Abstract
The thylakoid compartments of plant chloroplasts are a vital destination for copper. Copper is needed to form holo-plastocyanin, which must shuttle electrons between photosystems to convert light into biologically useful chemical energy. Copper can bind tightly to proteins, so it has been hypothesized that copper partitions onto ligand-exchange pathways to reach intracellular locations without inflicting damage en route. The copper metallochaperone Atx1 of chloroplast-related cyanobacteria (ScAtx1) engages in bacterial two-hybrid interactions with N-terminal domains of copper-transporting ATPases CtaA (cell import) and PacS (thylakoid import). Here we visualize copper delivery. The N-terminal domain PacSN has a ferredoxin-like fold that forms copper-dependent heterodimers with ScAtx1. Removal of copper, by the addition of the cuprous-ion chelator bathocuproine disulfonate, disrupts this heterodimer, as shown from a reduction of the overall tumbling rate of the protein mixture. The NMR spectral changes of the heterodimer versus the separate proteins reveal that loops 1, 3, and 5 (the carboxyl tail) of the ScAtx1 Cu(I) site switch to an apo-like configuration in the heterodimer. NMR data (2JNH couplings in the imidazole ring of 15N ScAtx1 His-61) also show that His-61, bound to copper(I) in [Cu(I)ScAtx1]2, is not coordinated to copper in the heterodimer. A model for the PacSN/Cu(I)/ScAtx1 complex is presented. Contact with PacSN induces change to the ScAtx1 copper-coordination sphere that drives copper release for thylakoid import. These data also elaborate on the mechanism to keep copper(I) out of the ZiaAN ATPase zinc sites.
Keywords: PacS, protein–protein interaction, ScAtx1, metallochaperone P-type ATPase
Most of the biosphere relies either directly or indirectly on effective copper delivery to the thylakoids of primary producers such as plants and cyanobacteria. Cyanobacterial thylakoids can be considered a bacterial precedent for copper trafficking to an internal compartment (1). Within the thylakoid lumen is located the copper protein plastocyanin, which is obligatory for photosynthetic electron transport within the chloroplasts of higher plants and optional in some cyanobacteria (2). Plastocyanin is imported into thylakoids as an unfolded protein (3), necessitating a separate copper supply to form the holoenzyme. Some cyanobacterial thylakoids also contain a caa3-type cytochrome c6 oxidase that requires three atoms of copper (1). The enzymatic demand for copper in thylakoids sets cyanobacteria apart from other bacteria, in which there is currently no unequivocally documented metabolic need for this metal to traverse the cytosol. Other bacteria do have copper requirements located outside or embedded within the plasma membrane. Ongoing studies of copper supply to particulate methane monooxygenase and for molybdopterin biosynthesis are also searching for evidence of cytosolic copper demand in other bacterial groups (4).
Analyses of mutants of the cyanobacterium Synechocystis PCC 6803 established that two copper-transporting P1-type ATPases, PacS and CtaA, plus a small soluble copper metallochaperone, Atx1 (ScAtx1), are required for normal photosynthetic electron transfer via plastocyanin and for the activity of a second, thylakoid-located copper protein, a caa3-type cytochrome oxidase (5, 6). In common with related orthologues from other bacteria, yeast, and human (7–10), ScAtx1 directly interacts in two hybrid assays (5) and in vitro studies (11) with the soluble, N-terminal domains of P1-type copper ATPases. However, unlike eukaryotic copper metallochaperones, which interact with a single ATPase, ScAtx1 associates with two such ATPases, PacS and CtaA. In Synechococcus PCC 7942, PacS is located in thylakoid membranes (12), whereas CtaA is thought to import copper at the plasma membrane (13). The phenotypes of ΔctaA and ΔpacS mutants of Synechocystis PCC 6803 are consistent with both ATPases transporting copper in an inward direction, one into the cytosol and then the other into the thylakoid lumen (5). This pathway provides an attractive system for studying the process of copper transfer between a copper metallochaperone and its partners.
There is considerable interest in understanding the processes at a molecular level by which copper is handed between partner proteins in cellular trafficking pathways and intracellular metal release is avoided. Recently, the NMR solution structure of ScAtx1 (11) and extended x-ray absorption fine structure data (14) revealed that loop 1, containing Cys ligands, and loop 5, containing His-61, are involved in copper coordination, forming a symmetrical copper-bridged [Cu(I)ScAtx1]2 homodimer. The third imidazole ligand is absent in related proteins from other organisms (15), and it is noted that the absence of a final β strand means that loop 5 becomes a carboxyl tail in ScAtx1 (11). In yeast, it has been proposed that the observed flexibility in loops 1 and 5 of Atx1 may provide a trigger for copper release and allow the metallochaperone to adapt to its two partners: downstream of CTR1 and upstream of Ccc2 (10). In the CopZ/Cu(I)/CopAb complex (CopAb being the second N-terminal domain of the CopA ATPase from Bacillus subtilis), loop 1 of CopZ takes the conformation of the apo form rather than that of the copper form (7), which indicates that copper is released from the metallochaperone to the ATPase. ΔcopZ mutants were subsequently shown to be copper-sensitive (8), consistent with a role in export rather than import. It is anticipated that any analogous mechanism for ScAtx1 has somehow been adapted to force copper transfer to the metallochaperone from one P1-type ATPase and release to another, and His-61 has been suggested to play a significant role to drive the two distinct copper-transfer mechanisms (1). Indeed, conversion of His-61 to Arg altered two-hybrid interaction with PacS but not with CtaA, implying that the residue at position His-61 can interact differently with the complementary surfaces of the two different copper ATPases (14).
Here we have determined the solution structure of the N-terminal region of PacS (PacSN) in its apo form and studied protein–protein interaction between apoPacSN and Cu(I)ScAtx1 to investigate the role of His-61 in the copper-transfer mechanism. We establish that apoPacSN is organized into a ferredoxin-like fold comprising residues 1–72, whereas residues 74–95, constituting the linker between the ferredoxin-like domain and the first transmembrane helix of PacS ATPase, are not structured and flexible. We also establish that apoPacSN forms a heterodimeric complex with Cu(I)ScAtx1 by breaking the homodimeric state of [Cu(I)ScAtx1]2. The copper ion is associated with PacSN in the protein complex rather than solely ScAtx1, with His-61 ceasing to contribute toward copper coordination and Glu-13 assisting intermolecular metal migration.
Results and Discussion
Solution Structure and Dynamics of apoPacSN.
The apo form of PacSN (95 aa) is essentially in a folded state, as indicated by the 1H-15N heteronuclear single quantum correlation (HSQC) spectrum showing a good spreading of NH signals. However, a number of peaks are clustered in the spectral region, which is typical of unfolded polypeptides (amide proton resonances clustered between 8 and 8.5 ppm). Of the expected 91 15N backbone amide resonances, 78 were observed and assigned. The missing resonances are located in loop regions and in the C-terminal tail. Resonance assignments are available at the BioMagResBank (www.bmrb.wisc.edu).
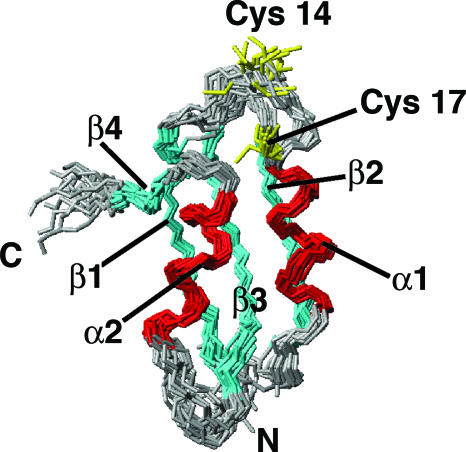
The solution structure of apoPacSN, as obtained by using 1,356 meaningful upper-distance limits and 86 angle restraints, has, after energy minimization, an rms deviation to the mean structure for protein backbone (for residues 4–70) of 0.63 Å (with a variability of 0.19 Å) and 1.08 Å (with a variability of 0.22 Å) for the heavy atoms. A statistical analysis of the apoPacSN structure is reported in Table 2, which is published as supporting information on the PNAS web site. The structure (shown in Fig. 1) has the classical ferredoxin-like fold, typical of metallochaperones and soluble N-terminal domains of heavy metal ATPases (16–19), featuring the secondary structure elements β1 (3–10), α1 (17–26), β2 (31–37), β3 (41–48), α2 (54–62), and β4 (66–68).
Fig. 1.
Solution structure of apoPacSN; backbone of the 20 lowest-energy conformers of apoPacSN (residues 1–72). The secondary structure elements are indicated: β-strands in cyan and α-helices in red. Copper-binding residues (Cys-14 and Cys-17) are shown in yellow.
The correlation time for molecular reorientation (τm), as estimated from the R2/R1 ratio, is 5.6 ± 0.4 ns, as expected for a protein of this size in a monomeric state (10). The heteronuclear relaxation parameters [R1, R2, and 1H-15N nuclear Overhauser effect (NOE) (Fig. 5, which is published as supporting information on the PNAS web site)] are mostly homogeneous along the polypeptide sequence from residues 1–70, indicating the presence of an essentially rigid and compact structure, with the exception of a few residues, mainly located in loops, which show the presence of conformational equilibria. The average relaxation values are 2.00 s−1 (with a variability of 0.17 s−1) for R1, 8.44 s−1 (with a variability of 1.71 s−1) for R2, and 0.75 s−1 (with a variability of 0.09 s−1) for 1H-15N NOEs, calculated excluding the C-terminal tail (residues 71–95). Conversely, the C-terminal tail, encompassing residues 71–95, is characterized by very low or negative 1H-15N NOE values. The spectral density-function analysis (Fig. 6, which is published as supporting information on the PNAS web site) confirms that apoPacSN protein behaves as a rigid body up to residue 70, while it shows that the C-terminal region is flexible, experiencing local motions on a nanosecond–picosecond time scale (faster than the overall protein-tumbling rate), as indicated by J(ωH) values significantly higher than average. In addition, because all of the NH signals of the C-terminal tail (residues 74–95) are clustered in the spectral region typical of unfolded polypeptides, the C-terminal tail of the protein must be fluctuating in solution with random coil conformations. Some motions on both nanosecond–picosecond and millisecond–microsecond time scales [as indicated by J(0) and J(ωH) values higher than the average (Fig. 6)] are also observed for a few residues located close to the metal-binding site (loops 1 and 3 and at the beginning of helix α1).
Interaction of apoPacSN with Copper(I).
ApoPacSN is able to bind 1 eq of copper(I), forming the Cu(I)PacSN adduct. However, this form is soluble only at low protein concentrations (<0.5 mM), whereas it precipitates at higher concentrations. The NMR spectra indicate that the protein maintains the same structure also in the presence of copper(I), with residues 1–70 in a folded environment, while the C-terminal tail remains in an unfolded state. The analysis of the average chemical shifts ‖ (Fig. 7, which is published as supporting information on the PNAS web site) reveals that the changes, still relatively small, are confined to residues 13–20, which contains the metal-binding ligands Cys-14 and Cys-17, whereas all of the other residues have negligible chemical-shift differences. The τm value of the copper-bound protein was found to be 6.7 ± 0.5 ns from the R2/R1 ratios, consistent with a slight aggregation of PacSN molecules after metal binding (τm of apoPacSN, 5.6 ± 0.4 ns).
Interaction Between ScAtx1, PacSN, and Copper(I).
Dynamical aspects.
The interaction between the copper chaperone ScAtx1 and the N-terminal metal-binding domain PacSN has been investigated by recording 1H-15N HSQC spectra on Cu(I)15NScAtx1 and apo15NPacSN samples at increasing concentrations (up to doubling) of the unlabeled partners, apoPacSN and Cu(I)ScAtx1, respectively. The two titrations show that (i) the apparent concentration of labeled protein decreases as its signals decrease in intensity, and (ii) a new set of signals appear and increase in intensity, indicating the presence of another species or of a mixture of species in rapid exchange. When the two proteins are mixed, the following equilibria can be operative:
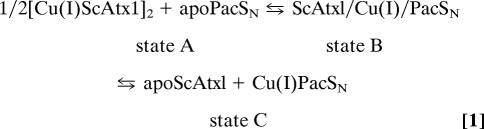 |
where Cu(I)ScAtx1 is in a homodimeric form. The first equilibrium in Eq. 1 is slow on the NMR time scale, because the intensity of the signals of Cu(I)ScAtx1 and apoPacSN decrease along both titrations [Cu(I)15NScAtx1 with unlabeled apoPacSN and apo15NPacSN with unlabeled Cu(I)ScAtx1], and those of the new set of signals increase. The second equilibrium of Eq. 1 must be fast on the NMR time scale, because the new sets of signals differ from those of the isolated apoScAtx1, Cu(I)ScAtx1, Cu(I)PacSN, or apoPacSN. Overall, therefore, the spectral changes and the relaxation data (see below) suggest that (i) ScAtx1 is interacting with PacSN and (ii) the new species represents a mixture of the ScAtx1/Cu(I)/PacSN heterodimer in fast exchange with the free apoScAtx1 and Cu(I)PacSN (states B + C). The lifetime of the adduct between Cu(I)ScAtx1 and apoPacSN is long relative to the difference between the chemical shift of the NHs of the mixture of states B and C on one side and those of the free forms of [Cu(I)ScAtx1]2 or apoPacSN on the other and has a lower limit in the range of milliseconds. This limit is obtained considering the maximum difference between chemical shifts of a given resonance, not affected by a significant line broadening, in the free protein with respect to that in the complex and was calculated from the apo15NPacSN/Cu(I)ScAtx1 titration. In the case of the interaction between yeast Atx1 and its partner Ccc2a, all of the equilibria in Eq. 1 are fast on the NMR time scale (10), with a lifetime <1 ms. The different behavior between yeast Atx1 interacting with Ccc2a compared with ScAtx1 interacting with PacSN (i.e., fast versus slow exchange) can be rationalized on the basis that copper is more easily accessible in the monomeric state of yeast Atx1, whereas it is largely buried in the homodimeric state of ScAtx1, which is maintained at concentrations as low as 0.1 mM (11). It remains to be established whether the protein is dimeric at concentrations found inside cells.
The relative intensity of the signals provides quantitative information on the distribution of the species involved in the first equilibrium. From the relative intensity of the two sets of signals, it is calculated that ≈70% of the proteins are in the states B + C when the two proteins are 1 mM and in a 1:1 ratio. Information on the species distribution in the second equilibrium can be obtained from relaxation data. Because the 15N longitudinal (R1) and transverse (R2) relaxation rates are very sensitive to the protein-tumbling rate (i.e., the correlation time τm), they provide precise information on the average molecular size. If the signals of ScAtx1 in the B + C mixture are analyzed, the relative amounts of apoScAtx1 and ScAtx1/Cu(I)/PacSN can be obtained. The experimental value of the average τm of ScAtx1 in states B + C is 7.1 ± 0.4 ns (Table 1), whereas a τm of ≈10 ns is expected for ScAtx1/Cu(I)/PacSN, because it results from the sum of the two individual τm values (4.3 + 5.6 ns). Therefore, the complex ScAtx1/Cu(I)/PacSN is present as 55% of the mixture B+C ** , whereas 45% is constituted by the free apoScAtx1 and Cu(I)PacSN proteins. The same values are obtained when the signals of PacSN in the B + C mixture are analyzed (Table 1). If the distribution of the species in the first equilibrium of Eq. 1 is also considered, at a 1:1 ratio ≈40% of each protein is in the ScAtx1/Cu(I)/PacSN complex and 60% in the isolated ScAtx1 and PacSN proteins (states A + C), half of which are metallated.
Table 1.
Average R1 and R2 relaxation rates (s−1) for amide 15N nuclei measured for apo and Cu(I)ScAtxl and apoPacSN from Synechocystis PCC 6803 and for Cu(I)ScAtxl in the presence of unlabeled apoPacSN and apoPacSN in the presence of unlabeled Cu(I)ScAtxl
| R1, s−1 | R2, s−1 | τm, ns | |
|---|---|---|---|
| ApoScAtxl (0.6 mM)* | 2.46 ± 0.07 | 7.09 ± 0.19 | 4.3 ± 0.3 |
| Cu(I)ScAtxl (0.8 mM)* | 1.66 ± 0.14 | 10.98 ± 0.40 | 7.6 ± 0.7 |
| ApoPacSN (1 mM)* | 2.00 ± 0.17 | 8.44 ± 1.71 | 5.6 ± 0.4 |
| Cu(I)ScAtxl in presence of apoPacSN at 1:1 ratio† | 2.42 ± 0.18 | 10.90 ± 0.22 | 7.1 ± 0.4 |
| Cu(I)ScAtxl in presence of apoPacSN at 1:1 ratio + 1.0 mM BCS† | 2.62 ± 0.20 | 7.36 ± 0.35 | 4.1 ± 0.4 |
| apoPacSN in presence of Cu(I)ScAtxl at 1:1 ratio* | 1.84 ± 0.41 | 11.65 ± 2.05 | 7.9 ± 0.9 |
Correlation time for molecular tumbling τm (ns) as estimated from the R2/R1 ratio is also reported.
*Performed at 600 MHz.
†Performed at 500 MHz.
Copper binding can be reversed by the addition of bathocuproine disulfonate, a strong Cu(I) chelator that is capable of extracting copper from the 1:1 ScAtx1/Cu(I)/PacSN complex. In the presence of 3 eq of bathocuproine disulfonate, the complex was converted into the two isolated apo proteins, resulting in colored bathocuproine disulfonate/copper(I) complex. The 2D 1H-15N HSQC spectrum of ScAtx1 is almost identical to the apo form, and the correlation time τm is similar to that of the monomeric apo state (Table 1), indicating that the two apo species are not interacting with each other once copper is sequestrated by a strong copper(I) chelator. This copper dependency is analogous to observations on yeast and human homologues in which copper is necessary for complex formation (20).
Structural aspects.
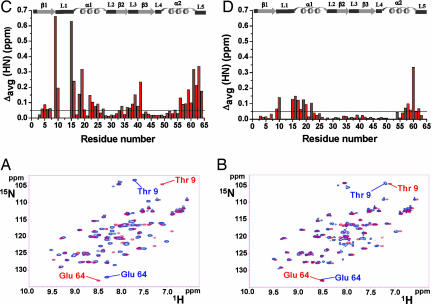
In both proteins the resonances of the residues closer to the metal-binding sites broaden beyond detection [in Cu(I)ScAtx1 residues 11–14 and in apoPacSN residues 13, 14, 17, and 19–21], suggesting that conformational equilibria are operative in loop 1 and helix α1 of both proteins, as already observed for other homologous interacting pairs (7, 10). From a comparison of the 1H-15N HSQC of free Cu(I)15NScAtx1 (Fig. 2A) or apo15NScAtx1 (Fig. 2B) (blue contours) with that of the ScAtx1/Cu(I)/PacSN, apoScAtx1, and Cu(I)PacSN mixture (red contours), it seems that the chemical-shift changes are mainly localized in stretches of 9–25, 38–41, and 56–64 residues, which constitute loops 1 and 3, helices α1 and α2, and the C terminus. The corresponding regions of yeast Atx1 and bacterial CopZ are involved in protein–protein contacts with their partners (7, 10). Furthermore, the 1H and 15N resonances of ScAtx1 in the complex are closer to those of the apoScAtx1 form than to those of Cu(I)ScAtx1. This is clearly evident from the NH chemical shifts of loop 3 (residues 38–41), located in close contact with the metal-binding site, which in the complex are very similar to those observed in apoScAtx1 but different from those of Cu(I)ScAtx1 (Fig. 2). These data suggest that ScAtx1 in the complex is structurally similar to the apo form.
Fig. 2.
Cu(I)15NScAtx1 protein interaction with unlabeled apoPacSN. (A and B) Superposition of 1H-15N HSQC spectra at 298 K of Cu(I)ScAtx1 (A) or apoScAtx1 (B) (blue contours) with ScAtx1/Cu(I)/PacSN, apoScAtx1, and Cu(I)PacSN mixture (red contours) at a 1:1 ratio. (C and D) NHs of Thr-9 and Glu-64 close in sequence to loop 1 and loop 5, respectively, are indicated. The weighted-average chemical-shift differences Δavg(HN) (see Results and Discussion) between Cu(I)ScAtx1 (C) or apoScAtx1 (D) and the 1:1 ScAtx1/Cu(I)/PacSN, apoScAtx1, and Cu(I)PacSN mixture are shown. Chemical-shift differences are not reported for residues 11–14, because their 1H-15N crosspeaks are not observed in the presence of PacSN. The secondary structure elements of Cu(I)ScAtx1 are reported along the top.
This picture is confirmed when the other partner, PacSN, is analyzed. Fig. 8, which is published as supporting information on the PNAS web site, shows the 1H-15N HSQC spectra of the isolated apo15NPacSN protein or Cu(I)15NPacSN (blue contours) overlaid onto that of the ScAtx1/Cu(I)/PacSN, apoScAtx1, and Cu(I)PacSN mixture containing equal concentrations of the two proteins (red contours). Unlike ScAtx1, no dramatic differences between the 1H and 15N resonances of PacSN in the complex and those of free apo or Cu(I)PacSN forms are observed. However, the major changes are still localized in loop 1 and helix α1 (residues 13–24) and helix α2 and loop 5 (residues 61–64) (Fig. 8), similar to what was observed in the yeast Atx1/Ccc2a complex (10). In particular, the chemical shifts of Ala-18, which is the only residue close to the metal-binding motif whose NH is still detectable in the complex, differs in the complex from that of the isolated apo and Cu(I)PacSN (Fig. 8).
The copper-binding site in ScAtx1 involves two Cys (12 and 15, located in loop 1 and helix α1, respectively) and His-61 in loop 5 (the carboxyl tail). This was established on the basis of the solution NMR structure and 2J 1H-15N HSQC experiments focused on His-61 (11), in agreement with previous extended x-ray absorption fine structure data (14). The solution structure also indicated a dimeric state for the copper form, where the coordinating residues of two copper ions are provided by both protein molecules of the dimer (11). Addition of apoPacSN to Cu(I)ScAtx1 produces the detachment of His-61 from copper as revealed in 2JNH 1H-15N HSQC experiments performed during a titration. In a 1:1 protein mixture, a broad pattern in the 2JNH 1H-15N HSQC spectrum appears, the chemical shift of which is similar to that observed for His-61 in apoScAtx1 (Fig. 9, which is published as supporting information on the PNAS web site). These data suggest that His-61 of ScAtx1 ceases to be involved in copper coordination in the ScAtx1/Cu(I)/PacSN complex, consistent with the interaction of copper(I) with PacSN in the complex.
haddock Structural Model.
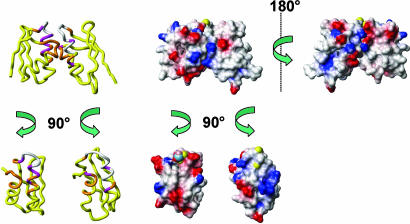
On the basis of the spectral changes observed after interaction of one protein with the partner, a structural model of the complex was calculated (Fig. 3) using Cu(I)ScAtx1 and apoPacSN as starting structures. This adduct can be taken as the first step in the interaction between the two proteins. In the complex, ScAtx1 and PacSN maintain their global fold with an average value of buried-contact surface area ranging from 1,320 to 1,050 Å2 within all calculated structures. The interacting region involves mainly loops 1 and 5 and helices α1 and α2 on both proteins. From the model (Fig. 4), it clearly emerges that copper(I) is shared by Cys residues of PacSN and ScAtx1, whereas His-61, which is not involved in metal coordination, moves very far from copper (at 6.8 Å) as a consequence of the presence of the interacting PacSN. Intermolecular Cys-bridged copper binding, together with the concomitant release of His-61, is therefore one of the key driving forces for the interaction between the two proteins and copper transfer.
Fig. 3.
A model for the ScAtx1-PacSN heterodimer exposes a mechanism of copper release from ScAtx1 to PacSN. (Left) The structure reports the spectral changes after protein interaction. The color code is as follows: magenta, Δavg(HN) > 0.1 ppm; orange, 0.05 < Δavg(HN) < 0.1 ppm; yellow, Δavg(HN) < 0.05 ppm; white, not observed. (Right) The electrostatic surface of the Cu(I) form of ScAtx1 and apoPacSN. The positively charged, negatively charged, and neutral amino acids are represented in blue, red, and white, respectively. Two views of the molecules rotated by 180° and 90° are shown to allow for viewing of the interaction surfaces. The Sγ sulfurs of Cys residues and copper(I) ion are shown in yellow and blue, respectively.
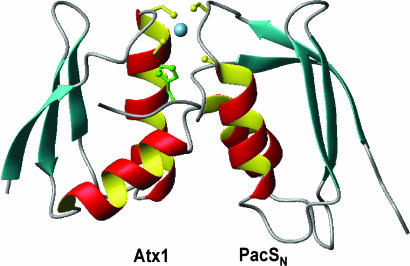
Fig. 4.
The lowest-energy structure of the ScAtx1-PacSN complex showing the copper(I) ligands. His-61 of ScAtx1 is shown in green. It is evident that His-61 moves away from the copper site in the complex.
In addition to copper binding, the complex is stabilized further by a network of intermolecular hydrogen bonds and salt bridges, involving both charged and polar amino acids at the interface between the two proteins. The identity and the number of less conserved hydrogen bonds may vary from one run to another, but some electrostatic interactions are consistently found in the 20 lowest-energy structures. These interactions occur between the side chains of Asn-25, Glu-26, Lys-21, and Ser-58 of ScAtx1 with the side chains of Lys-27, Asp-58, and Arg-62 of PacSN and between the backbone carbonyl of Ala-57 and Ser-58 of ScAtx1 with the side chains of Arg-23 and Arg-62 of PacSN. These interactions may optimize the relative orientation of the two proteins to allow a close contact between the two metal-binding regions for efficient metal transfer. In this respect, the interaction mode of ScAtx1 and PacSN is similar to the analogous yeast complex (21), in which there is a similar average contact area, ranging from 1,302 to 1,123 Å2. However, in the Atx1/Ccc2a model, the Eelec contribution (−480 kcal·mol−1) to Einter intermolecular energy is much higher than in ScAtx1/PacSN (Table 3, which is published as supporting information on the PNAS web site). The complementary charge on the surfaces of the yeast homologues (10) is indeed much more prominent with respect to the bacterial ScAtx1 and PacSN (Fig. 3), thus contributing more to the stabilization of the protein–protein complex. This distinction is consistent with a greater proportion of heterodimer with the yeast proteins. An intermolecular hydrogen bond was consistently detected between Oε of Glu-13 in ScAtx1 and SHγ of PacSN Cys-14. Substitution of Glu-13 in ScAtx1 with a residue that does not form hydrogen bonds (Ala) or that will not abstract a proton (Gln) impairs two-hybrid interactions (bacterial two-hybrid assays are described in Supporting Text and Fig. 10, which are published as supporting information on the PNAS web site).
Conclusions
We have structurally characterized the N-terminal metal-binding region of the copper(I)-ATPase PacS in its apo state and found that it forms a ferredoxin-like fold typical of this class of proteins (Fig. 1). Conformational changes that occur after formation of ScAtx1/Cu(I)/PacSN heterodimers have been monitored in both partners, and they are most extensive for ScAtx1 (Figs. 2 and 8). The chemical environments of residues forming loops 1, 3, and 5 (the carboxyl tail) of Cu(I)ScAtx1, the three regions that are proximal to the copper(I)-binding site, undergo the most remarkable change. In the heterodimer, loops 1 and 3 of ScAtx1 are more similar in conformation to those of apoScAtx1, consistent with Cu(I)ScAtx1 switching to an apo-like form after contact with the soluble domain of the ATPase, PacS. Critically, it emerges from NMR signals (Fig. 9) and the adduct model (Fig. 4) that His-61 of ScAtx1 is removed from the metal coordination sphere after interaction with PacSN as a steric consequence of the interaction with PacSN. In addition, Glu-13 of ScAtx1 forms a hydrogen bond with Cys-14 of PacSN in the complex. Displacement of His-61 imidazole provides a trigger for copper transfer.
These data imply that there is a molecular mechanism to produce a favored vector for copper transfer that is from the metallochaperone to the PacS ATPase. In other metallochaperone–ATPase interactions that have been studied, displacement of loop 5 has been observed, and it has been proposed that it could open the metal-binding site to aid ligand invasion from the presumed acceptor (10). Unlike Atx1 from Synechocystis PCC 6803 these proteins do not have a third ligand on loop 5. It is now clearly shown how displacement of His-61 provides a mechanism for copper release from the metallochaperone and donation to the ATPase (Fig. 4). This more extreme variant of the release mechanism could allow this metallochaperone to deliver copper to one ATPase and also acquire it from another. A prediction is that His-61 will not be displaced from the ligand sphere during interaction with CtaAN if the vector for transfer is to be reversed such that copper is loaded onto ScAtx1 from this ATPase. CtaAN has proved refractory to purification, so this latter hypothesis remains untested.
In addition to PacS and CtaA, Synechocystis PCC 6803 also contains a P1-type ATPase, ZiaA, that transports zinc and not copper (22) but has an N-terminal domain with higher affinity for copper(I) than for zinc (23). A subdomain of ZiaA (ZiaAN) is predicted to form a ferredoxin-like fold analogous to the N-terminal regions of the two copper transporters, but ScAtx1 gave no detectable two-hybrid interaction with neither this subdomain of ZiaA (23) nor the entire soluble N-terminal region of ZiaA (5). Provided that there is no freely available cytosolic copper in Synechocystis PCC 6803, as implied for Escherichia coli (24), the lack of ScAtx1–ZiaAN interaction constitutes a kinetic barrier to discourage formation of otherwise thermodynamically favored copper-ZiaAN complexes (23) while copper traffics to the thylakoid. It is notable that the correlation time for molecular tumbling τm decreases when copper is removed by bathocuproine disulfonate (Table 1), implying that copper encourages interaction between ScAtx1 and PacSN, and it is predicted that this will not apply to ScAtx1–ZiaAN. We are now aware of another level of complexity, because if ZiaAN were to acquire copper from ScAtx1, it would not only have to form a complex but it would also need to (i) induce the conformational change caused by PacSN to displace His-61 from the ligand sphere, to convert ScAtx1 into an apo-like form, and (ii) precisely position Glu-13 to allow hydrogen bonding to one of the accepting thiol ligands. Notably, Asp-16 on ZiaAN is likely to repulse Glu-13 such that it will not orientate proximally to ZiaAN thiols.
The concept that metallochaperones deliver copper to intracellular destinations has been widely reported and reviewed (4, 25–27). In eukaryotes, the copper chaperone for superoxide dismutase 1 (CCS) is thought to deliver copper to superoxide dismutase 1 (SOD1), COX17 to mitochondria, and ATOX1 to the trans-Golgi network. Evidence that yeast SOD1 has femtomolar affinity for copper but remains inactive in the absence of CCS supports the notion that copper is supplied by the metallochaperone (28). However, it was established more recently that CCS activates SOD1, at least in part, via formation of an essential disulfide (29, 30), and CCS is not obligatory for SOD1 activation in some species (31). Furthermore, COX17 is now known to be functional when tethered to the mitochondrial inner membrane (32). Thus, some of the evidence for intracellular copper delivery by metallochaperones has become more equivocal. The data presented here have established that ScAtx1 undergoes subtle conformational changes after interaction with its presumptive copper-accepting partner, which is fully consistent with copper release from the metallochaperone, providing timely support for the hypothesis that this protein has evolved to deliver copper to the thylakoid copper importer.
Materials and Methods
PacSN and ScAtx1 Protein Expression and Purification.
Recombinant PacSN protein was expressed in E. coli BL21(DE3) cells with M9 minimal medium supplemented with [13C]glucose and/or (15NH4)2SO4 used to generate labeled protein. PacSN protein was purified by using a 5-ml HiTrap Q XL column eluted with 25 mM Tris (pH 9)/50 mM NaCl. PacSN was concentrated and further purified on a HiLoad 16/60 Superdex 75 column equilibrated in 25 mM Tris (pH 9). Proteins were kept in anaerobic conditions, and buffer was changed by ultrafiltration against sodium phosphate (pH 7) with the addition of reducing agent DTT to 4 mM. Protein purification was monitored through gel electrophoresis. The Cu(I)PacSN protein was prepared by adding 1 eq of [Cu(I)-(CH3CN)4]PF6 in CH3CN to a solution of the reduced apoprotein in 100 mM phosphate buffer (pH 7). No exogenous thiols were added to the sample buffer. Isotopic labeling, protein expression, purification, and metallation of ScAtx1 were performed as described in ref. 11.
NMR Experiments and Structure Calculations.
The NMR experiments, collected at fields ranging from 18.8 to 11.7 T (from 800 to 500 MHz) (reported in Table 4, which is published as supporting information on the PNAS web site), were recorded on 13C/15N- and 15N-labeled samples of apoPacSN at 298 K. An automated candid approach (33) was used to assign the ambiguous NOE crosspeaks. Structure calculations then were performed through iterative cycles of dyana (34) followed by restrained energy minimization with amber 8.0 (35) applied to each member of the family. The quality of the structures was evaluated by using the program procheck-nmr (36).
NMR Titration of the Two Proteins.
Titrations of Cu(I)15NScAtx1 with unlabeled apoPacSN and apo15NPacSN with unlabeled Cu(I)ScAtx1 were performed following the NMR spectral changes in the 1H-15N HSQC spectra after the addition of increasing amounts of the unlabeled partner. Aliquots were added in a Coy chamber under a nitrogen atmosphere at 298 K. Two-dimensional total correlation spectroscopy and NOESY and three-dimensional NOESY-15N HSQC experiments were recorded on the 15NScAtx1/Cu(I)/PacSN and 15NPacSN/Cu(I)/ScAtx1 mixtures with protein concentration ratios of 1:0.5 and 1:1, respectively. To identify the coordination mode of copper(I)-binding histidine during the titration steps of Cu(I)15NScAtx1 with unlabeled apoPacSN, a 1H-15N HSQC experiment was performed for measuring 2JNεHδ, 2JNεHε, 2JNδHε, and 3JNδHδ coupling constants (37). In this experiment, the INEPT (insensitive nuclei enhanced by polarization transfer) delay was set to 22 ms.
Relaxation Measurements and Analysis.
15N R1, R2, and steady-state heteronuclear NOE measurements were performed at 14.1 and 11.7 T (600 and 500 MHz, respectively) at 298 K using already-reported pulse sequences (38) on the isolated apo and Cu(I)PacSN and the 15NScAtx1/Cu(I)/PacSN and ScAtx1/Cu(I)/15NPacSN mixtures. In all experiments, the water signal was suppressed with a “water flip-back” scheme (39). Relaxation rates R1 and R2 were determined by fitting the crosspeak intensities measured as a function of the delay within the pulse sequence to a single-exponential decay. Errors on the rates were estimated through a Monte Carlo approach. The heteronuclear NOE values were obtained from the ratio of the peak intensity for 1H-saturated and unsaturated spectra. The experimental relaxation rates were used to map the spectral density-function values, J(ωH), J(ωN), and J(0), following a procedure as described (40).
The overall rotational correlation time τm values were estimated from the R2/R1 ratio by using the program quadric_diffusion (41, 42). The relaxation data of those NHs having an exchange contribution to the R2 value or exhibiting large-amplitude internal motions, as monitored by low NOE values, were excluded from this analysis, because inclusion of these data would bias the calculated tensor parameters (43, 44).
haddock Docking Calculations.
haddock calculations were performed with standard protocols [as described at www.nmr.chem.uu.nl/haddock/haddock.html (45)] to obtain a model of the complex between PacSN and ScAtx1. The “active” residue and “passive” residues were selected on the basis of mapping the backbone chemical-shift changes following a procedure similar to that reported in ref. 21. Solvent-accessible residues whose crosspeaks in NMR spectra disappeared after complex formation were also considered active. Eight amino acids of ScAtx1 and seven amino acids of PacSN were used as active ambiguous interaction restraints. Around these amino acids, we defined five passive amino acids for ScAtx1 and seven for PacSN. The copper ion was linked to the sulfur atoms of ScAtx1 by including additional distance-unambiguous restraints of 2.3 ± 0.2 Å, whereas His-61 was not, because 2J 1H-15N HSQC experiments clearly show that it is detached from copper(I) coordination when ScAtx1 interacts with PacSN. The docking calculations were performed by using as input the average minimized NMR structures of the apoPacSN and the monomeric form of Cu(I)ScAtx1. The final 100 calculated structures results all converge to a single ensemble of conformations, displaying a pairwise rms deviation <1.5 Å. The buried interaction surface area was calculated by taking the difference between the solvent-accessible surface area for both individual partners and the solvent-accessible area of the complex. The 20 lowest-energy structures were analyzed in terms of intermolecular contacts (hydrogen bonds and nonbonded contacts).
Supplementary Material
Acknowledgments
G.A.S. and N.G.K. acknowledge the Leonardo Da Vinci Community Action Programme (University of Patras, 2003–2005) for financial support. This work was supported by European Community SPINE (Structural Proteomics in Europe) Grant QLG2-CT-2002-00988 and MIUR COFIN (Cofinanziamento del Ministero dell’Instruzione dell’ Università e della Ricerca) 2003 “Il ruolo degli ioni metallici nei processi metabolici.” N.J.R. received financial support from the Biotechnology and Biological Sciences Research Council.
Abbreviations
- HSQC
heteronuclear single quantum correlation
- NOE
nuclear Overhauser effect
- SOD1
superoxide dismutase 1
- CCS
copper chaperone for SOD1.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structural restraints for apoPacSN have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2GCF).
This paper was submitted directly (Track II) to the PNAS office.
The weighted-average chemical-shift differences were estimated by using the formula Δavg(HN) = {[(ΔH)2 + (ΔN/5)2]/2}½, where ΔH and ΔN are chemical-shift differences for 1H and 15N, respectively.
The species distribution calculation takes into account that the experimental τm value is given by [10 × fraction of ScAtx1/Cu(I)/PacSN + 4.3 × fraction of apoScAtx1].
References
- 1.Cavet J. S., Borrelly G. P., Robinson N. J. FEMS Microbiol. Rev. 2003;27:165–181. doi: 10.1016/S0168-6445(03)00050-0. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L., McSpadden B., Pakrasi H. B., Whitmarsh J. J. Biol. Chem. 1992;267:19054–19059. [PubMed] [Google Scholar]
- 3.Bogsch E., Brink S., Robinson C. EMBO J. 1997;16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tottey S., Harvie D. R., Robinson N. J. Acc. Chem. Res. 2005;38:775–783. doi: 10.1021/ar0300118. [DOI] [PubMed] [Google Scholar]
- 5.Tottey S., Rondet S. A., Borrelly G. P., Robinson P. J., Rich P. R., Robinson N. J. J. Biol. Chem. 2002;277:5490–5497. doi: 10.1074/jbc.M105857200. [DOI] [PubMed] [Google Scholar]
- 6.Tottey S., Rich P. R., Rondet S. A. M., Robinson N. J. J. Biol. Chem. 2001;276:19999–20004. doi: 10.1074/jbc.M011243200. [DOI] [PubMed] [Google Scholar]
- 7.Banci L., Bertini I., Ciofi-Baffoni S., Del Conte R., Gonnelli L. Biochemistry. 2003;42:1939–1949. doi: 10.1021/bi027096p. [DOI] [PubMed] [Google Scholar]
- 8.Radford D. S., Kihlken M. A., Borrelly G. P. M., Horwood C. R., Le Brun N. E., Cavet J. S. FEMS Microbiol. Lett. 2003;220:105–112. doi: 10.1016/S0378-1097(03)00095-8. [DOI] [PubMed] [Google Scholar]
- 9.Wernimont A. K., Huffman D. L., Lamb A. L., O’Halloran T. V., Rosenzweig A. C. Nat. Struct. Biol. 2000;7:766–771. doi: 10.1038/78999. [DOI] [PubMed] [Google Scholar]
- 10.Arnesano F., Banci L., Bertini I., Cantini F., Ciofi-Baffoni S., Huffman D. L., O’Halloran T. V. J. Biol. Chem. 2001;276:41365–41376. doi: 10.1074/jbc.M104807200. [DOI] [PubMed] [Google Scholar]
- 11.Banci L., Bertini I., Borrelly G. P. M., Ciofi-Baffoni S., Robinson N. J., Su X. C. J. Biol. Chem. 2004;279:27502–27510. doi: 10.1074/jbc.M402005200. [DOI] [PubMed] [Google Scholar]
- 12.Kanamaru K., Kashiwagi S., Mizuno T. Mol. Microbiol. 1994;13:369–377. doi: 10.1111/j.1365-2958.1994.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 13.Phung L. T., Ajlani G., Haselkorn R. Proc. Natl. Acad. Sci. USA. 1994;91:9651–9654. doi: 10.1073/pnas.91.20.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrelly G. P. M., Blindauer C. A., Schmid R., Butler C. S., Cooper C. E., Harvey I., Sadler P. J., Robinson N. J. Biochem. J. 2004;378:293–297. doi: 10.1042/BJ20031669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnesano F., Banci L., Bertini I., Ciofi-Baffoni S., Molteni E., Huffman D. L., O’Halloran T. V. Genome. Res. 2002;12:255–271. doi: 10.1101/gr.196802. [DOI] [PubMed] [Google Scholar]
- 16.Banci L., Bertini I., Ciofi-Baffoni S., Huffman D. L., O’Halloran T. V. J. Biol. Chem. 2001;276:8415–8426. doi: 10.1074/jbc.M008389200. [DOI] [PubMed] [Google Scholar]
- 17.Arnesano F., Banci L., Bertini I., Huffman D. L., O’Halloran T. V. Biochemistry. 2001;40:1528–1539. doi: 10.1021/bi0014711. [DOI] [PubMed] [Google Scholar]
- 18.Rosenzweig A. C., Huffman D. L., Hou M. Y., Wernimont A. K., Pufahl R. A., O’Halloran T. V. Structure (London) 1999;7:605–617. doi: 10.1016/s0969-2126(99)80082-3. [DOI] [PubMed] [Google Scholar]
- 19.Gitschier J., Moffat B., Reilly D., Wood W. I., Fairbrother W. J. Nat. Struct. Biol. 1998;5:47–54. doi: 10.1038/nsb0198-47. [DOI] [PubMed] [Google Scholar]
- 20.van Dongen E. M., Klomp L. W., Merkx M. Biochem. Biophys. Res. Commun. 2004;323:789–795. doi: 10.1016/j.bbrc.2004.08.160. [DOI] [PubMed] [Google Scholar]
- 21.Arnesano F., Banci L., Bertini I., Bonvin A. M. Structure (London) 2004;12:669–676. doi: 10.1016/j.str.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Thelwell C., Robinson N. J., Turner-Cavet J. S. Proc. Natl. Acad. Sci. USA. 1998;95:10728–10733. doi: 10.1073/pnas.95.18.10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borrelly G. P. M., Rondet S. A., Tottey S., Robinson N. J. Mol. Microbiol. 2004;53:217–227. doi: 10.1111/j.1365-2958.2004.04106.x. [DOI] [PubMed] [Google Scholar]
- 24.Changela A., Chen K., Xue Y., Holshen J., Outten C. E., O’Halloran T. V., Mondragon A. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 25.O’Halloran T. V., Culotta V. C. J. Biol. Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 26.Harrison M. D., Jones C. E., Solioz M., Dameron C. T. Trends Biochem. Sci. 2000;25:29–32. doi: 10.1016/s0968-0004(99)01492-9. [DOI] [PubMed] [Google Scholar]
- 27.Rosenzweig A. C. Acc. Chem. Res. 2001;34:119–128. doi: 10.1021/ar000012p. [DOI] [PubMed] [Google Scholar]
- 28.Rae T., Schmidt P. J., Pufahl R. A., Culotta V. C., O’Halloran T. V. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 29.Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., O’Halloran T. V. J. Biol. Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 30.Furukawa Y., Torres A. S., O’Halloran T. V. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen L. T., Culotta V. C. J. Biol. Chem. 2005;280:41373–41379. doi: 10.1074/jbc.M509142200. [DOI] [PubMed] [Google Scholar]
- 32.Maxfield A. B., Heaton D. N., Winge D. R. J. Biol. Chem. 2004;279:5072–5080. doi: 10.1074/jbc.M311772200. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann T., Güntert P., Wüthrich K. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 34.Guntert P., Mumenthaler C., Wüthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 35.Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Wang B., Pearlman D. A., et al. amber. San Francisco: Univ. of California; 2004. Version 8.0. [Google Scholar]
- 36.Laskowski R. A., Rullmann J. A. C., MacArthur M. W., Kaptein R., Thornton J. M. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 37.Eijkelenboom A. P., Van den Ent F. M., Vos A., Doreleijers J. F., Hard K., Tullius T. D., Plasterk R. H., Kaptein R., Boelens R. Curr. Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 38.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 39.Grzesiek S., Bax A. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 40.Peng J. W., Wagner G. J. Magn. Reson. 1992;98:308–332. [Google Scholar]
- 41.Mandel M. A., Akke M., Palmer A. G., III J. Mol. Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 42.Brüschweiler R., Liao X., Wright P. E. Science. 1995;268:886–889. doi: 10.1126/science.7754375. [DOI] [PubMed] [Google Scholar]
- 43.Tjandra N., Feller S. E., Pastor R. W., Bax A. J. Am. Chem. Soc. 1995;117:12562–12566. [Google Scholar]
- 44.Kay L. E., Torchia D. A., Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez C., Boelens R., Bonvin A. M. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.