nonradioactive chase. Aliquots were removed at the indicated times and precipitated with trichloroacetic acid. The samples were then immunoprecipitated with antiserum to API, as described in MATERIALS AND METHODS. After resolution of the immunoprecipitated samples by SDS-PAGE, the amounts of both prAPI and mature API were quantified with the use of a STORM PhosphorImager. The percentage of mature API (%mAPI) was calculated as mature API divided by prAPI plus mature API at each time point. The dotted line corresponds to apg5ts cells shifted from nonpermissive to permissive temperatures, as described in C below. (B) CPY sorting is unaffected at the nonpermissive temperature in the apg5ts mutant. apg5ts cells were preincubated and labeled as described for A at 38°C and subjected to nonradioactive chase for the indicated times. Samples were then immunoprecipitated with antiserum to API or CPY, resolved by SDS-PAGE, and visualized with the use of a STORM PhosphorImager. (C) The apg5ts mutant is thermally reversible. apg5ts cells were preincubated at 38°C for 5 min, labeled for 10 min, subjected to a 30-min nonradioactive chase to accumulate prAPI, and then shifted to 24°C for 120 min of additional chase. Aliquots were removed during the chase at the permissive temperature at the indicated times and immunoprecipitated with antiserum to API, and the proteins were resolved by SDS-PAGE. API was detected with a STORM PhosphorImager. Quantification of the import kinetics is presented in A for comparison (dotted line).
Nitrogen-starvation Analysis
Yeast strains were examined for their ability to survive when deprived of a nitrogen source. Cultures were grown to exponential phase in SMD, harvested by centrifugation, washed twice in SD(−N), and resuspended in SD(−N) at a final concentration of 1.0 OD600 unit/ml. The cultures were then incubated at 30°C, and samples were removed at d 0, 2, 5, and 8 and spread onto SMD or SMD(−Trp) plates. Replica samples of each culture were plated, and colonies were counted after 2 d of growth at 30°C. The viability of the cells in each sample was expressed as the percentage of colonies produced by the same volume of culture at d 0.
Generation and Analysis of a Temperature-sensitive apg5 Mutant
APG5 was mutagenized by the method of Muhlrad et al. (1992). Briefly, full-length APG5 was amplified under mutagenic PCR conditions. The mutagenized gene was co-transformed, along with a gapped plasmid containing homology to both ends of the PCR product, into a yeast strain harboring a deletion of the chromosomal copy of APG5 (MGY101). Cell lysates from steady-state cultures were initially screened by Western blot for the appearance of mature API when grown at room temperature and for an accumulation of prAPI when cultures were grown at 38°C. Mutants that passed the initial screening criterion were then analyzed by pulse-chase labeling, as described above, for the kinetics of prAPI delivery to the vacuole at permissive and nonpermissive temperatures. A mutant, apg5-43, which displayed a strong block in prAPI import at the nonpermissive temperature, was chosen for further analysis.
The reversibility of the thermal effects on Apg5p function was analyzed by preincubating the apg5ts strain at 38°C for 5 min. The culture was then pulse labeled for 10 min and chased for 30 min at 38°C. Immediately after the 38°C chase, the culture was transferred to a 24°C water bath. Samples were then removed at the indicated times (Figure 4) and immunoprecipitated as described above.
Subcellular Fractionations and Analysis of Apg5p Membrane Association
apg5Δ cells harboring pCAPG5-HA or pMAPG5-GFP were grown to 1.0 OD600 unit/ml, and 50 OD600 units of cells were collected by centrifugation. Spheroplasts were then generated in SMD and lysed osmotically, as described previously (Scott and Klionsky, 1995), by resuspension in physiological salts lysis buffer [20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, 100 mM potassium acetate, 50 mM potassium chloride, 5 mM magnesium chloride] containing 100 mM sorbitol and protease inhibitors. After centrifugation at 300 × g for 2 min, the supernatant (T, total lysate) was fractionated by 13,000 × g centrifugation for 10 min, generating a pellet (P13) and a supernatant (S13) fraction. The S13 fraction was further centrifuged at 100,000 × g for 30 min to generate a pellet (P100) and a supernatant (S100) fraction. Each fraction was subjected to SDS-PAGE and immunoblotting with the use of anti-HA, anti-PGK, anti-ALP, anti-Pep12p, or anti-GFP antibody/antiserum.
To biochemically characterize membrane association, spheroplasts were prepared and osmotically lysed as described above in PS200 buffer (20 mM PIPES, pH 6.8, 200 mM sorbitol) containing 5 mM MgCl2 and protease inhibitors. Lysed spheroplasts were centrifuged at 13,000 × g for 10 min and separated into supernatant and pellet fractions. Aliquots of the pellet fraction were subsequently resuspended in lysis buffer alone as a control and in lysis buffer containing 1 M KCl, 0.1 M Na2CO3, pH 10.5, 3 M urea, or 1% Triton X-100. The treated pellet fractions were incubated on ice for 10 min and then centrifuged at 13,000 × g for 10 min to separate the supernatant and pellet fractions. All samples were precipitated by adding trichloroacetic acid to a final concentration of 10% and analyzed by Western blotting.
Protease Sensitivity and API-binding Experiments
Spheroplasts were prepared, pulse labeled, and chased as described previously (Scott and Klionsky, 1995). At the indicated times (Figures 5 and 6), spheroplasts were recovered by centrifugation and lysed osmotically by gentle resuspension in PS200 with or without 5 mM MgCl2, as indicated. Supernatant and pellet fractions were collected by centrifugation at 5000 × g for 5 min. The percentage of bound prAPI was calculated by dividing the amount recovered in the pellet fraction by the sum of prAPI recovered in the supernatant and pellet fractions. Protease treatment was performed by resuspending 2-OD600-unit cell equivalents of the pellet fraction in 100 μl of PS200 containing 5 mM MgCl2 and digesting with 50 μg/ml proteinase K for 15 min on ice in the presence or absence of 0.2% Triton X-100. The percentage of protease-protected precursor was calculated by dividing the amount of prAPI or precursor carboxypeptidase Y (CPY) remaining after protease digestion by the total amount recovered in the pellet fraction.
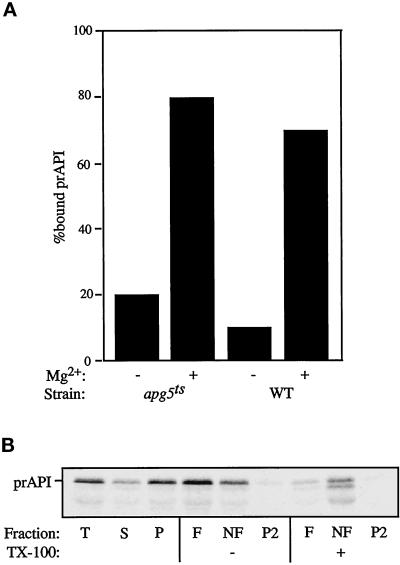
Figure 5.
(A) prAPI assembles into higher-order, pelletable Cvt complexes in apg5ts. Spheroplasts were shifted to 38°C for 5 min, pulse labeled for 5 min, and subjected to a cold chase for 20 min. The labeled spheroplasts were lysed by resuspension in PS200 with or without 5 mM MgCl2 and then separated into a soluble fraction and a membrane-containing pellet fraction, as described in MATERIALS AND METHODS. The percentage of prAPI recovered in the pellet fraction is represented. (B) prAPI is membrane associated in the apg5ts mutant. apg5ts spheroplasts were pulse labeled for 5 min, chased for 60 min, and subjected to fractionation into total (T), supernatant (S), and pellet (P) fractions, as described in MATERIALS AND METHODS. The P fraction was resuspended in 60% sucrose in the absence or presence of 1% Triton X-100 (TX-100) and overlaid with 55% sucrose followed by 35% sucrose. After centrifugation at 100,000 × g for 60 min, float (F), nonfloat (NF), and pellet (P2) fractions were collected and API was recovered by immunoprecipitation. Samples were analyzed by SDS-PAGE, and API was detected with a STORM PhosphorImager.
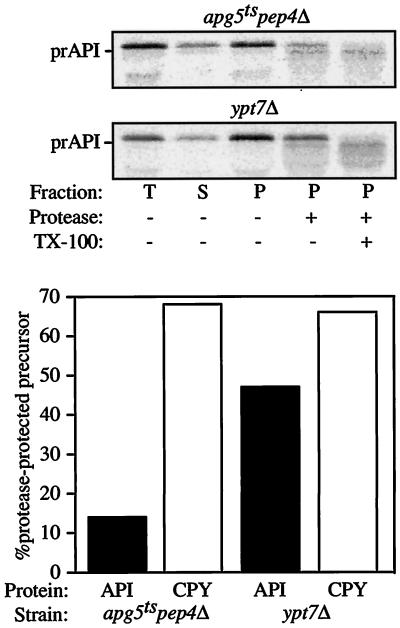
Figure 6.
prAPI is protease accessible in the apg5ts mutant. Spheroplasts isolated from apg5ts pep4Δ and ypt7Δ cells were shifted to 38°C for 5 min, labeled for 5 min, and then chased for 60 min. A portion of the sample was removed and immunoprecipitated as a total (T) control. After lysis in PS200 with 5 mM MgCl2 and centrifugation at 5000 × g, supernatant (S) and pellet (P) fractions were collected and subjected to protease treatment, as indicated. The resulting fractions were subsequently immunoprecipitated with antiserum against API, CPY, and hexokinase (cytosolic marker). Hexokinase was recovered 78 and 81% in the supernatant fraction in apg5ts pep4Δ and ypt7Δ cells, respectively, indicating efficient lysis of the spheroplasts. The graph represents the percentage of prAPI and p2CPY recovered in the pellet fraction that is protease resistant (lane 4 divided by lane 3). TX-100, Triton X-100.
Flotation Experiments
Membrane flotation experiments were performed by lysing spheroplasts in GB (gradient buffer; 20 mM PIPES, 5 mM MgCl2, 1× complete EDTA-free protease inhibitor cocktail). One-percent Triton X-100 was added to the lysis buffer as indicated. Five OD600 equivalents of lysate were then mixed with an equal volume (100 μl) of GB containing 30% Ficoll. One milliliter of 13% Ficoll in GB and 200 μl of 2% Ficoll in GB were then overlaid, and the resulting step gradient was subjected to centrifugation at 13,000 × g for 10 min at room temperature. Fractions were collected from the top. The first 300 μl is the float fraction (F), the remaining 1.1 ml is the nonfloat fraction (NF), and the gradient pellet is the pellet fraction (P2). The resulting fractions were subjected to either immunoprecipitation or immunoblotting, as indicated.
To examine prAPI flotation in the apg5ts strain, spheroplasts were pulse labeled for 5 min and subjected to a nonradioactive chase for 60 min. A portion of the sample was removed as a total (T) control. The spheroplasts were lysed in PS200 buffer containing 5 mM MgCl2 and centrifuged at 5000 × g to generate supernatant (S) and pellet (P) fractions. The P fraction was resuspended in 60% sucrose in the absence or presence of 1% Triton X-100 and overlaid with 55% sucrose followed by 35% sucrose. After centrifugation at 100,000 × g for 60 min, float (F), nonfloat (NF), and pellet (P2) fractions were collected as described above and prAPI was recovered by immunoprecipitation, resolved by SDS-PAGE, and visualized with a STORM PhosphorImager.
Electron Microscopy
For the apg5ts mutant, cells were grown in YPD at 30°C and shifted to 38°C for 3 h before sample preparation. The apg5Δ strain was grown in YPD and shifted to SD(−N) for 3 h at 30°C before processing. Yeast cells were prepared for electron microscopy by the rapid freezing and freeze-substitution fixation method (Baba et al., 1997). Sections were stained with 4% uranyl acetate for 10 min and 0.4% lead citrate for 1 min. The preparation of samples for immunoelectron microscopy was done according to the procedures described previously (Baba et al., 1997) with slight modifications. In brief, cells attached to an aluminum disk were transferred to 0.01% osmium tetroxide in cold absolute acetone kept below −80°C. Substitution fixation was carried out at −80°C for 2 d. The samples were then transferred to −25°C and kept for 5 h, washed three times with cold absolute acetone, and then replaced stepwise to cold absolute ethanol at −25°C. The samples were infiltrated with LR White resin (London Resin, Hampshire, UK), kept at −25°C overnight, and then warmed gradually (at 4°C for 2 h, then at room temperature for 4 h). API was immunolabeled with affinity-purified API antibody (1:1000 dilution) for 90 min at room temperature. The sections were stained with 4% uranyl acetate for 10 min and with 0.02% lead citrate for 30 s.
Fluorescence Microscopy
Samples were examined on a Leica (Deerfield, IL) DM IRB confocal microscope to visualize Apg5pGFP. Cultures subjected to nitrogen starvation were washed twice, resuspended in SD(−N) medium, and incubated for 2 h at 30°C before visualization.
RESULTS
HA-tagged Apg5p Retains Normal Function
The apg5 mutant was identified based on defects in macroautophagic protein uptake under starvation conditions (Tsukada and Ohsumi, 1993). Analysis of the APG5 gene indicated that it encodes a hydrophilic protein of 34 kDa (Kametaka et al., 1996). Recently, it was demonstrated that Apg5p is involved in a novel conjugation reaction (Mizushima et al., 1998a). Another protein required for macroautophagy, Apg12p, is covalently linked by a C-terminal glycine to an internal lysine residue of Apg5p. This conjugation reaction is required for both macroautophagy and import of prAPI through the Cvt pathway. To gain further insight into the function of Apg5p, we undertook a comprehensive characterization of this protein.
To identify Apg5p by immunodetection in cell lysates, the protein was tagged at its C terminus with a nine-amino acid HA epitope. To verify that the addition of the HA epitope to the C terminus of Apg5p did not adversely affect its function, the ability of the tagged protein to rescue apg5Δ-specific defects was analyzed. Under steady-state conditions in wild-type cells, most API was processed to its mature size (Figure 1A, lane 1), corresponding with its localization to the vacuole. In contrast, in an apg5 mutant or deletion strain, most API was in the higher-molecular-mass precursor form (prAPI), indicating a block in vacuolar delivery (Figure 1A, lanes 2 and 3). When Apg5p was expressed as a tagged protein (Apg5HAp) from a centromeric vector in an apg5 deletion strain, prAPI import was restored to wild-type levels (Figure 1A, lane 4). Similarly, the kinetics of prAPI delivery were essentially the same in both the wild-type strain and the apg5Δ strain expressing Apg5HAp (Figure 1B), confirming that the tagged protein was functioning properly within the Cvt pathway.
The ability of Apg5HAp to rescue the starvation phenotype of an apg5Δ strain was also analyzed. Cells from the wild-type and apg5Δ strains and apg5Δ expressing Apg5HAp from a centromeric plasmid were examined for starvation sensitivity, as described in MATERIALS AND METHODS. As shown in Figure 1C, the apg5Δ strain expressing Apg5HAp maintained viability at the level of the wild-type strain throughout the course of the experiment, whereas the viability of the apg5Δ strain was reduced by almost 90% after 5 d and was negligible after 8 d of nitrogen starvation. These results indicated that HA-tagged Apg5p maintained normal function within the autophagy pathway.
Apg5HAp and the Apg12p-Apg5HAp Conjugate Are Membrane Associated
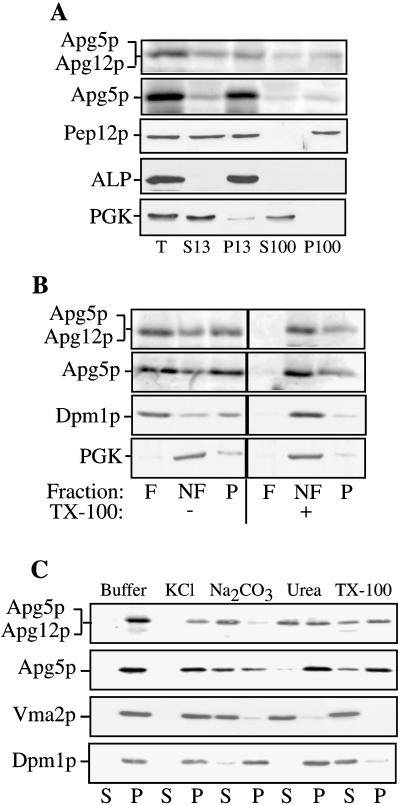
The predicted amino acid sequence of Apg5p suggests that it is a hydrophilic protein (Kametaka et al., 1996); however, both free Apg5p and the Apg12p-Apg5p conjugate were found primarily in a pelletable fraction (Mizushima et al., 1998a). We further examined the nature of the pelletable Apg5p through subcellular fractionation. The apg5Δ strain, expressing Apg5HAp, was grown to log phase, and spheroplasts were generated and lysed in a physiological salts buffer, as described in MATERIALS AND METHODS. The lysates were then centrifuged at 13,000 × g to generate supernatant (S13) and pellet (P13) fractions. The resulting supernatant fraction was centrifuged again at 100,000 × g to generate supernatant (S100) and pellet (P100) fractions. Under these lysis conditions, we consistently found that the pelletable pools of Apg5HAp and Apg12p-Apg5HAp conjugate were distributed between the P13 and P100 fractions (Figure 2). Quantifying the amount of Apg5HAp in each fraction over a set of four experiments indicated that an average of 87% of Apg5HAp and the Apg12p-Apg5HAp conjugate were pelletable. To confirm that our fractionation procedures faithfully separated soluble from pelletable proteins, we examined the distribution of the cytosolic marker PGK and the membrane-associated proteins Pep12p and ALP. As expected, PGK was recovered primarily in the supernatant fraction, whereas the vacuolar protein ALP was recovered exclusively in the P13 fraction. The endosomal marker Pep12p was distributed between the P13 and P100 fractions, as reported previously (Becherer et al., 1996).
Figure 2.
Subcellular fractionation and characterization of membrane association. (A) Apg5HAp is pelletable. Spheroplasts from the apg5Δ strain expressing Apg5HAp were lysed osmotically, as described in MATERIALS AND METHODS. After centrifugation at 500 × g for 5 min to remove debris and unlysed spheroplasts, the supernatant (T, total lysate) was fractionated by 13,000 × g centrifugation for 10 min, generating a pellet (P13) and a supernatant (S13) fraction. The supernatant fraction was further centrifuged at 100,000 × g for 30 min to generate a pellet (P100) and a supernatant (S100) fraction. Each fraction was subjected to SDS-PAGE and immunoblotting with the use of antibodies to the HA epitope (Apg5p), Pep12p (endosomal marker; P13 and P100), ALP (vacuolar marker; P13), or PGK (cytosolic marker; S13 or S100). (B) Apg5p is membrane associated. Spheroplasts expressing Apg5HAp were subjected to lysis with PS200 buffer containing 5 mM MgCl2. The lysate was loaded on the bottom of a Ficoll step gradient, as described in MATERIALS AND METHODS, in the absence or presence of 1% Triton X-100 (TX-100). The resulting gradients were centrifuged at 13,000 × g for 10 min. Membrane-containing float (F), nonfloat (NF), and pellet (P) fractions were collected and subjected to immunoblotting with antibodies to HA or Dpm1p (membrane protein marker) or antiserum to PGK (soluble protein marker), as indicated. A portion of Apg5HAp and the Apg12p-Apg5HAp conjugate are found in the membrane-associated float fraction. (C) Biochemicalcharacterization of pelletable Apg5p. Spheroplasts expressing Apg5HAp were lysed in PS200 buffer containing 5 mM MgCl2, as described in MATERIALS AND METHODS. The pellet fractions were resuspended in buffer alone or buffer containing 1 M KCl, 0.1 M Na2CO3, pH 10.5, 3 M urea, or 1% Triton X-100 (TX-100) and separated into supernatant (S) and pellet (P) fractions, as described in MATERIALS AND METHODS. Western blotting was performed with antibodies to HA or Dpm1p (integral membrane marker) or with antiserum to Vma2p (peripheral membrane marker).
It has recently been demonstrated that Apg5p interacts with the Apg16 protein (Mizushima et al., 1999). The presence of Apg5p in the pellet fraction could result from direct or indirect interaction with a pelletable membrane or through interaction with a pelletable protein complex. To differentiate between these possibilities, we performed a flotation experiment. Spheroplasts expressing Apg5HAp were lysed osmotically, and the resulting lysates were subjected to flotation through a Ficoll step gradient in the absence or presence of Triton X-100, as described in MATERIALS AND METHODS. In the absence of detergent, Apg5HAp was recovered in the float (F), nonfloat (NF), and pellet (P2) fractions (Figure 2A). In the presence of detergent, however, it was recovered exclusively in the nonfloat and pellet fractions. The Apg12p-Apg5HAp conjugate showed a similar distribution in the flotation analysis. These results suggest that a portion of Apg5HAp and the Apg12p-Apg5HAp conjugate are membrane associated. As controls for the flotation analysis, we examined the profile generated by the endoplasmic reticulum (ER) resident membrane protein Dpm1p (dolichol phosphate mannose synthase), as well as that of the cytosolic protein PGK. As expected, most of the Dpm1p was recovered in the float fraction in the absence of detergent, whereas PGK was recovered primarily in the nonfloat fraction independent of detergent treatment. These results confirm that only lipid- or membrane-associated proteins migrate in the float fraction in this assay. In addition to being membrane associated, populations of Apg5HAp and the Apg12p-Apg5HAp conjugate were present in a detergent-resistant pelletable complex. When membranes were solubilized with Triton X-100 before flotation, a portion of Apg5HAp and the conjugate were still recovered in a pellet fraction (Figure 2B).
We extended our examination of the membrane-associated Apg5p population through a biochemical analysis. Spheroplasts were lysed as described in MATERIALS AND METHODS in PIPES buffer containing 5 mM MgCl2, and the supernatant and pellet fractions were separated by centrifugation. The pelletable pool of Apg5HAp was then treated with various reagents. A concentration of 1 M KCl or 3 M urea had essentially no effect on Apg5HAp, but ∼50% of the conjugate was removed from the membrane after treatment with urea (Figure 2C). Alkaline conditions stripped essentially all of the conjugate and >50% of the Apg5HAp from the membrane. As we observed in our flotation analysis, a significant portion of Apg5HAp was not solubilized by treatment with Triton X-100. To demonstrate that this result was specific to Apg5HAp, we also analyzed the peripheral membrane protein Vma2p (a component of the V1 sector of the vacuolar ATPase) and the integral ER membrane protein Dpm1p. As expected, detergent efficiently solubilized Dpm1p, whereas alkaline extraction and urea additionally removed Vma2p.
Apg5pGFP Accumulates in Punctate Structures in Specific apg/cvt Mutants
The data presented above indicated that a significant pool of Apg5p was membrane associated. Subcellular fractionation by sucrose density gradient in previous studies demonstrated that Apg5p and Apg12p-Apg5p did not co-migrate with vacuole, Golgi complex, or ER marker proteins (Mizushima et al., 1998a). To gain further insight into the location of Apg5p function, we examined its subcellular location in vivo. The jellyfish GFP was fused to the C terminus of Apg5p, and the fusion protein was observed by fluorescence microscopy. At both centromeric and 2μ expression levels, the Apg5pGFP construct was capable of complementing the prAPI import defect in the apg5Δ strain (data not shown), indicating that the fusion protein maintained normal function. Furthermore, when lysates from cells expressing Apg5pGFP at the 2μ level were subjected to fractionation by differential centrifugation, the distribution of both Apg5pGFP and the Apg12p-Apgp5GFP conjugate between the P13, P100, and S100 fractions was essentially the same as we observed in our fractionation studies of Apg5HAp (Figure 2 and data not shown).
Apg5pGFP expressed from a multicopy plasmid in wild-type cells did not appear to accumulate significantly at any distinct subcellular location (Figure 3). Instead, we observed a random staining pattern throughout the cytosol. In 3–5% of the cells, larger punctate structures could be observed (data not shown). Although the significance of this staining pattern is not yet known, the observations were in agreement with our biochemical data, which indicated that a population of Apg5p is pelletable and membrane associated under physiological conditions.
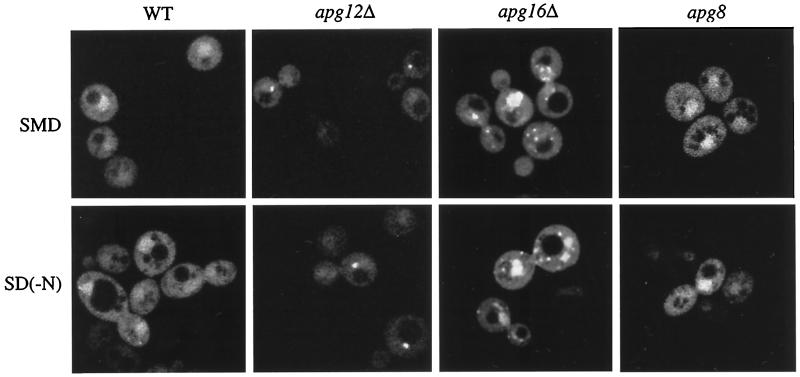
Figure 3.
Apg5pGFP is localized to punctate staining structures. Cells from the apg5Δ strain (WT) and from the mutant strains apg8, apg12Δ, and apg16Δ expressing Apg5pGFP from a 2μ plasmid were grown in SMD or shifted to SD(−N), as described in MATERIALS AND METHODS, and observed on a Leica DM IRB confocal microscope with the use of 510- to 525-nm filter settings. The fluorescent punctate pattern of Apg5pGFP in apg12Δ was essentially the same as that observed for apg7Δ.
We next examined the localization of Apg5pGFP in different apg/cvt mutant strains to determine if blocks in the import pathway affected localization. In addition to the staining pattern observed in the wild-type strain, Apg5pGFP displayed large punctate staining patterns in several of the mutant strains, including apg7Δ, apg12Δ, and apg16Δ (Figure 3). There were typically one to five punctate structures per cell, and these could be detected in at least 50% of the fluorescing cells. The punctate staining observed in these mutants was not due to a general defect in the autophagy pathway, because this pattern was not detected in other mutant strains, including apg8, which is blocked in both the Cvt pathway and autophagy (Figure 3 and data not shown).
Because Apg5p is also required for macroautophagy, we wanted to determine if the localization pattern of Apg5pGFP changed under starvation conditions. The indicated strains expressing Apg5pGFP were grown to log phase and then shifted to medium lacking a nitrogen source for 2 h, as described in MATERIALS AND METHODS. The Apg5pGFP staining patterns observed during growth in rich medium in each of the indicated strains did not change during nitrogen starvation (Figure 3). In all of the strains examined, essentially identical results were obtained with a centromeric Apg5pGFP construct when a clear signal could be detected (data not shown). Detection of the fluorescent signal at the level produced by the centromeric construct, however, was problematic.
Apg5p Has a Direct Role in the Cvt Pathway
The biochemical analyses of Apg5p in this study indicated that a significant fraction of the protein is membrane associated. The in vivo studies with Apg5pGFP revealed a punctate distribution that may correspond to association with vesicular structures that have been shown to play a role in macroautophagy and the Cvt pathway (Baba et al., 1997; Scott et al., 1997). To precisely determine the site of action of Apg5p in these pathways, we decided to carefully examine the state of API in an apg5 mutant. To determine the immediate consequence of a loss of Apg5p function, we generated a conditional mutant. The original apg5 mutant was isolated based on its inability to survive starvation conditions (Tsukada and Ohsumi, 1993). With null mutants, however, there is a possibility that observed defects are indirect and result from a chronic loss of function. Accordingly, we used a molecular genetic approach to first determine if a loss of Apg5p function had a direct effect on the Cvt pathway. Using a PCR-based strategy (Muhlrad et al., 1992), we introduced random mutations into APG5, as described in MATERIALS AND METHODS. Examining prAPI maturation at permissive and restrictive temperatures identified a temperature-sensitive apg5 strain (apg5ts).
At the permissive temperature, the wild-type and apg5ts strains exhibited essentially identical kinetics for import of prAPI (Figure 4A). In contrast, at the nonpermissive temperature, there was a rapid and virtually complete block in processing of prAPI in the apg5ts strain. The import of CPY, a vacuolar hydrolase delivered through a portion of the secretory pathway, showed normal kinetics at 38°C, indicating that the defect in prAPI import was not a pleiotropic effect of incubating the culture at an elevated temperature (Figure 4B). The rapid block in prAPI import seen upon shifting to the nonpermissive temperature suggested that Apg5p functions directly within the Cvt pathway.
To provide further evidence that Apg5p has a direct role in the import of prAPI, the ability to reverse the thermal effects in the apg5ts strain was examined. Radiolabeled prAPI was accumulated for 30 min at the nonpermissive temperature, followed by a chase under permissive conditions. A time-course analysis revealed maturation of prAPI with essentially the same kinetics as seen when import was carried out exclusively at a permissive temperature (Figure 4, A and C). These data suggested that the block in import at 38°C was a direct effect of the conditional mutation in Apg5p. Moreover, the relatively rapid import of accumulated prAPI after switching from nonpermissive to permissive temperatures is a strong indication that the precursor protein was blocked in the authentic import pathway and not diverted into a nonphysiological dead-end route.
Apg5p Functions in Vesicle Formation
In light of the evidence that Apg5p has a direct role in both the Cvt and macroautophagy pathways, an investigation was undertaken to identify the step(s) in prAPI import requiring Apg5p function. prAPI was used as a marker protein to analyze the functional state of the Cvt pathway. During import, prAPI oligomerizes, assembles into a large complex, and is sequestered within cytoplasmic vesicles (Baba et al., 1997; Kim et al., 1997; Scott et al., 1997). The various steps in the Cvt pathway can be characterized by distinct biochemical parameters, allowing us to define specific stages of the uptake process.
After oligomerization, prAPI assembles into higher-order Cvt complexes that are pelletable from cell lysates in a salt-dependent manner (Baba et al., 1997; Kim et al., 1997). To determine if Apg5p is involved in the formation of this pelletable complex or in a later step in the targeting pathway, the apg5ts strain was used in subcellular fractionation experiments. The effect of inactivation of Apg5p on the assembly of Cvt complexes was examined by shifting apg5ts spheroplasts to 38°C for 5 min. The spheroplasts were then pulse labeled for 5 min, followed by a nonradioactive chase of 20 min to allow prAPI to oligomerize and form Cvt complexes. The labeled spheroplasts were then lysed osmotically in the absence or presence of 5 mM MgCl2, which fulfills the salt requirement for maintaining the pelletability of Cvt complexes. prAPI was pelletable in a salt-dependent manner in both the wild-type and apg5ts strains (Figure 5A), despite the fact that vacuolar delivery of prAPI was almost completely blocked in the apg5ts strain at the nonpermissive temperature. These results indicate that the oligomerization and complex-formation steps of the import process do not require Apg5p function and that a step subsequent to these events must require this protein.
Immunoelectron microscopy combined with biochemical studies show that the next stage of the Cvt pathway involves the enclosure of the Cvt complexes containing prAPI within double-membrane vesicles (Baba et al., 1997; Kim et al., 1997). These studies also indicate that partially membrane-enclosed structures exist when prAPI is overexpressed. These are likely to be the intermediate stages of the membrane-wrapping process that results in the formation of Cvt vesicles and autophagosomes. To determine if the pelletable prAPI that accumulates in the apg5ts mutant is membrane associated, flotation experiments were performed. Spheroplasts were lysed in the presence of MgCl2 and centrifuged to collect pelletable prAPI, which was then subjected to flotation analysis, as described in MATERIALS AND METHODS. In the apg5ts mutant at the nonpermissive temperature, the majority of prAPI was recovered in the float fraction in the absence of detergent, indicating that it is membrane associated (Figure 5B). When membranes were solubilized by the addition of detergent, prAPI was recovered in the nonfloat fraction.
Mutants defective in the next stage of the Cvt pathway, the fusion of Cvt vesicles with the vacuole, accumulate cytosolic vesicles containing prAPI (Scott et al., 1997). Ypt7p is a rab GTPase known to be required for homotypic vacuole fusion (Haas et al., 1995). We previously showed that cells bearing a chromosomal deletion of YPT7 were also defective in prAPI delivery (Kim et al., 1999). This defect is seen in both nutrient-rich and nutrient-starvation conditions (Kirisako et al., 1999), suggesting that Ypt7p is required for delivery of both Cvt vesicles and autophagosomes to the vacuole. The ypt7Δ strain serves as a control for protease protection of prAPI that is enclosed within a cytosolic vesicle. Spheroplasts were prepared from ypt7Δ cells and subjected to osmotic lysis under conditions that retain the integrity of subcellular organelles. Protease was then added in the presence or absence of detergent to determine the accessibility of p2CPY traveling through the Vps pathway and prAPI traveling via the Cvt pathway. In ypt7Δ spheroplasts, both p2CPY and prAPI were trapped in a pelletable, protease-protected compartment (Figure 6), consistent with a role for Ypt7p in the step of the Cvt/Apg pathways involving vesicle fusion with the vacuole, as well as a similar role in the Vps pathway.
To determine if Apg5p is also involved in the fusion step of Cvt vesicles with the vacuole, a similar experiment was carried out with the use of the agp5ts strain. To have an internal control for a protease-protected compartment, we constructed a pep4Δ apg5ts strain. As a result of the pep4 mutation, p2CPY accumulates in the vacuole and will be protected from protease digestion if the integrity of the vacuole membrane is retained during spheroplast lysis. When spheroplasts from the pep4Δ apg5ts double mutant were fractionated after a 5-min pulse and a 60-min chase at the nonpermissive temperature, the majority of prAPI was recovered in the pellet fraction (Figure 6). However, unlike in ypt7Δ spheroplasts, the majority of this material was protease sensitive. In contrast, p2CPY remained protease protected in the pep4Δ apg5ts double mutant, verifying the integrity of subcellular compartments during the lysis procedure. Immunoprecipitation of the cytosolic marker hexokinase suggested that the small amount of protease-protected prAPI detected in the pep4Δ apg5ts strain is the result of unlysed spheroplasts (data not shown). The observation that prAPI-containing complexes that accumulate in the apg5ts strain are protease sensitive indicates that Apg5p acts at a step before vesicle fusion with the vacuole and is likely involved in the formation of Cvt vesicles/autophagosomes.
prAPI Is Associated with Membrane Structures in the Absence of Apg5p Function
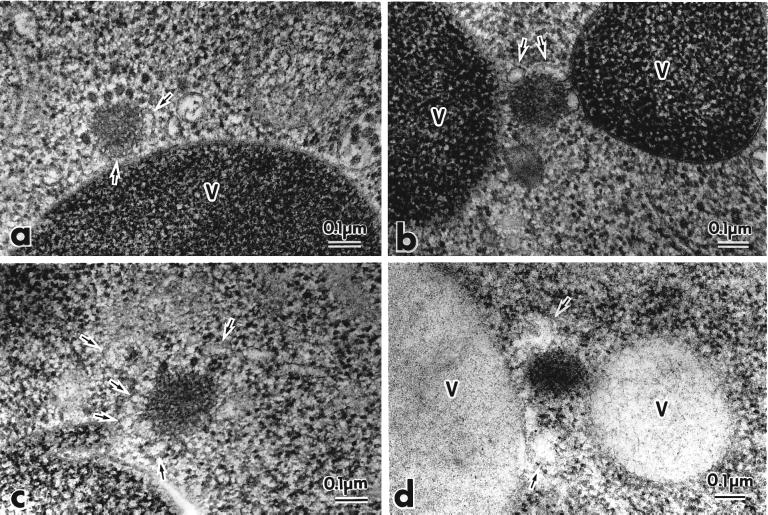
Our analyses of prAPI binding, protease sensitivity, and flotation indicated a role for Apg5p in the formation/completion of Cvt vesicles and autophagosomes. To gain further insight into the function of Apg5p, we examined the state of prAPI in the apg5ts and apg5Δ mutants by electron microscopy with the use of freeze-substitution fixation to ensure optimal preservation of intracellular membranes (Figure 7). The Cvt complex containing prAPI appears as an electron-dense core surrounded by spherical particles (Baba et al. 1997). In the apg5ts mutant, the Cvt complex looks similar to the complex detected in wild-type cells but appears to be slightly larger (Baba et al., 1997) (Figure 7a). When the apg5ts mutant was grown in vegetative conditions and shifted to the nonpermissive temperature, membrane sac–like structures were seen associated with or partly covering the Cvt complex (Figure 7, a–c). These membrane structures were not present as single continuous sheets but rather as several discontinuous sacs. In addition, we were unable to detect completely wrapped Cvt complexes in the form of Cvt vesicles. To confirm that the electron-dense cores corresponded to the prAPI region of the Cvt complex, we immunostained samples with the use of anti-API antibodies (Figure 7d). The electron-dense region surrounded by membranous structures was heavily stained by immunogold particles, indicating that it contained prAPI.
Figure 7.
Electron microscopic analysis reveals that prAPI is associated with membrane sac structures in the apg5ts mutant. The apg5ts strain was grown to log phase in YPD and shifted to 38°C for 3 h. Samples were prepared for electron microscopy by rapid freezing and freeze-substitution fixation and stained with lead citrate for 1 min (a–c) or stained with lead citrate for 30 s and immunostained with anti-API antibodies (d), as described in MATERIALS AND METHODS. The arrows mark membranous structures around the Cvt complex. V, vacuole.
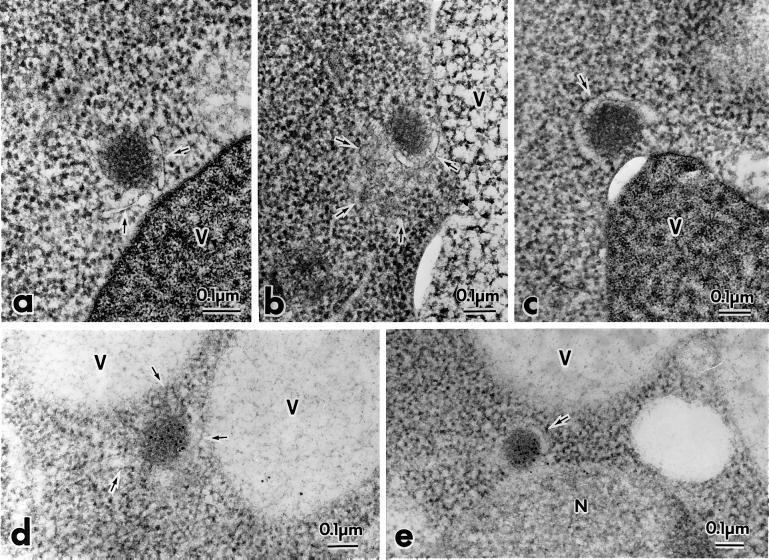
We extended the electron microscopy analysis by examining the apg5Δ mutant under starvation conditions to induce autophagy. Similar to the results with the apg5ts mutant that lacked Cvt vesicles in vegetative conditions, no enclosed autophagosomes were detected in the cytosol after starvation (Figure 8). The Cvt complexes again appeared to be slightly larger than those seen in wild-type cells. These complexes were mostly associated with membrane sacs (Figure 8, a and c). Occasionally, the membrane sacs were not directly apposed to the Cvt complex (Figure 8b). As with the apg5ts mutant, we verified the presence of prAPI in the Cvt complexes through immunostaining (Figure 8, d and e). The observation of structures that may correspond to premature autophagosomes in the apg5Δ strain in starvation conditions and to incomplete Cvt vesicles in the apg5ts strain are in agreement with our biochemical studies, which indicated that prAPI is in the form of a Cvt complex that is protease sensitive and membrane associated. Together, these results suggest that Apg5p functions at the step of Cvt vesicle or autophagosome formation.
Figure 8.
prAPI is membrane associated in the apg5Δ mutant. The apg5Δ strain was grown to log phase in YPD and shifted to SD(−N) medium for 3 h to induce autophagy. Samples were prepared for electron microscopy with the use of rapid freezing and freeze-substitution fixation and stained with lead citrate for 1 min (a–c) or stained with lead citrate for 30 s and immunostained with anti-API antibodies (d and e), as described in MATERIALS AND METHODS. The arrows mark membranous structures around the Cvt complex. V, vacuole; N, nucleus.
DISCUSSION
Macroautophagy is a unique pathway that is essential for yeast cells during starvation. It involves the sequestration of cytoplasm by the formation of a double-membrane sequestering vesicle. At present, few details of the mechanism of vesicle formation are understood. Topologically, the sequestration event in macroautophagy or the Cvt pathway is the opposite of vesicle-budding reactions used for protein transport through the secretory pathway. With Cvt vesicles/autophagosomes, the cytosolic face of the forming vesicle is in contact with the cargo. In contrast, the cargo contacts the lumenal side of vesicles budding from secretory pathway organelles. The origin of the sequestering membrane is not known, nor is the mechanism by which this membrane enwraps cytosol and/or a Cvt complex. Genetic analyses of both API import and autophagy have led to the identification of a series of mutants defective in one or both processes, and many of the complementing genes have recently been cloned. Molecular biological analyses of the gene products have started to provide insights into these novel pathways.
Apg5p was shown to be one of the components in a unique nonubiquitin conjugation reaction that is required for prAPI import and autophagy (Mizushima et al., 1998a). By tagging Apg5p with the HA epitope, we have demonstrated through subcellular fractionation and flotation analysis that Apg5HAp may exist in distinct biochemical forms in the cell. A population of the protein is membrane associated, but a portion of Apg5p also appears to exist as part of a pelletable complex that is presumably devoid of membrane (Figure 2). In addition, a fraction of the protein is soluble. It remains to be determined whether these species correspond to different functional forms or stages of Apg5p.
In vivo studies with an Apg5pGFP hybrid protein revealed two distinct types of staining patterns (Figure 3). In wild-type cells and many of the apg/cvt mutant strains (apg8 is shown as an example), Apg5pGFP displayed a random staining pattern, and no distinct subcellular location could be identified. This staining pattern was observed during growth in rich medium and during nitrogen-starvation conditions (Figure 3).
In some apg/cvt mutant strains, including apg7Δ, apg12Δ, and apg16Δ (Figure 3), Apg5pGFP displayed punctate staining in addition to the staining pattern observed in wild-type cells. These punctate structures were often in close proximity to the vacuole. However, subcellular fractionation of the apg12Δ and apg7Δ strains by density gradient indicated that Apg5p did not co-localize substantially with the vacuolar membrane marker ALP or other organellar markers (Mizushima et al., 1998a) (data not shown). The punctate Apg5pGFP structures are not the result of a general block in the Apg/Cvt pathways because punctate fluorescence was not observed in all of the apg or cvt strains (e.g., see apg8 in Figure 3).
To gain further insight into the function of Apg5p in autophagy and the Cvt pathway, we generated a temperature-sensitive allele of APG5, the first conditional mutant in either pathway. Import of prAPI was rapidly and specifically blocked in the apg5ts strain at the nonpermissive temperature (Figure 4). We used this mutant to determine the site of action of Apg5p by assessing the stage of prAPI import immediately after shifting cells to nonpermissive conditions. prAPI was membrane associated (Figure 5B) but protease sensitive (Figure 6) in the apg5ts mutant, indicating that Apg5p is required for completion of the vesicle-sequestration step.
We performed electron microscopy with the use of freeze-substitution fixation and immunostaining to gain further insight into the state of prAPI in the absence of functional Apg5p. In both the apg5ts mutant (Figure 7) and the apg5Δ strain (Figure 8), we detected prAPI in the form of a Cvt complex, in agreement with our studies indicating normal assembly of the complex (Figure 5A). These complexes were partially wrapped or associated with membrane sacs. This result agrees with our biochemical observations that prAPI is protease sensitive yet membrane associated. We cannot rule out the possibility that these sac-like structures are a terminal phenotype resulting from the apg5 mutation and do not represent the situation in wild-type cells. The fact that similar structures are seen in the apg5ts mutant, however, suggests that they are not caused by chronic loss of Apg5p. In general, the membrane sac structures in the apg5Δ strain appeared to be in less direct contact with the Cvt complex than was seen with the apg5ts mutant. These membrane sacs may represent intermediates in Cvt vesicle or autophagosome formation.
The data generated in these studies have allowed us to identify the role of Apg5p in the Cvt and macroautophagy pathways. We have used this new information to expand our current understanding of how yeast cells package and deliver cytosolic proteins and organelles to the vacuole. Based on the biochemical and morphological data, we propose a model in which Apg5p functions in sequestering prAPI (as well as bulk cytosol during nutrient starvation) into double-membrane vesicles (Figure 9). It is likely that this sequestration step involves several other Apg/Cvt proteins, including the conjugation machinery such as Apg7p and additional proteins such as Aut7p (Kirisako et al., 1999; Huang et al., 2000). The predicted role of Apg5p also agrees with the biochemical and in vivo data indicating that it is a membrane-associated protein that may be associated with vesicular structures. Recently, the GSA7 gene of Pichia pastoris was cloned and shown to be homologous to APG7 of S. cerevisiae (Kim et al., 1999; Tanida et al., 1999; Yuan et al., 1999). The gsa7 mutant is defective in a late stage of peroxisome sequestration during pexophagy and is characterized by partially engulfed peroxisomes. This observation further implicates Apg5p and the associated conjugation machinery in some aspect of vesicle formation/completion during autophagy and the Cvt pathway. Additional proteins, including at least Apg16p, are needed to complete the cycle of import (Mizushima et al., 1999).
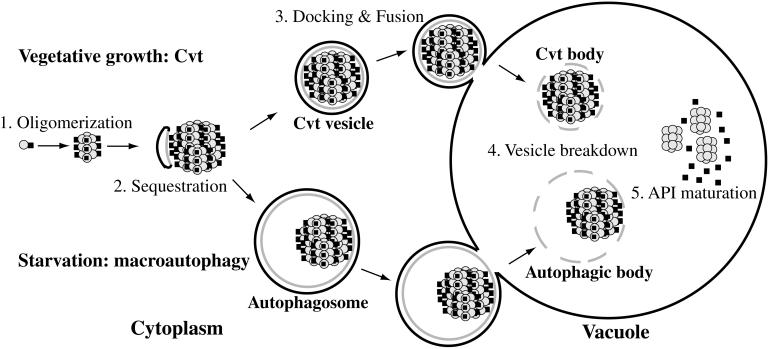
Figure 9.
Two-pathway model for Cvt/Apg transport to the vacuole. (1) prAPI rapidly oligomerizes into a dodecameric conformation after translation. (2) The dodecamers assemble into Cvt complexes, which are subsequently sequestered and wrapped by double-membrane Cvt vesicles and autophagosomes in a process requiring Apg5p. (3) The vesicles are then targeted to the vacuole, where subsequent docking and fusion of the outer membrane results in the release of the inner vesicle into the lumen. (4) The inner membrane is degraded within the lumen, allowing vacuolar hydrolases access to the cargo. (5) The N-terminal propeptides of the prAPI molecules are proteolytically removed in a PEP4-dependent manner to generate mature API dodecamers.
Recent studies of mammalian cells suggest that macroautophagy in yeast could be connected to the metazoan process of programmed cell death. A human gene was recently identified that is homologous to the APG5 gene in yeast (Hammond et al., 1998). The human Apg5p homologue is an apoptosis-specific protein that is highly expressed during apoptosis. Furthermore, the human Apg12p homologue is conjugated to human Apg5p, suggesting that the process in conserved throughout eukaryotes (Mizushima et al., 1998b). Given our current understanding of the intracellular events that occur during apoptosis and macroautophagy, the discovery of a possible evolutionary relationship is not entirely surprising. For example, it is known that during macroautophagy, bulk cytoplasm, including organelles, is degraded in response to specific environmental cues (Takeshige et al., 1992). Similarly, a variety of cytosolic enzymes and regulatory factors, as well as nuclear lamins, have been shown to be degraded in apoptotic cells (Lazebnik et al., 1994; Neamati et al., 1995; Jänicke et al., 1996). More recently, it has been demonstrated in mammalian cells that autophagy is a transient early stage in response to apoptosis-inducing cytotoxic factors (Prins et al., 1998) and that inhibition of autophagy in human T-lymphoblastic leukemic cells acts to prevent tumor necrosis factor-α–induced apoptosis (Jia et al., 1997). Thus, it seems likely that some apoptosis-specific processes in higher eukaryotes involve evolutionarily conserved mechanisms common to both the Cvt pathway and macroautophagy in yeast.
Characterization of the gene products involved in the Cvt pathway and macroautophagy has been under way for several years, yet our current models of how these pathways function at the molecular level are still incomplete and speculative. For example, although several proteins involved in the sequestration step have now been identified, the exact mechanisms that result in the formation of a double-membrane vesicle are still unknown. Further studies of the Apg/Cvt proteins and their interactions will be necessary to provide essential information about the complex processes involved in the packaging and delivery of proteins and organelles from the cytoplasm to the vacuole.
ACKNOWLEDGMENTS
We thank John Kim for helpful discussion and critical reading of the manuscript and Dr. Masako Osumi (Japan Women's University) for the use of electron microscopy facilities. This work was supported by Public Health Service grant GM-53396 from the National Institutes of Health to D.J.K., a National Institutes of Health biotechnology training grant to M.D.G., an American Cancer Society, California Division, senior postdoctoral fellowship to S.V.S., and Grants-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan to Y.O.
Abbreviations used:
- ALP
alkaline phosphatase
- API
aminopeptidase I
- CPY
carboxypeptidase Y
- Cvt
cytoplasm-to-vacuole targeting
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HA
hemagglutinin
- PGK
phosphoglycerate kinase
- PIPES
piperazine-N,N′-bis(2-ethanesulfonic acid)
- prAPI
precursor aminopeptidase I
REFERENCES
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytosol to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process: autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;4:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A, Scheglmann D, Lazar T, Gallwitz D, Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond EM, Brunet CL, Johnson GD, Parkhill J, Milner AE, Brady G, Gregory CD, Grand RJ. Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett. 1998;425:391–395. doi: 10.1016/s0014-5793(98)00266-x. [DOI] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.-P., Scott, S.V., Kim, J., and Klionsky, D.J. (2000). The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. (in press). [DOI] [PubMed]
- Irie K, Takase M, Lee KS, Levin DE, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänicke RU, Walker PA, Lin XY, Porter AG. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J. 1996;15:6969–6978. [PMC free article] [PubMed] [Google Scholar]
- Jia L, Dourmashkin RR, Allen PD, Gray AB, Newland AC, Kelsey SM. Inhibition of autophagy abrogates tumor necrosis factor α induced apoptosis in human T-lymphoblastic leukemic cells. Br J Haematol. 1997;98:673–685. doi: 10.1046/j.1365-2141.1997.2623081.x. [DOI] [PubMed] [Google Scholar]
- Kametaka S, Matsuura A, Wada Y, Ohsumi Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene. 1996;178:139–143. doi: 10.1016/0378-1119(96)00354-x. [DOI] [PubMed] [Google Scholar]
- Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998a;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human: the counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998b;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Neamati N, Fernandez A, Wright S, Kiefer J, McConkey DJ. Degradation of lamin B1 precedes oligonucleosomal DNA fragmentation in apoptotic thymocytes and isolated thymocyte nuclei. J Immunol. 1995;154:3788–3795. [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Prins JB, Ledgerwood EC, Ameloot P, Vandenabeele P, Paulo PR, Bright NA, O'Rahilly S, Bradley JR. Tumor necrosis factor-induced cytotoxicity is not related to rates of mitochondrial morphological abnormalities or autophagy-changes that can be mediated by TNFR-I or TNFR-II. Biosci Rep. 1998;18:329–340. doi: 10.1023/a:1020261316486. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. In vitro reconstitution of cytoplasm to vacuole protein targeting in yeast. J Cell Biol. 1995;131:1727–1735. doi: 10.1083/jcb.131.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5341. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: a novel protein activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tomashek JJ, Sonnenburg JL, Artimovich JM, Klionsky DJ. Resolution of subunit interactions and cytoplasmic subcomplexes of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1996;271:10397–10404. doi: 10.1074/jbc.271.17.10397. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Yuan W, Stromhaug PE, Dunn WA., Jr Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol Biol Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]