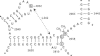Abstract
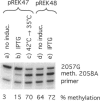
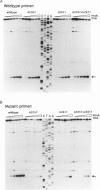
The ErmE methyltransferase confers resistance to MLS antibiotics by specifically dimethylating adenine 2058 (A2058, Escherichia coli numbering) in bacterial 23S rRNA. To define nucleotides in the rRNA that are part of the motif recognized by ErmE, we investigated both in vivo and in vitro the effects of mutations around position A2058 on methylation. Mutagenizing A2058 (to G or U) completely abolishes methylation of 23S rRNA by ErmE. No methylation occurred at other sites in the rRNA, demonstrating the fidelity of ErmE for A2058. Breaking the neighboring G2057-C2611 Watson-Crick base pair by introducing either an A2057 or a U2611 mutation, greatly reduces the rate of methylation at A2058. Methylation remains impaired after these mutations have been combined to create a new A2057-U2611 Watson-Crick base interaction. The conformation of this region in 23S rRNA was probed with chemical reagents and it was shown that the A2057 and U2611 mutations alone and in combination alter the reactivity of A2058 and adjacent bases. However, mutagenizing position G-->A2032 in an adjacent loop, which has been implicated to interact with A2058, alters neither the ErmE methylation at A2058 nor the accessibility of this region to the chemical reagents. The data indicate that a less-exposed conformation at A2058 leads to reduction in methylation by ErmE. Nucleotide G2057 and its interaction with C2611 maintain the conformation at A2058, and are thus important in forming the structural motif that is recognized by the ErmE methyltransferase.
Full text
PDF

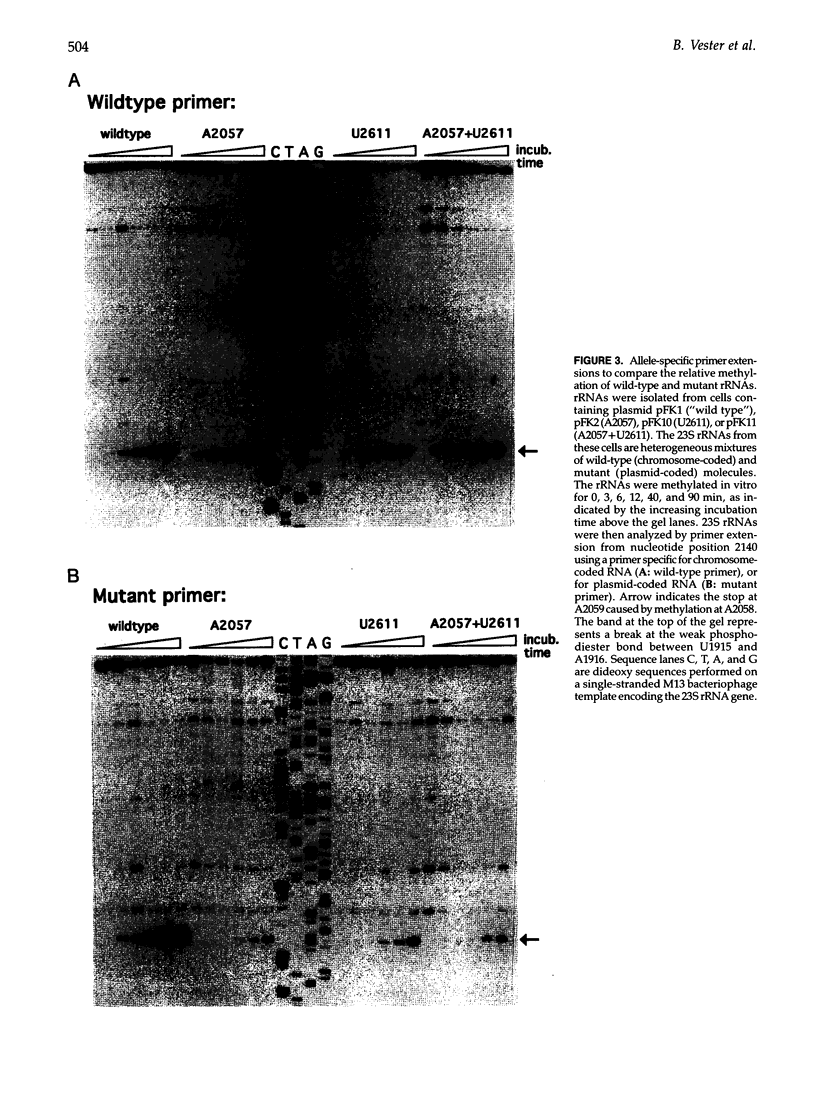
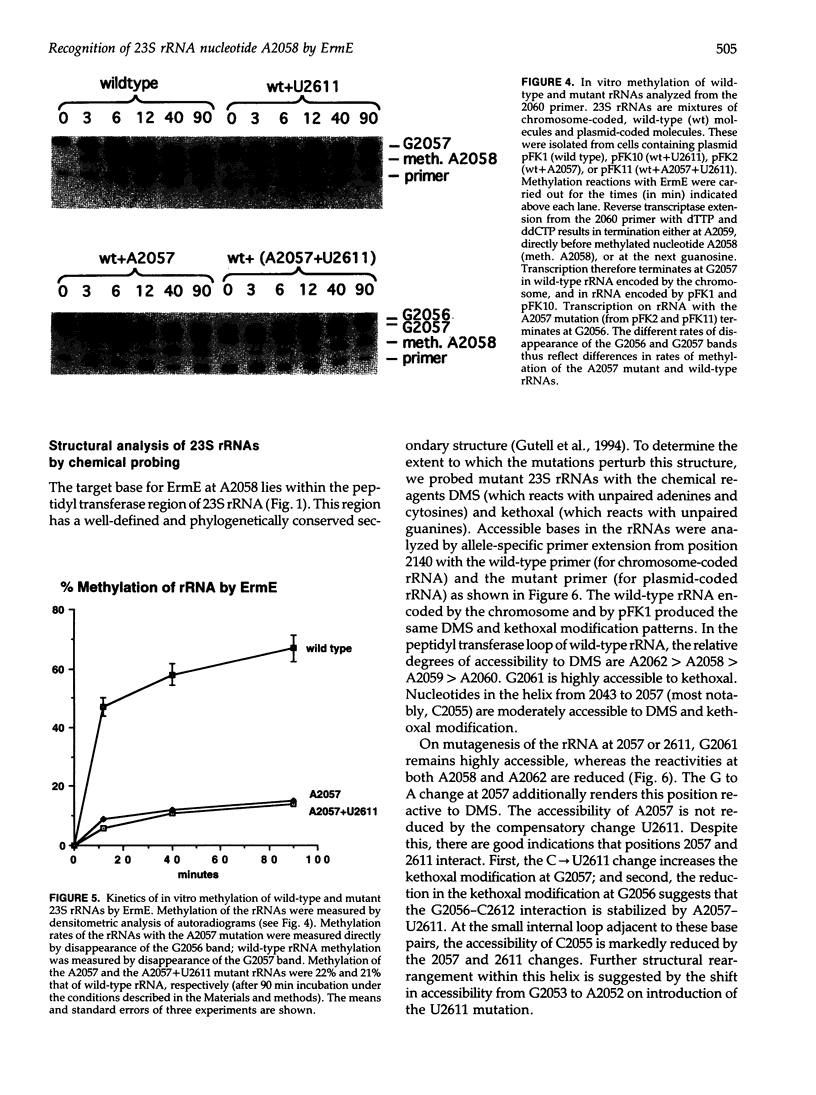




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aagaard C., Rosendahl G., Dam M., Powers T., Douthwaite S. Specific structural probing of plasmid-coded ribosomal RNAs from Escherichia coli. Biochimie. 1991 Dec;73(12):1439–1444. doi: 10.1016/0300-9084(91)90176-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Cseplö A., Etzold T., Schell J., Schreier P. H. Point mutations in the 23 S rRNA genes of four lincomycin resistant Nicotiana plumbaginifolia mutants could provide new selectable markers for chloroplast transformation. Mol Gen Genet. 1988 Oct;214(2):295–299. doi: 10.1007/BF00337724. [DOI] [PubMed] [Google Scholar]
- Dhillon N., Leadlay P. F. A repeated decapeptide motif in the C-terminal domain of the ribosomal RNA methyltransferase from the erythromycin producer Saccharopolyspora erythraea. FEBS Lett. 1990 Mar 26;262(2):189–193. doi: 10.1016/0014-5793(90)80186-m. [DOI] [PubMed] [Google Scholar]
- Douthwaite S., Aagaard C. Erythromycin binding is reduced in ribosomes with conformational alterations in the 23 S rRNA peptidyl transferase loop. J Mol Biol. 1993 Aug 5;232(3):725–731. doi: 10.1006/jmbi.1993.1426. [DOI] [PubMed] [Google Scholar]
- Douthwaite S. Functional interactions within 23S rRNA involving the peptidyltransferase center. J Bacteriol. 1992 Feb;174(4):1333–1338. doi: 10.1128/jb.174.4.1333-1338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S. Interaction of the antibiotics clindamycin and lincomycin with Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 1992 Sep 25;20(18):4717–4720. doi: 10.1093/nar/20.18.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring T., Greuer B., Brimacombe R. The three-dimensional folding of ribosomal RNA; localization of a series of intra-RNA cross-links in 23S RNA induced by treatment of Escherichia coli 50S ribosomal subunits with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res. 1991 Jul 11;19(13):3517–3524. doi: 10.1093/nar/19.13.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J., Garrett R. A. Binding sites of the antibiotics pactamycin and celesticetin on ribosomal RNAs. Biochimie. 1991 Jul-Aug;73(7-8):1145–1149. doi: 10.1016/0300-9084(91)90158-w. [DOI] [PubMed] [Google Scholar]
- Ettayebi M., Prasad S. M., Morgan E. A. Chloramphenicol-erythromycin resistance mutations in a 23S rRNA gene of Escherichia coli. J Bacteriol. 1985 May;162(2):551–557. doi: 10.1128/jb.162.2.551-557.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M. Y., Weisblum B. 23S ribosomal ribonucleic acid of macrolide-producing streptomycetes contains methylated adenine. J Bacteriol. 1979 Mar;137(3):1464–1467. doi: 10.1128/jb.137.3.1464-1467.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Larsen N., Woese C. R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994 Mar;58(1):10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalic D., Giannattasio R. B., Jin H. J., Weisblum B. 23S rRNA domain V, a fragment that can be specifically methylated in vitro by the ErmSF (TlrA) methyltransferase. J Bacteriol. 1994 Nov;176(22):6992–6998. doi: 10.1128/jb.176.22.6992-6998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Larsen J. E., Gerdes K., Light J., Molin S. Low-copy-number plasmid-cloning vectors amplifiable by derepression of an inserted foreign promoter. Gene. 1984 Apr;28(1):45–54. doi: 10.1016/0378-1119(84)90086-6. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987 Aug;69(8):879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Borden A., Morgan E. A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Morgan E. A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984 Jun 11;12(11):4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner R., Cundliffe E., Schmidt F. J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem. 1983 Oct 25;258(20):12702–12706. [PubMed] [Google Scholar]
- Thompson C. J., Kieser T., Ward J. M., Hopwood D. A. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene. 1982 Nov;20(1):51–62. doi: 10.1016/0378-1119(82)90086-5. [DOI] [PubMed] [Google Scholar]
- Uchiyama H., Weisblum B. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene. 1985;38(1-3):103–110. doi: 10.1016/0378-1119(85)90208-2. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. Keeping RNA happy. RNA. 1995 Mar;1(1):4–6. [PMC free article] [PubMed] [Google Scholar]
- Vannuffel P., Di Giambattista M., Morgan E. A., Cocito C. Identification of a single base change in ribosomal RNA leading to erythromycin resistance. J Biol Chem. 1992 Apr 25;267(12):8377–8382. [PubMed] [Google Scholar]
- Vester B., Douthwaite S. Domain V of 23S rRNA contains all the structural elements necessary for recognition by the ErmE methyltransferase. J Bacteriol. 1994 Nov;176(22):6999–7004. doi: 10.1128/jb.176.22.6999-7004.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalacain M., Cundliffe E. Methylation of 23S rRNA caused by tlrA (ermSF), a tylosin resistance determinant from Streptomyces fradiae. J Bacteriol. 1989 Aug;171(8):4254–4260. doi: 10.1128/jb.171.8.4254-4260.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]