Abstract
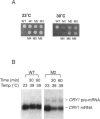
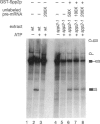
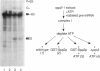
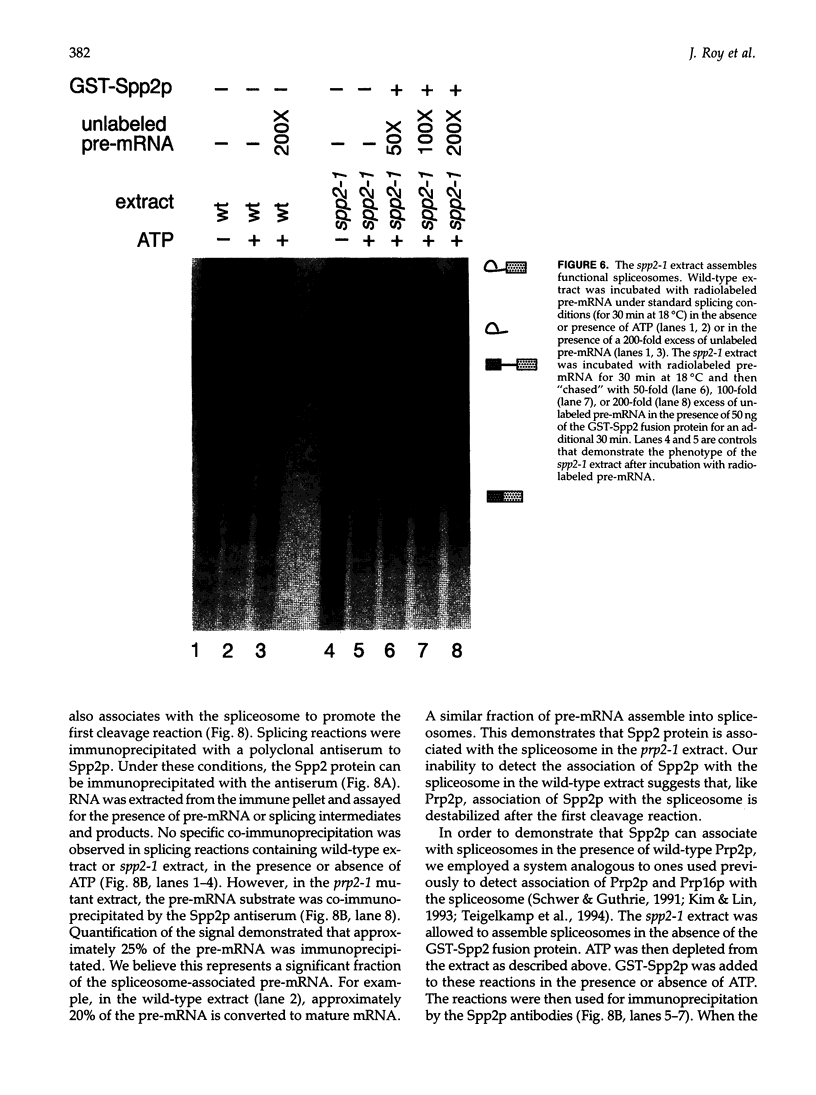
Pre-mRNA processing occurs by assembly of splicing factors on the substrate to form the spliceosome followed by two consecutive RNA cleavage-ligation reactions. The Prp2 protein hydrolyzes ATP and is required for the first reaction (Yean SL, Lin RJ, 1991, Mol Cell Biol 11:5571-5577; Kim SH, Smith J, Claude A, Lin RJ, 1992, EMBO J 11:2319-2326). The Saccharomyces cerevisiae SPP2 gene was previously identified as a high-copy suppressor of temperature-sensitive prp2 mutants (Last RL, Maddock JR, Woolford JL Jr, 1987, Genetics 117:619-631). We have characterized the function of Spp2p in vivo and in vitro. Spp2p is an essential protein required for the first RNA cleavage reaction in vivo. Depletion of Spp2p from yeast cells results in accumulation of unspliced pre-mRNAs. A temperature-sensitive spp2-1 mutant accumulates pre-mRNAs in vivo and is unable to undergo the first splicing reaction in vitro. However, spliceosomal complexes are assembled in extracts prepared from the mutant. We show that Spp2p function is required after spliceosome assembly but prior to the first reaction. Spp2p associates with the spliceosome before the first RNA cleavage reaction and is likely to be released from the spliceosome following ATP hydrolysis by Prp2p. The Prp2 and Spp2 proteins are capable of physically interacting with each other. These results suggest that Spp2p interacts with Prp2p in the spliceosome prior to the first cleavage-ligation reaction. Spp2p is the first protein that has been found to interact with a DEAD/H box splicing factor.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arenas J. E., Abelson J. N. The Saccharomyces cerevisiae PRP21 gene product is an integral component of the prespliceosome. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6771–6775. doi: 10.1073/pnas.90.14.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Couto J. R., Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990 Mar 9;60(5):705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- Chen J. H., Lin R. J. The yeast PRP2 protein, a putative RNA-dependent ATPase, shares extensive sequence homology with two other pre-mRNA splicing factors. Nucleic Acids Res. 1990 Nov 11;18(21):6447–6447. doi: 10.1093/nar/18.21.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Chien C. T., Bartel P. L., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991 Feb 7;349(6309):487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Coppolecchia R., Buser P., Stotz A., Linder P. A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J. 1993 Oct;12(10):4005–4011. doi: 10.1002/j.1460-2075.1993.tb06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto J. R., Tamm J., Parker R., Guthrie C. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987 Jul;1(5):445–455. doi: 10.1101/gad.1.5.445. [DOI] [PubMed] [Google Scholar]
- Dalbadie-McFarland G., Abelson J. PRP5: a helicase-like protein required for mRNA splicing in yeast. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M., Tsay Y. F., Paulovich A. G., Woolford J. L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993 May;13(5):2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Roeder G. S. MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol Cell Biol. 1990 May;10(5):2379–2389. doi: 10.1128/mcb.10.5.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994 Aug;4(8):271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Grifo J. A., Abramson R. D., Satler C. A., Merrick W. C. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984 Jul 10;259(13):8648–8654. [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989 Jun 15;339(6225):562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- Horowitz D. S., Abelson J. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 1993 Feb;7(2):320–329. doi: 10.1101/gad.7.2.320. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo M., Dever T. E., Merrick W. C., Sonenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol Cell Biol. 1991 Dec;11(12):5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Lin R. J. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):888–892. doi: 10.1073/pnas.90.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Smith J., Claude A., Lin R. J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992 Jun;11(6):2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D. S., Beggs J. D. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Nov 25;18(22):6559–6564. doi: 10.1093/nar/18.22.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Last R. L., Maddock J. R., Woolford J. L., Jr Evidence for related functions of the RNA genes of Saccharomyces cerevisiae. Genetics. 1987 Dec;117(4):619–631. doi: 10.1093/genetics/117.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last R. L., Stavenhagen J. B., Woolford J. L., Jr Isolation and characterization of the RNA2, RNA3, and RNA11 genes of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2396–2405. doi: 10.1128/mcb.4.11.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Lustig A. J., Lin R. J., Abelson J. The yeast RNA gene products are essential for mRNA splicing in vitro. Cell. 1986 Dec 26;47(6):953–963. doi: 10.1016/0092-8674(86)90810-x. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- McEachern M. J., Filutowicz M., Helinski D. R. Mutations in direct repeat sequences and in a conserved sequence adjacent to the repeats result in a defective replication origin in plasmid R6K. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1480–1484. doi: 10.1073/pnas.82.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick W. C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992 Jun;56(2):291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Pulaski B. A., Woolford J. L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991 Nov;11(11):5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T. W. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell. 1994 Jul 15;78(1):1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Plumpton M., McGarvey M., Beggs J. D. A dominant negative mutation in the conserved RNA helicase motif 'SAT' causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994 Feb 15;13(4):879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Roy J., Zheng B., Rymond B. C., Woolford J. L., Jr Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol Cell Biol. 1995 Jan;15(1):445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990 Mar;10(3):1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S. R., Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992 Feb;6(3):283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Schwer B., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991 Feb 7;349(6309):494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Matunis M. J., Michael W. M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993 Mar 11;21(5):1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Staudinger J., Perry M., Elledge S. J., Olson E. N. Interactions among vertebrate helix-loop-helix proteins in yeast using the two-hybrid system. J Biol Chem. 1993 Mar 5;268(7):4608–4611. [PubMed] [Google Scholar]
- Tarn W. Y., Lee K. R., Cheng S. C. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol. 1993 Mar;13(3):1883–1891. doi: 10.1128/mcb.13.3.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S., McGarvey M., Plumpton M., Beggs J. D. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994 Feb 15;13(4):888–897. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. RNA splicing. Alive with DEAD proteins. Nature. 1991 Feb 7;349(6309):463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- Yean S. L., Lin R. J. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol Cell Biol. 1991 Nov;11(11):5571–5577. doi: 10.1128/mcb.11.11.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]