Abstract
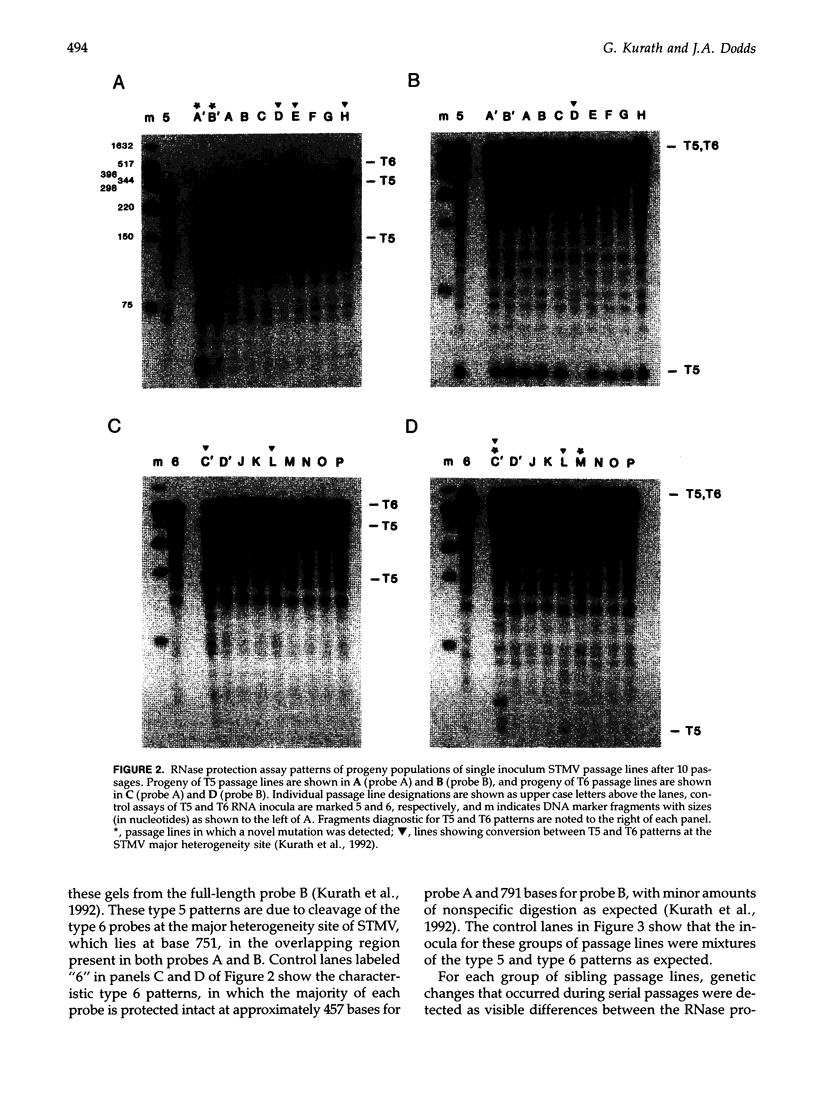
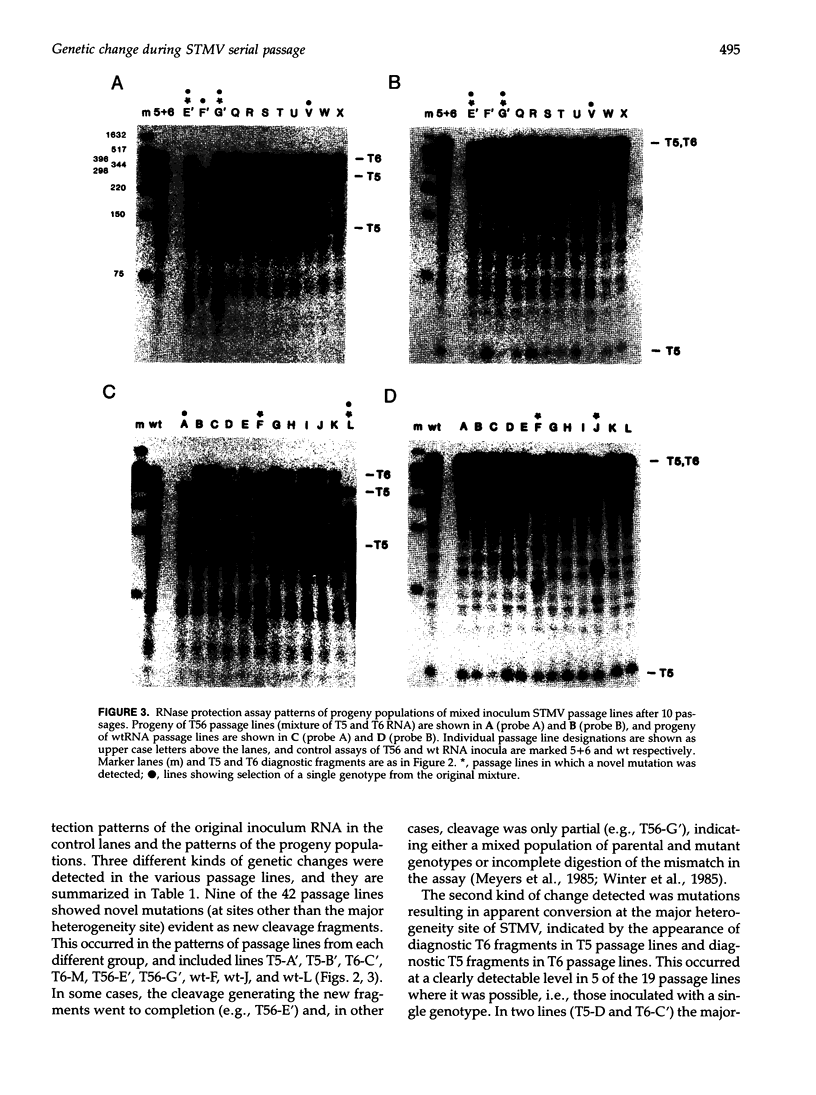
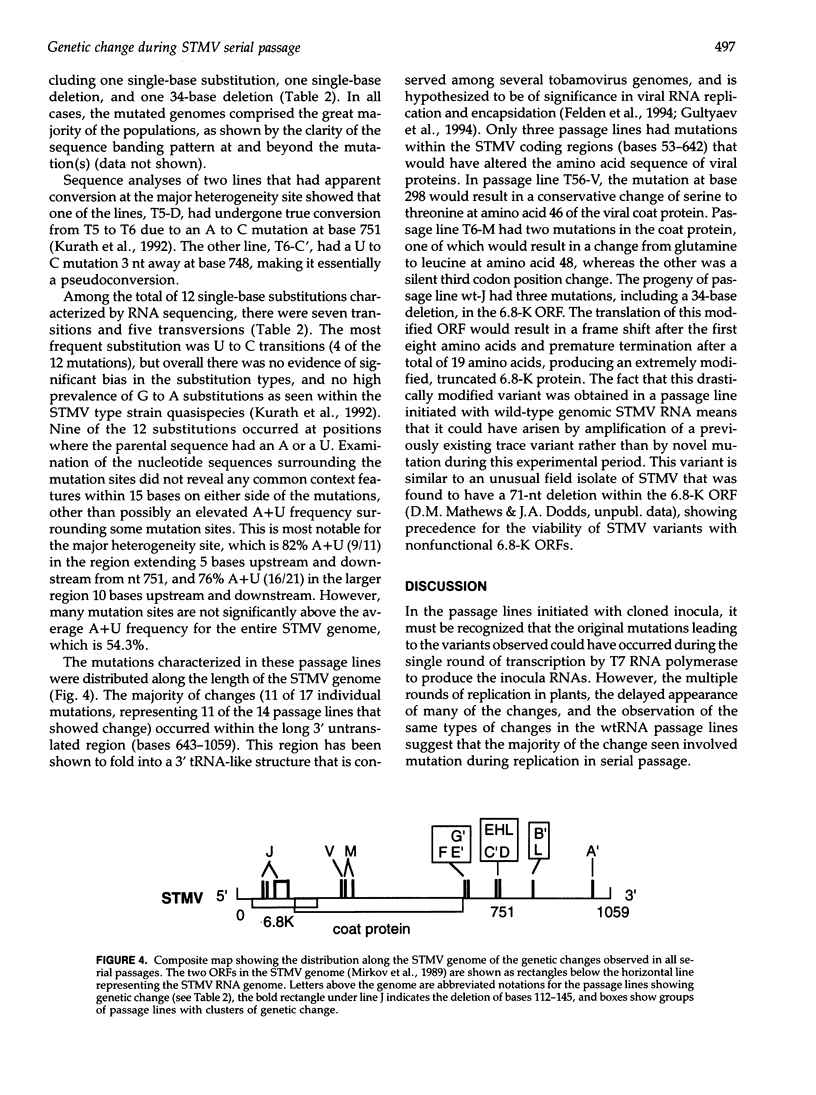
The high level of genetic diversity and rapid evolution of viral RNA genomes are well documented, but few studies have characterized the rate and nature of ongoing genetic change over time under controlled experimental conditions, especially in plant hosts. The RNA genome of satellite tobacco mosaic virus (STMV) was used as an effective model for such studies because of advantageous features of its genome structure and because the extant genetic heterogeneity of STMV has been characterized previously. In the present study, the process of genetic change over time was studied by monitoring multiple serial passage lines of STMV populations for changes in their consensus sequences. A total of 42 passage lines were initiated by inoculation of tobacco plants with a helper tobamovirus and one of four STMV RNA inocula that were transcribed from full-length infectious STMV clones or extracted from purified STMV type strain virions. Ten serial passages were carried out for each line and the consensus genotypes of progeny STMV populations were assessed for genetic change by RNase protection analyses of the entire 1,059-nt STMV genome. Three different types of genetic change were observed, including the fixation of novel mutations in 9 of 42 lines, mutation at the major heterogeneity site near nt 751 in 5 of the 19 lines inoculated with a single genotype, and selection of a single major genotype in 6 of the 23 lines inoculated with mixed genotypes. Sequence analyses showed that the majority of mutations were single base substitutions. The distribution of mutation sites included three clusters in which mutations occurred at or very near the same site, suggesting hot spots of genetic change in the STMV genome. The diversity of genetic changes in sibling lines is clear evidence for the important role of chance and random sampling events in the process of genetic diversification of STMV virus populations.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldaoud R., Dawson W. O., Jones G. E. Rapid, random evolution of the genetic structure of replicating tobacco mosaic virus populations. Intervirology. 1989;30(4):227–233. doi: 10.1159/000150096. [DOI] [PubMed] [Google Scholar]
- Baier M., Dittmar M. T., Cichutek K., Kurth R. Development of vivo of genetic variability of simian immunodeficiency virus. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8126–8130. doi: 10.1073/pnas.88.18.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Díez J., Martínez M. A., Hernández J., Holguín A., Borrego B., Mateu M. G. New observations on antigenic diversification of RNA viruses. Antigenic variation is not dependent on immune selection. J Gen Virol. 1993 Oct;74(Pt 10):2039–2045. doi: 10.1099/0022-1317-74-10-2039. [DOI] [PubMed] [Google Scholar]
- Domingo E., Martínez-Salas E., Sobrino F., de la Torre J. C., Portela A., Ortín J., López-Galindez C., Pérez-Breña P., Villanueva N., Nájera R. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance--a review. Gene. 1985;40(1):1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- Felden B., Florentz C., McPherson A., Giegé R. A histidine accepting tRNA-like fold at the 3'-end of satellite tobacco mosaic virus RNA. Nucleic Acids Res. 1994 Aug 11;22(15):2882–2886. doi: 10.1093/nar/22.15.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gultyaev A. P., van Batenburg E., Pleij C. W. Similarities between the secondary structure of satellite tobacco mosaic virus and tobamovirus RNAs. J Gen Virol. 1994 Oct;75(Pt 10):2851–2856. doi: 10.1099/0022-1317-75-10-2851. [DOI] [PubMed] [Google Scholar]
- Kearney C. M., Donson J., Jones G. E., Dawson W. O. Low level of genetic drift in foreign sequences replicating in an RNA virus in plants. Virology. 1993 Jan;192(1):11–17. doi: 10.1006/viro.1993.1002. [DOI] [PubMed] [Google Scholar]
- Keese P., Gibbs A. Plant viruses: master explorers of evolutionary space. Curr Opin Genet Dev. 1993 Dec;3(6):873–877. doi: 10.1016/0959-437x(93)90007-c. [DOI] [PubMed] [Google Scholar]
- Kurath G., Heick J. A., Dodds J. A. RNase protection analyses show high genetic diversity among field isolates of satellite tobacco mosaic virus. Virology. 1993 May;194(1):414–418. doi: 10.1006/viro.1993.1278. [DOI] [PubMed] [Google Scholar]
- Kurath G., Palukaitis P. Serial passage of infectious transcripts of a cucumber mosaic virus satellite RNA clone results in sequence heterogeneity. Virology. 1990 May;176(1):8–15. doi: 10.1016/0042-6822(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Kurath G., Rey M. E., Dodds J. A. Analysis of genetic heterogeneity within the type strain of satellite tobacco mosaic virus reveals variants and a strong bias for G to A substitution mutations. Virology. 1992 Jul;189(1):233–244. doi: 10.1016/0042-6822(92)90699-p. [DOI] [PubMed] [Google Scholar]
- Kurath G., Rey M. E., Dodds J. A. Tobamovirus helper specificity of satellite tobacco mosaic virus involves a domain near the 5' end of the satellite genome. J Gen Virol. 1993 Jul;74(Pt 7):1233–1243. doi: 10.1099/0022-1317-74-7-1233. [DOI] [PubMed] [Google Scholar]
- Luo G. X., Chao M., Hsieh S. Y., Sureau C., Nishikura K., Taylor J. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol. 1990 Mar;64(3):1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkov T. E., Mathews D. M., Du Plessis D. H., Dodds J. A. Nucleotide sequence and translation of satellite tobacco mosaic virus RNA. Virology. 1989 May;170(1):139–146. doi: 10.1016/0042-6822(89)90361-9. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Rao A. L., Hall T. C. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J Virol. 1993 Feb;67(2):969–979. doi: 10.1128/jvi.67.2.969-979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cerezo E., García-Arenal F. Genetic heterogeneity of the RNA genome population of the plant virus U5-TMV. Virology. 1989 Jun;170(2):418–423. doi: 10.1016/0042-6822(89)90432-7. [DOI] [PubMed] [Google Scholar]
- Skotnicki M. L., Mackenzie A. M., Gibbs A. J. Turnip yellow mosaic virus variants produced from DNA clones encoding their genomes. Arch Virol. 1992;127(1-4):25–35. doi: 10.1007/BF01309572. [DOI] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- Winter E., Yamamoto F., Almoguera C., Perucho M. A method to detect and characterize point mutations in transcribed genes: amplification and overexpression of the mutant c-Ki-ras allele in human tumor cells. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7575–7579. doi: 10.1073/pnas.82.22.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]




