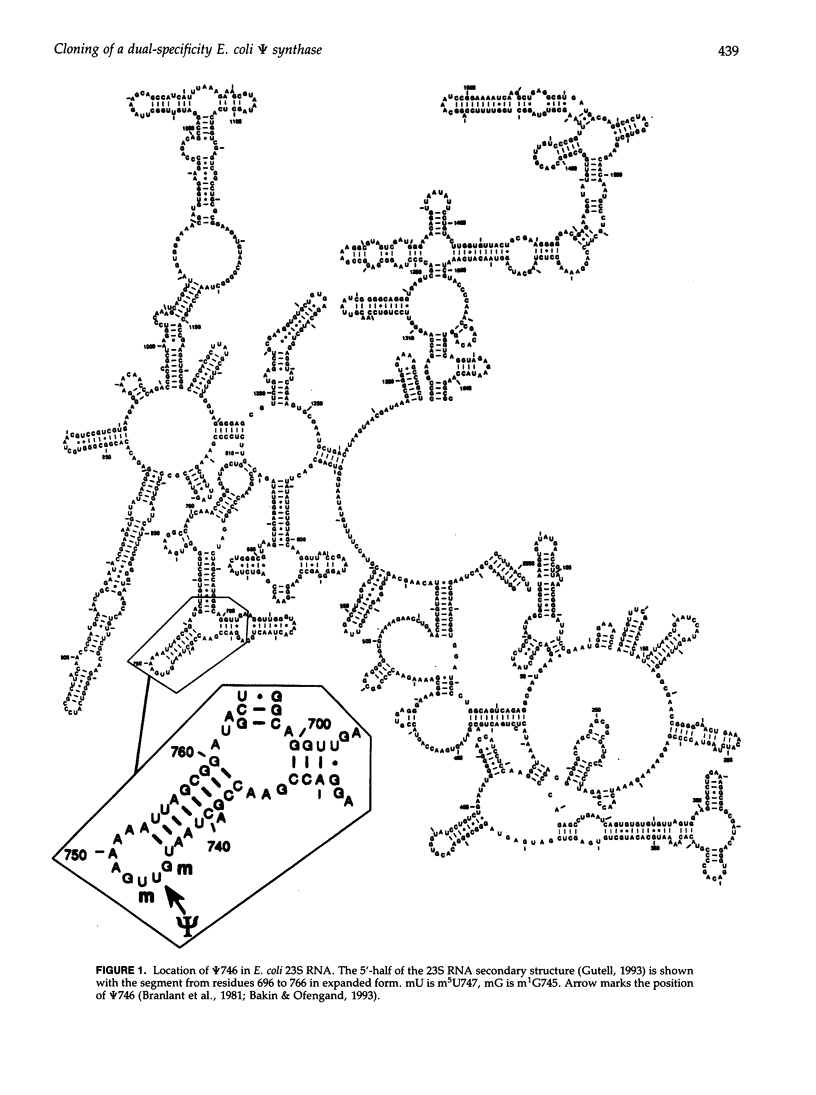
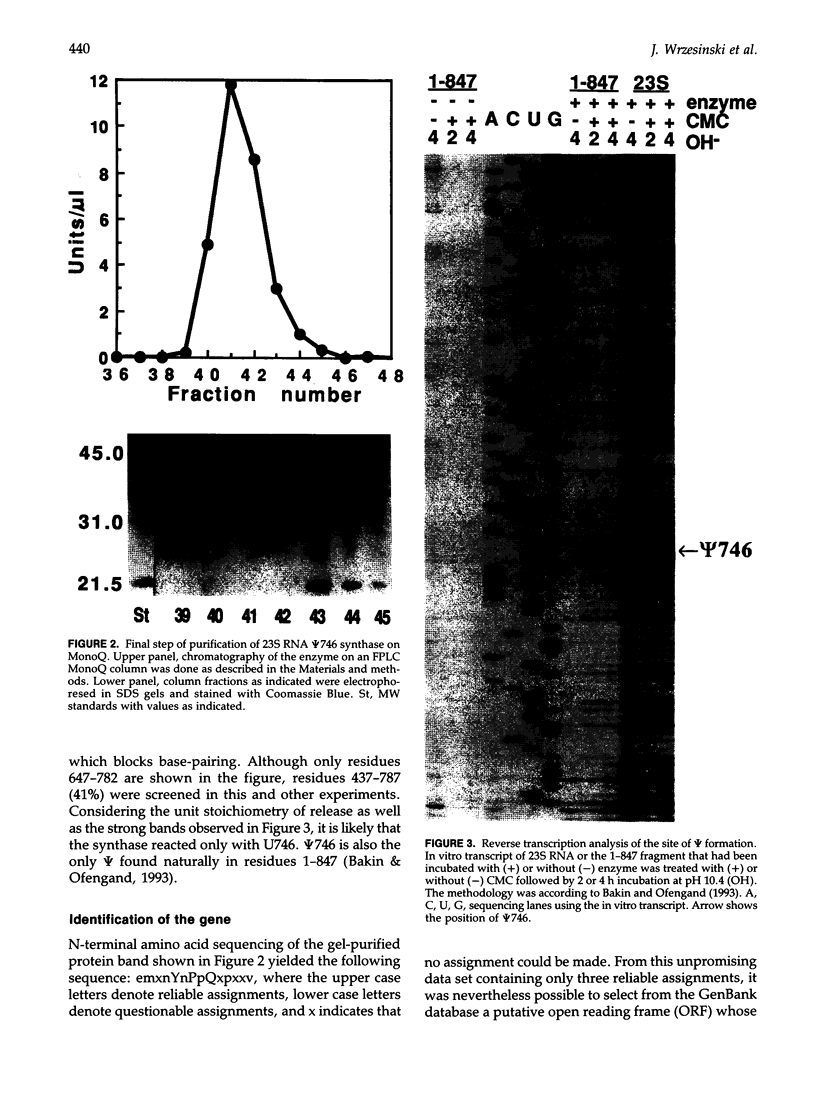
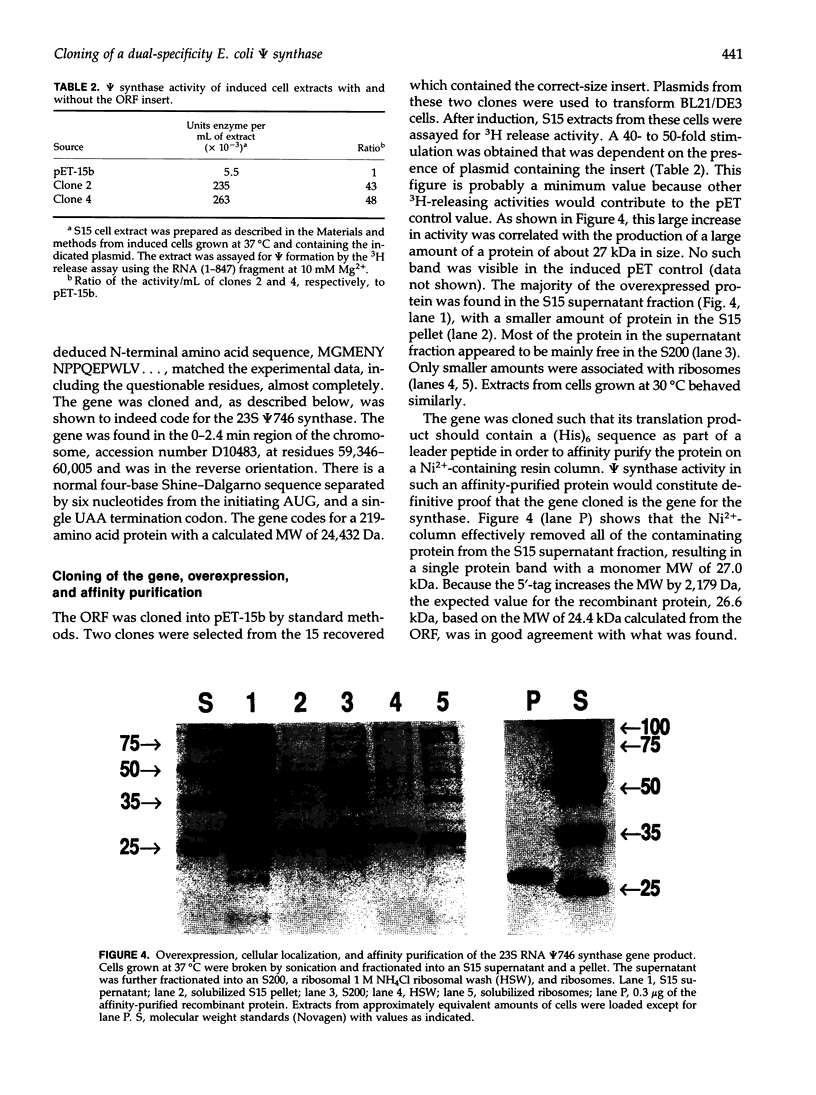
Abstract
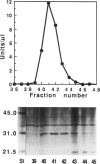
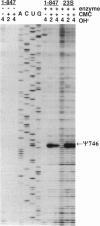
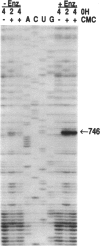
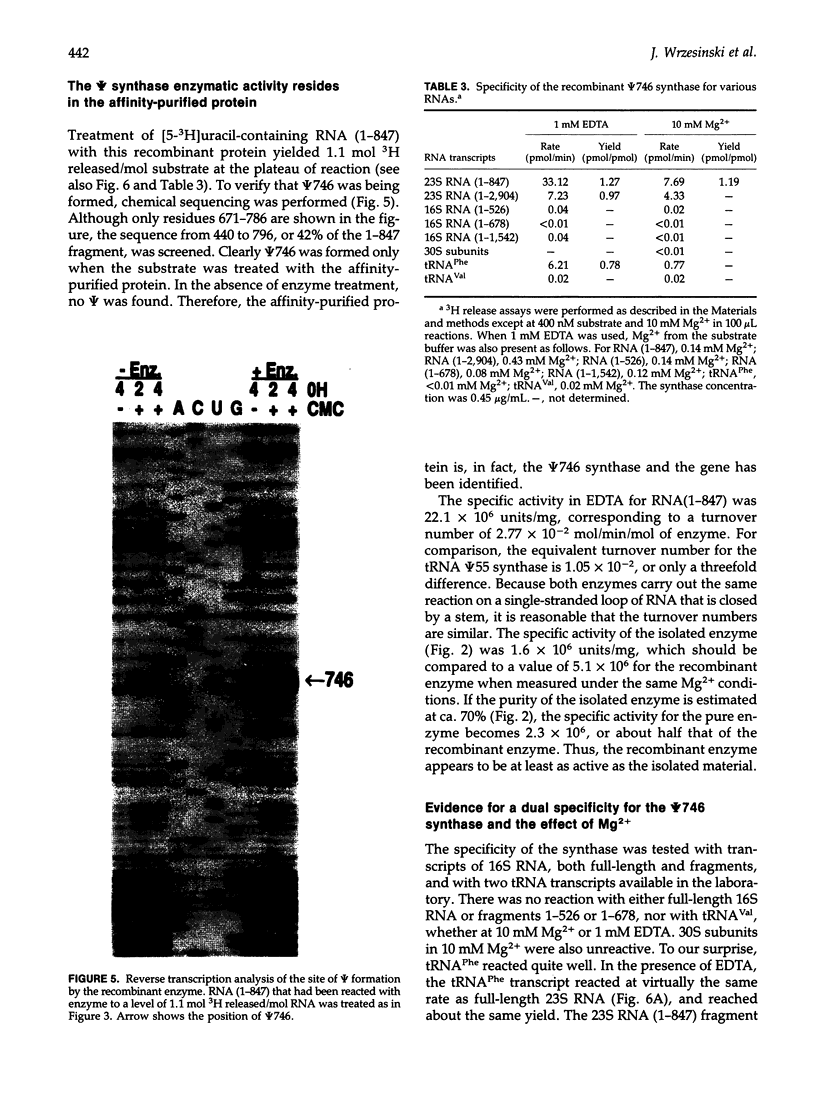
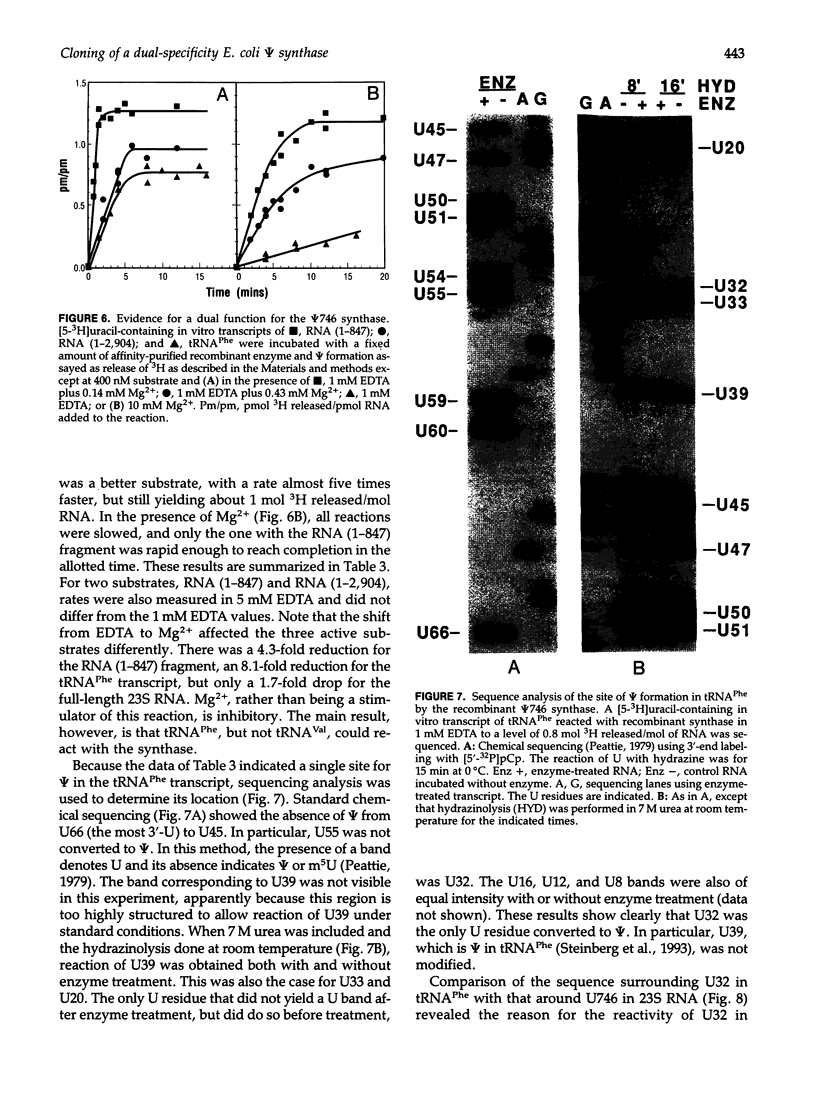
An Escherichia coli pseudouridine (psi) synthase, which forms both psi 746 in E. coli 23S ribosomal RNA and psi 32 in tRNA(Phe), has been isolated and cloned. The enzyme contains 219 amino acids and has a calculated MW of 24,432 Da. Amino acid sequence comparison with the three other psi synthases that have been cloned to date, two for tRNA and one for 16S RNA, did not reveal any common sequence motifs, despite the catalysis of a common reaction. The gene was cloned behind a (His)6 leader for affinity purification. Upon overexpression, most of the enzyme remained soluble in the cell cytoplasm and could be purified to homogeneity on a Ni(2+)-containing resin. The enzyme reacted with both full-length 23S RNA or a fragment from residues 1-847, forming 1 mol psi/mol RNA at position 746, a normal site for psi. The enzyme has no dependence on Mg2+. The same yield was obtained in 1 mM EDTA as in 10 mM Mg2+, and the rate was faster in EDTA than in Mg2+. Full-length 16S RNA or fragments 1-526 or 1-678, as well as tRNA(Val) transcripts, were not modified in either EDTA or Mg2+. tRNA(Phe) transcripts, however, were modified with a yield of 1 mol psi/mol transcript at a rate in EDTA like that of 23S RNA. Sequencing showed all of the psi to be at position 32, a normal site for psi in this tRNA. Both 23S rRNA psi 746 and tRNA psi 32 occur in single-stranded segments of the same sequence, psi UGAAAA, closed by a stem. Therefore, this synthase may require for recognition only a short stretch of primary sequence 3' to the site of pseudouridylation. This is the first example of a dual-specificity modifying enzyme for RNA, that is, one which is specific for a single site in one RNA, and equally site-specific in a second class of RNA. The essentiality of these psi residues can now be assessed by disruption of the synthase gene.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arps P. J., Marvel C. C., Rubin B. C., Tolan D. A., Penhoet E. E., Winkler M. E. Structural features of the hisT operon of Escherichia coli K-12. Nucleic Acids Res. 1985 Jul 25;13(14):5297–5315. doi: 10.1093/nar/13.14.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A., Kowalak J. A., McCloskey J. A., Ofengand J. The single pseudouridine residue in Escherichia coli 16S RNA is located at position 516. Nucleic Acids Res. 1994 Sep 11;22(18):3681–3684. doi: 10.1093/nar/22.18.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin A., Lane B. G., Ofengand J. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry. 1994 Nov 15;33(45):13475–13483. doi: 10.1021/bi00249a036. [DOI] [PubMed] [Google Scholar]
- Bakin A., Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993 Sep 21;32(37):9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe R., Mitchell P., Osswald M., Stade K., Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993 Jan;7(1):161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- Cortese R., Kammen H. O., Spengler S. J., Ames B. N. Biosynthesis of pseudouridine in transfer ribonucleic acid. J Biol Chem. 1974 Feb 25;249(4):1103–1108. [PubMed] [Google Scholar]
- Denman R., Weitzmann C., Cunningham P. R., Nègre D., Nurse K., Colgan J., Pan Y. C., Miedel M., Ofengand J. In vitro assembly of 30S and 70S bacterial ribosomes from 16S RNA containing single base substitutions, insertions, and deletions around the decoding site (C1400). Biochemistry. 1989 Feb 7;28(3):1002–1011. doi: 10.1021/bi00429a013. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1993 Jul 1;21(13):3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammen H. O., Marvel C. C., Hardy L., Penhoet E. E. Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. J Biol Chem. 1988 Feb 15;263(5):2255–2263. [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Lane B. G., Ofengand J., Gray M. W. Pseudouridine and O2'-methylated nucleosides. Significance of their selective occurrence in rRNA domains that function in ribosome-catalyzed synthesis of the peptide bonds in proteins. Biochimie. 1995;77(1-2):7–15. doi: 10.1016/0300-9084(96)88098-9. [DOI] [PubMed] [Google Scholar]
- Lane B. G., Ofengand J., Gray M. W. Pseudouridine in the large-subunit (23 S-like) ribosomal RNA. The site of peptidyl transfer in the ribosome? FEBS Lett. 1992 May 4;302(1):1–4. doi: 10.1016/0014-5793(92)80269-m. [DOI] [PubMed] [Google Scholar]
- Limbach P. A., Crain P. F., McCloskey J. A. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994 Jun 25;22(12):2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Nurse K., Wrzesinski J., Bakin A., Lane B. G., Ofengand J. Purification, cloning, and properties of the tRNA psi 55 synthase from Escherichia coli. RNA. 1995 Mar;1(1):102–112. [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993 Jul 1;21(13):3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Weitzmann C. J., Cunningham P. R., Ofengand J. Cloning, in vitro transcription, and biological activity of Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 1990 Jun 25;18(12):3515–3520. doi: 10.1093/nar/18.12.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]