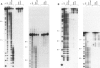Abstract
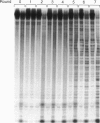
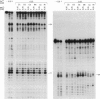
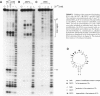
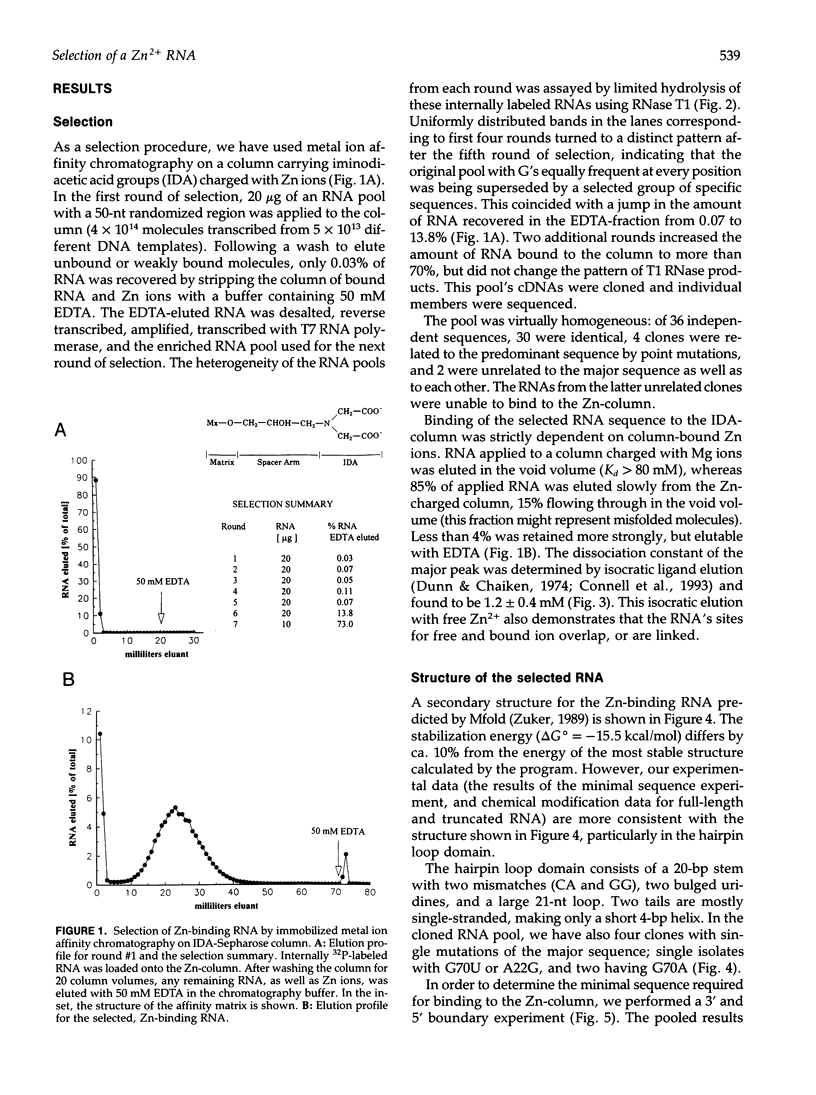
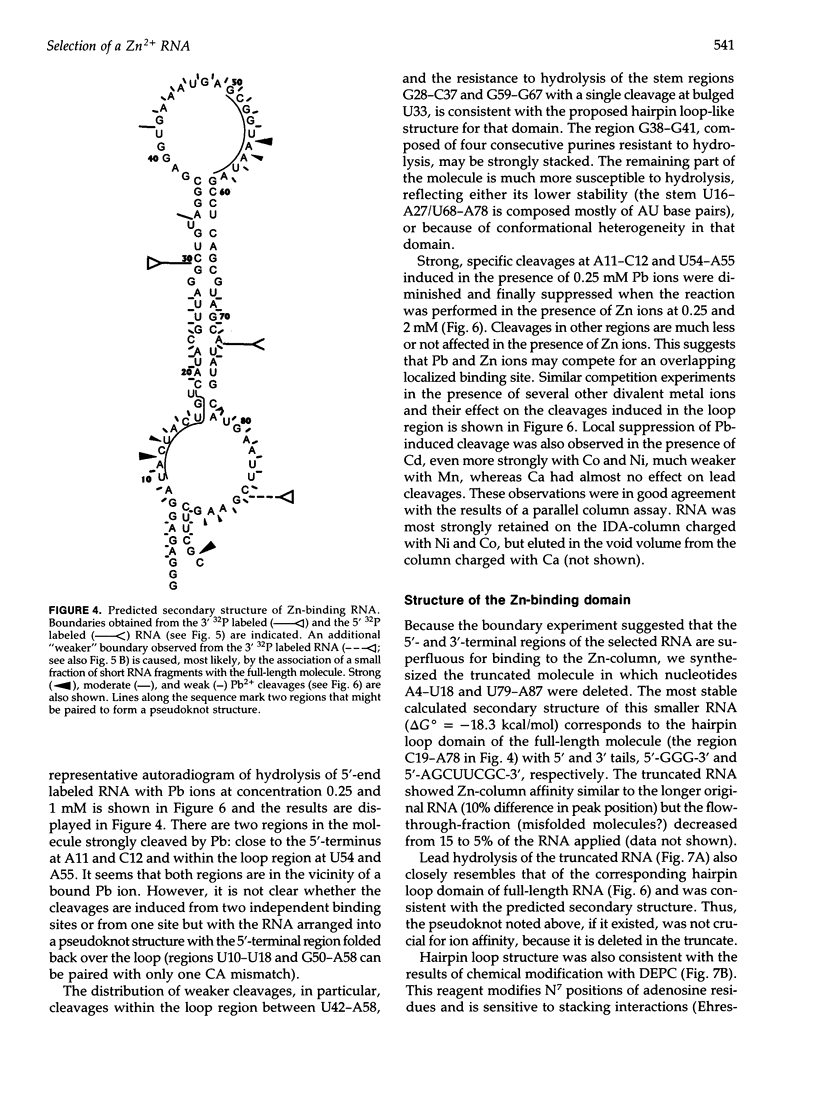
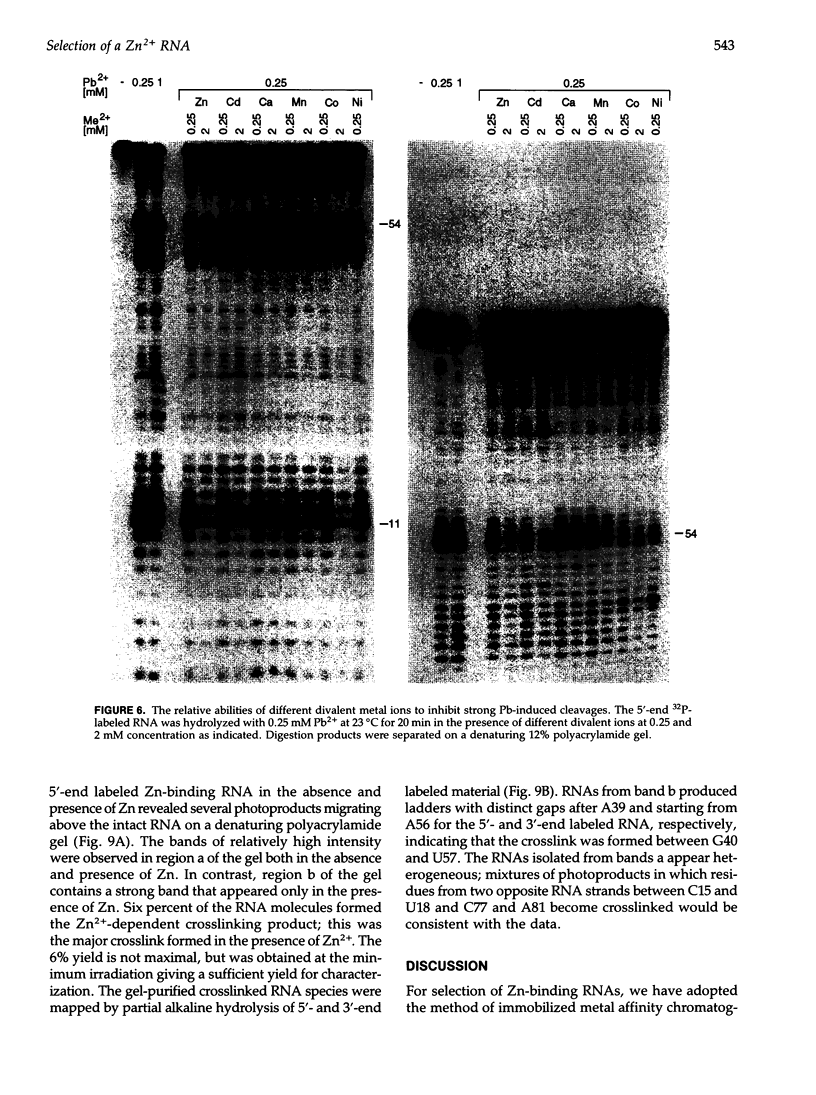
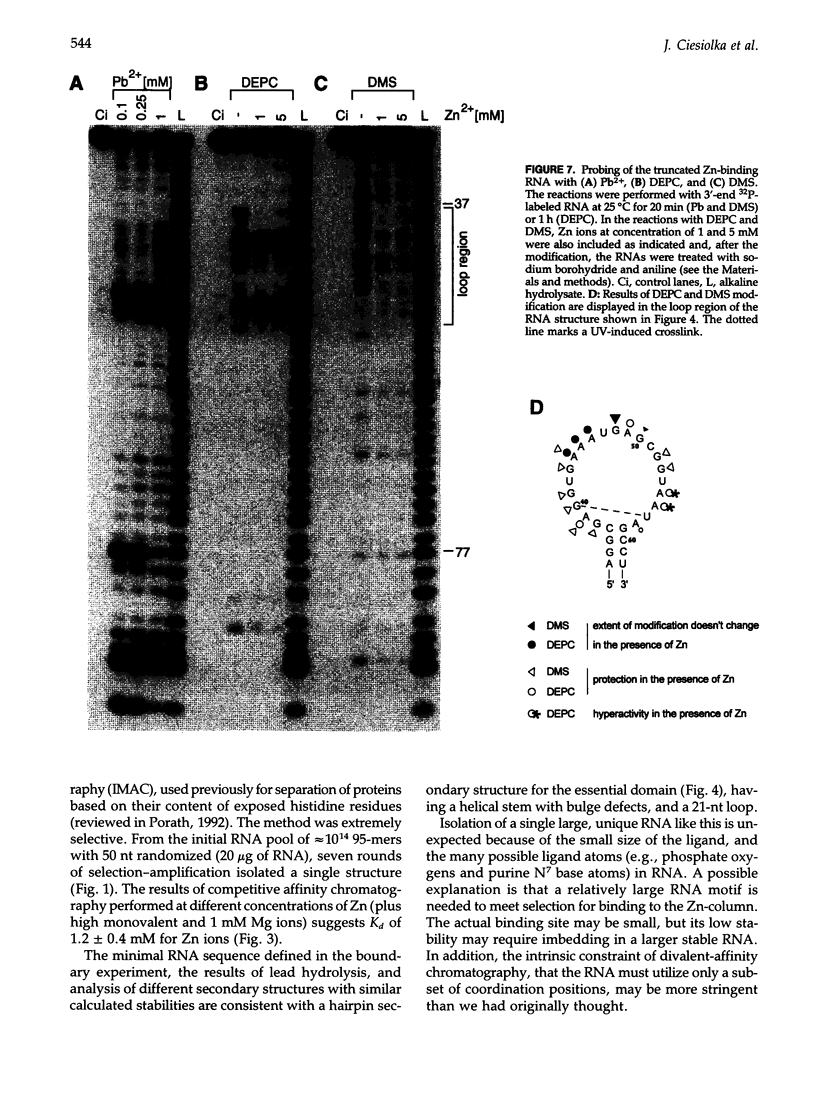
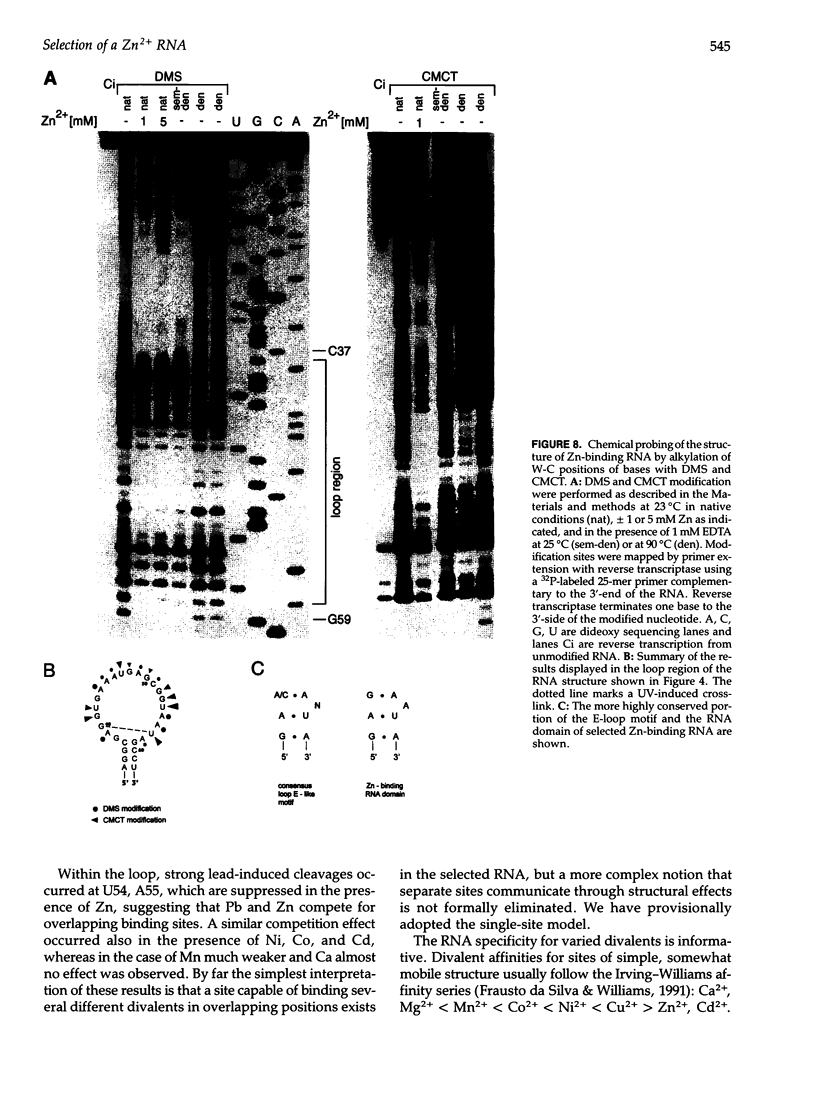
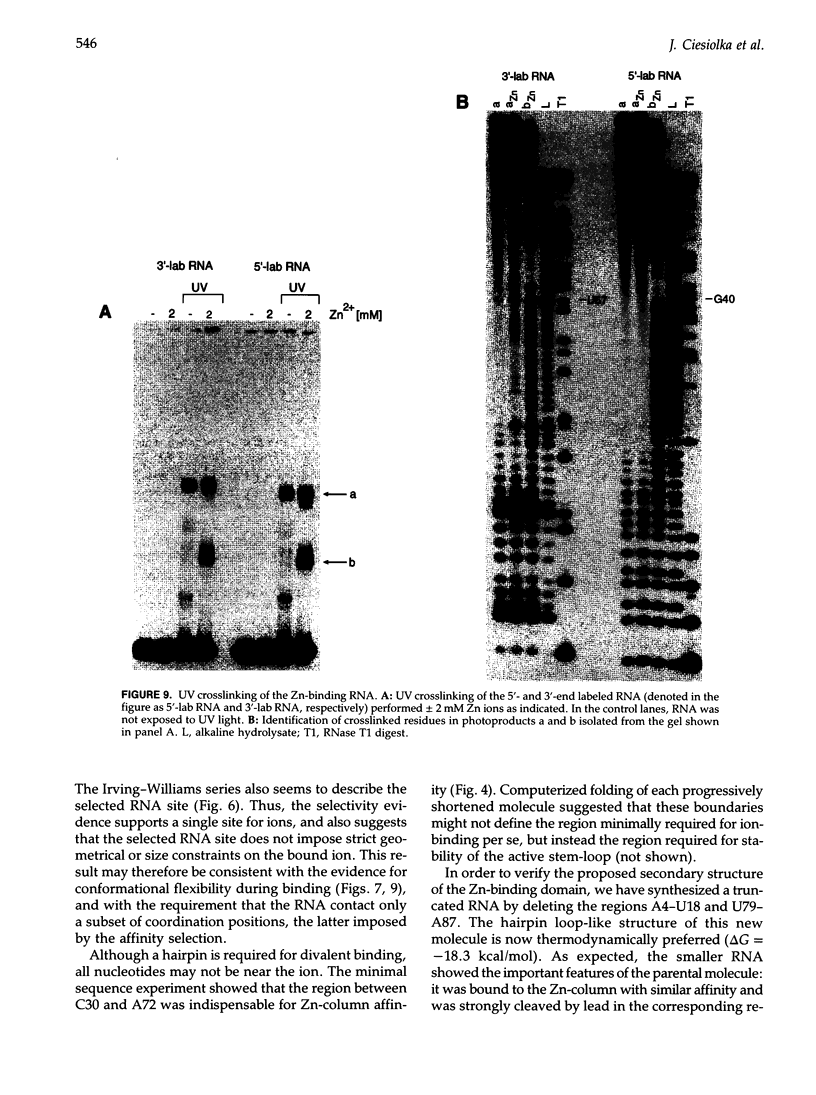
We have selected an RNA that depends on zinc for affinity to a column, starting from a pool of ribooligonucleotides with 50 randomized positions. This RNA's chemical sensitivities, calculated folding thermodynamics, and activity when fragmented suggest that an ion binding site lies within a complex 21-nt hairpin loop, near the junction with an imperfect helical stem. This RNA site has an unselected selectivity among divalents, preferring nickel, cobalt, and cadmium to calcium, magnesium, and manganese, as expected for a simple site of chelation. A moderate zinc-dependent change in loop structure accompanies divalent binding and can be detected by chemical probing and zinc-dependent UV-induced crosslinking. The latter also demonstrates the apposition of loop sequences to make a structure that may be related to the E-loop motif found in a number of other RNA molecules; the E-loop motif, accordingly, may be a divalent site.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allain F. H., Varani G. Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res. 1995 Feb 11;23(3):341–350. doi: 10.1093/nar/23.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlen L. S., Sampson J. R., DiRenzo A. B., Uhlenbeck O. C. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry. 1990 Mar 13;29(10):2515–2523. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Robertson H. D. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6590–6594. doi: 10.1073/pnas.82.19.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. S., Hingerty B. E., Dewan J. C., Klug A. Pb(II)-catalysed cleavage of the sugar-phosphate backbone of yeast tRNAPhe--implications for lead toxicity and self-splicing RNA. Nature. 1983 Jun 9;303(5917):543–546. doi: 10.1038/303543a0. [DOI] [PubMed] [Google Scholar]
- Butcher S. E., Burke J. M. A photo-cross-linkable tertiary structure motif found in functionally distinct RNA molecules is essential for catalytic function of the hairpin ribozyme. Biochemistry. 1994 Feb 1;33(4):992–999. doi: 10.1021/bi00170a018. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991 Jan 25;251(4992):401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
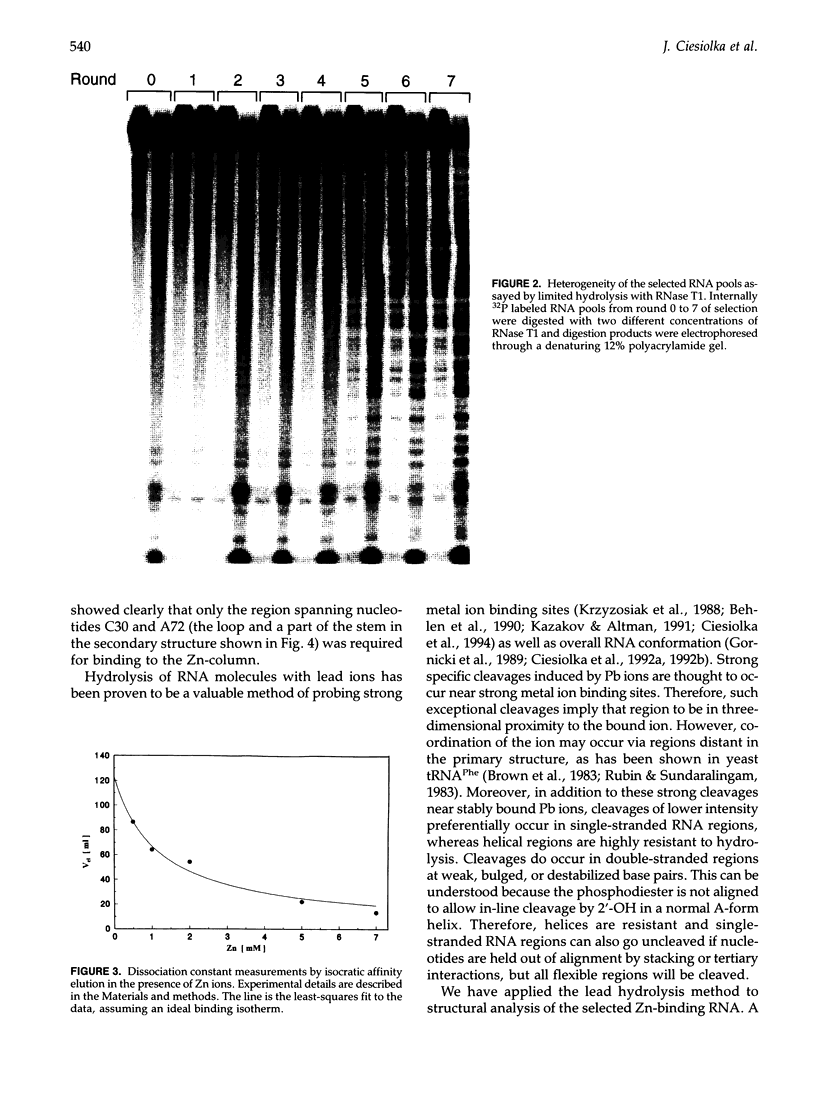
- Chowrira B. M., Berzal-Herranz A., Burke J. M. Ionic requirements for RNA binding, cleavage, and ligation by the hairpin ribozyme. Biochemistry. 1993 Feb 2;32(4):1088–1095. doi: 10.1021/bi00055a014. [DOI] [PubMed] [Google Scholar]
- Christian E. L., Yarus M. Metal coordination sites that contribute to structure and catalysis in the group I intron from Tetrahymena. Biochemistry. 1993 May 4;32(17):4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- Ciesiolka J., Hardt W. D., Schlegl J., Erdmann V. A., Hartmann R. K. Lead-ion-induced cleavage of RNase P RNA. Eur J Biochem. 1994 Jan 15;219(1-2):49–56. doi: 10.1111/j.1432-1033.1994.tb19913.x. [DOI] [PubMed] [Google Scholar]
- Ciesiołka J., Lorenz S., Erdmann V. A. Structural analysis of three prokaryotic 5S rRNA species and selected 5S rRNA--ribosomal-protein complexes by means of Pb(II)-induced hydrolysis. Eur J Biochem. 1992 Mar 1;204(2):575–581. doi: 10.1111/j.1432-1033.1992.tb16670.x. [DOI] [PubMed] [Google Scholar]
- Ciesiołlka J., Lorenz S., Erdmann V. A. Different conformational forms of Escherichia coli and rat liver 5S rRNA revealed by Pb(II)-induced hydrolysis. Eur J Biochem. 1992 Mar 1;204(2):583–589. doi: 10.1111/j.1432-1033.1992.tb16671.x. [DOI] [PubMed] [Google Scholar]
- Connell G. J., Illangesekare M., Yarus M. Three small ribooligonucleotides with specific arginine sites. Biochemistry. 1993 Jun 1;32(21):5497–5502. doi: 10.1021/bi00072a002. [DOI] [PubMed] [Google Scholar]
- Connell G. J., Yarus M. RNAs with dual specificity and dual RNAs with similar specificity. Science. 1994 May 20;264(5162):1137–1141. doi: 10.1126/science.7513905. [DOI] [PubMed] [Google Scholar]
- Dahm S. C., Uhlenbeck O. C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991 Oct 1;30(39):9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Dunn B. M., Chaiken I. M. Quantitative affinity chromatography. Determination of binding constants by elution with competitive inhibitors. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2382–2385. doi: 10.1073/pnas.71.6.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990 Aug 30;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Gornicki P., Baudin F., Romby P., Wiewiorowski M., Kryzosiak W., Ebel J. P., Ehresmann C., Ehresmann B. Use of lead(II) to probe the structure of large RNA's. Conformation of the 3' terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J Biomol Struct Dyn. 1989 Apr;6(5):971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- Hertel K. J., Uhlenbeck O. C. The internal equilibrium of the hammerhead ribozyme reaction. Biochemistry. 1995 Feb 7;34(5):1744–1749. doi: 10.1021/bi00005a031. [DOI] [PubMed] [Google Scholar]
- Jenison R. D., Gill S. C., Pardi A., Polisky B. High-resolution molecular discrimination by RNA. Science. 1994 Mar 11;263(5152):1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Kazakov S., Altman S. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9193–9197. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Ohtsuka E. Effects of phosphorothioate and 2-amino groups in hammerhead ribozymes on cleavage rates and Mg2+ binding. Biochemistry. 1991 May 28;30(21):5145–5150. doi: 10.1021/bi00235a005. [DOI] [PubMed] [Google Scholar]
- Krol A., Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- Krzyzosiak W. J., Marciniec T., Wiewiorowski M., Romby P., Ebel J. P., Giegé R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988 Jul 26;27(15):5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- Lorsch J. R., Szostak J. W. In vitro selection of RNA aptamers specific for cyanocobalamin. Biochemistry. 1994 Feb 1;33(4):973–982. doi: 10.1021/bi00170a016. [DOI] [PubMed] [Google Scholar]
- Majerfeld I., Yarus M. An RNA pocket for an aliphatic hydrophobe. Nat Struct Biol. 1994 May;1(5):287–292. doi: 10.1038/nsb0594-287. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Uhlenbeck O. C. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+. Biochemistry. 1992 Apr 28;31(16):3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Altman S. Pathway of activation by magnesium ions of substrates for the catalytic subunit of RNase P from Escherichia coli. J Mol Biol. 1993 Apr 5;230(3):750–756. doi: 10.1006/jmbi.1993.1197. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Labuda D., Usman N., Yang J. H., Cedergren R. Relationship between 2'-hydroxyls and magnesium binding in the hammerhead RNA domain: a model for ribozyme catalysis. Biochemistry. 1991 Apr 23;30(16):4020–4025. doi: 10.1021/bi00230a029. [DOI] [PubMed] [Google Scholar]
- Pley H. W., Flaherty K. M., McKay D. B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994 Nov 3;372(6501):68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Porath J. Immobilized metal ion affinity chromatography. Protein Expr Purif. 1992 Aug;3(4):263–281. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- Prudent J. R., Uno T., Schultz P. G. Expanding the scope of RNA catalysis. Science. 1994 Jun 24;264(5167):1924–1927. doi: 10.1126/science.8009223. [DOI] [PubMed] [Google Scholar]
- Romaniuk P. J., de Stevenson I. L., Ehresmann C., Romby P., Ehresmann B. A comparison of the solution structures and conformational properties of the somatic and oocyte 5S rRNAs of Xenopus laevis. Nucleic Acids Res. 1988 Mar 25;16(5):2295–2312. doi: 10.1093/nar/16.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J. R., Sundaralingam M. Lead ion binding and RNA chain hydrolysis in phenylalanine tRNA. J Biomol Struct Dyn. 1983 Dec;1(3):639–646. doi: 10.1080/07391102.1983.10507471. [DOI] [PubMed] [Google Scholar]
- Rubin J. R., Wang J., Sundaralingam M. X-ray diffraction study of the zinc(II) binding sites in yeast phenylalanine transfer RNA. Preferential binding of zinc to guanines in purine-purine sequences. Biochim Biophys Acta. 1983 Mar 15;756(1):111–118. doi: 10.1016/0304-4165(83)90030-2. [DOI] [PubMed] [Google Scholar]
- Sassanfar M., Szostak J. W. An RNA motif that binds ATP. Nature. 1993 Aug 5;364(6437):550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- Smith D., Pace N. R. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993 May 25;32(20):5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Tomka M., Kierzek R., Bevilacqua P. C., Turner D. H. Effects of substrate structure on the kinetics of circle opening reactions of the self-splicing intervening sequence from Tetrahymena thermophila: evidence for substrate and Mg2+ binding interactions. Nucleic Acids Res. 1989 Jan 11;17(1):355–371. doi: 10.1093/nar/17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczak A. A., Moore P. B., Chang Y. L., Wool I. G. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Wimberly B. A common RNA loop motif as a docking module and its function in the hammerhead ribozyme. Nat Struct Biol. 1994 Nov;1(11):820–827. doi: 10.1038/nsb1194-820. [DOI] [PubMed] [Google Scholar]
- Wimberly B., Varani G., Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993 Feb 2;32(4):1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- Yarus M. How many catalytic RNAs? Ions and the Cheshire cat conjecture. FASEB J. 1993 Jan;7(1):31–39. doi: 10.1096/fasebj.7.1.8422972. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]