Abstract
Mouse peptide N-glycanase (mPNGase) cleaves the N-glycan chain from misfolded glycoproteins and glycopeptides. Previously, several proteins were found to directly interact with mPNGase; among them, both mHR23B and mS4 were found to link mPNGase to the proteasome. In this study, we found that the cytoplasmic protein mp97 participates in the formation of a ternary complex containing mouse autocrine motility factor receptor (mAMFR), mp97, and mPNGase. This assemblage recruits the cytosolic mPNGase close to the endoplasmic reticulum (ER) membrane, where the retrotranslocation of misfolded glycoproteins is thought to occur. In addition to the ER membrane-associated E3 ligase mAMFR, a cytosolic protein mY33K, containing both UBA and UBX domains, was found to also directly interact with mp97. Thus, a complex containing five proteins, mAMFR, mY33K, mp97, mPNGase, and mHR23B, is formed in close proximity to the ER membrane and serves to couple the activities of retrotranslocation, ubiquitination, and deglycosylation and, thereby, route misfolded glycoproteins to the proteasome.
Keywords: endoplasmic reticulum-associated degradation, retrotranslocation, proteasome, ubiquitination, deglycosylation
Eukaryotes have a stringent endoplasmic reticulum (ER) quality control system to ensure that only folded and fully functional proteins remain in the secretory pathway. Proteins that are misfolded or defective are retrotranslocated to the cytosol and degraded by the proteasome in a ubiquitin-dependent manner (ER-associated degradation) (1). The mechanism whereby the misfolded proteins are distinguished from the native and functional proteins is still unclear (2). As a prerequisite to degradation in the cytosol, the misfolded proteins undergo a process termed protein dislocation or retrotranslocation that routes them from the lumen of the ER to the cytosol (3), where they are degraded. Two possible retrotranslocons, the Sec61p-dependent complex and the Sec61-independent complex exist (2), but much work remains to be done to understand how these complexes function. The misfolded proteins may be extracted from or through the ER membrane driven by the AAA ATPase p97/VCP (valosin-containing protein) in vertebrates and its orthologue Cdc48 in yeast and are then degraded by the proteasome (2).
The action of the AAA ATPase p97 during the ER-associated degradation process has been extensively studied. This evolutionarily conserved protein is involved in a wide range of cellular functions, including protein degradation, regulation of gene expression, cell cycle, apoptosis, DNA repair, and vesicle membrane fusion (4). The specificity of p97-dependent processes is determined by a variety of cofactors. Two cofactors, Ufd1-Npl4 and p47, the UBX (ubiquitin regulatory X) domain protein family member, direct p97 to two different cellular functions, ubiquitin-dependent protein degradation and membrane fusion (4). Another UBX protein family member, UBX2, was found to interact with CDC48 and is hypothesized to recruit ubiquitinated substrates to the yeast CDC48Ufd1-Npl4 complex (5, 6). p97 also recruits the ubiquitin E3 ligases to the site of the retrotranslocation channel and couples the activities of polyubiquitination and retrotranslocation (7, 8).
Peptide N-glycanase (PNGase) is suspected to play important roles in the degradation of misfolded glycoproteins (for review, see ref. 9). Yeast and mammalian PNGase recognize and remove high-mannose N-glycans on denatured, but not native, glycoproteins (10–12). However, the rate of degradation of misfolded glycoproteins is not profoundly affected by a variety of conditions: by the deletion of the yeast PNGase gene, by inhibition of PNGase activity, by PNGase small interfering RNA, or by addition of the inhibitor Z-VAD-fmk (13–15). All these observations suggest that the presence of N-glycans in the misfolded glycoproteins is not a principal obstacle to the proteasome-mediated proteolysis for some glycoproteins, although removal of the glycan might facilitate the more efficient unfolding of specific domains of glycoproteins by the proteasome (10). In contrast, the degradation of ricin A was recently reported to be PNGase-dependent (16), which indicates that, in this case, PNGase is essential for the degradation of this and perhaps other glycoproteins.
In this study, we established the formation of three ternary complexes, mouse autocrine motility factor receptor (mAMFR)(c)–mp97–mPNGase, mAMFR(c)–mp97–mY33K, and mY33K–mp97–mPNGase and a quintenary complex containing mAMFR(c), mY33K, mp97, mPNGase, and mHR23B. Based on these complexes, we propose a model coupling the activities of retrotranslocation, ubiquitination, deglycosylation, and degradation.
Results
The N Terminus of mPNGase Interacts with the Cytosolic Tail of mAMFR.
In an earlier study, we showed, using the yeast two-hybrid system, that mAMFR was one of the candidate proteins that interacted with mPNGase (17). In this study, further yeast two-hybrid experiments were carried out to determine which region of mPNGase mediates the interaction with mAMFR. A set of constructs containing different fragments of mPNGase was prepared in the pBTM116 vector, namely residues 1–130, 1–181, 1–471, 1–651, 181–471, 181–651, and 471–651. A variant of mAMFR, mAMFR(c), lacking the transmembrane domain at its N terminus, was prepared in the pACTII vector. The constructs of mPNGase variants and mAMFR(c) were cotransformed into L4O cells. Assays were performed to examine β-galactosidase activity and His auxotrophy. As summarized in Table 1, colonies containing pACTII-mAMFR(c) and one of four mPNGase fragments (residues 1–130, 1–181, 1–471, and 1–651) grew on −Trp–Leu–His plates and showed strong β-galactosidase activity, indicating an interaction between mAMFR(c) and these mPNGase variants. All other colonies containing pACTII-mAMFR(c) and one of the other mPNGase variants (residues 181–651, 181–471, and 471–651) had no interaction with mAMFR(c). These results, obtained by using the yeast two-hybrid system, clearly establish that the N terminus of mPNGase is sufficient for the interaction with mAMFR(c).
Table 1.
β-Galactosidase activity assay in the yeast two-hybrid analysis shows that binding of mAMFR(c) depends on the N-terminal region of mPNGase
| Fragment of mPNGase | Interaction shown with yeast two-hybrid system |
|---|---|
| 1–130 | + |
| 1–181 | + |
| 1–471 | + |
| 1–651 | + |
| 181–471 | − |
| 471–651 | − |
| 181–651 | − |
mAMFR Forms a Complex in Vivo with mPNGase in COS1 Cells.
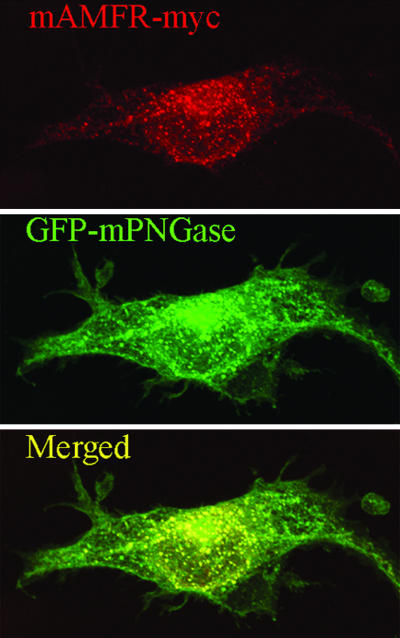
To detect whether mAMFR forms a complex with mPNGase in vivo, two mammalian expression constructs, pEGFP-C1-mPNGase, with GFP fused to the N terminus of mPNGase, and pcDNA4.1-mAMFRc-myc, with a myc tag at the C terminus of mAMFR, were prepared. The plasmids were transfected alone or together into COS1 cells, and the proteins were transiently expressed. The cells were fixed in paraformaldehyde, and the mouse monoclonal anti-Myc Ab and the Alexa Fluor 568-conjugated goat anti-mouse IgG were used to detect mAMFR-myc. Colocalization of GFP-mPNGase and mAMFR-myc were observed in the region of the ER localized around the nucleus but not in the region distal from the nucleus (Fig. 1, Bottom).
Fig. 1.
Colocalization of mAMFR-myc and GFP-mPNGase. COS1 cells transiently expressing GFP-mPNGase (green) and mAMFR-myc (red) were assessed for colocalization by confocal microscopy after staining with anti-myc Ab. The merged panel shows colocalization (yellow) of mAMFR and GFP-mPNGase in the ER-enriched perinuclear region of the cell.
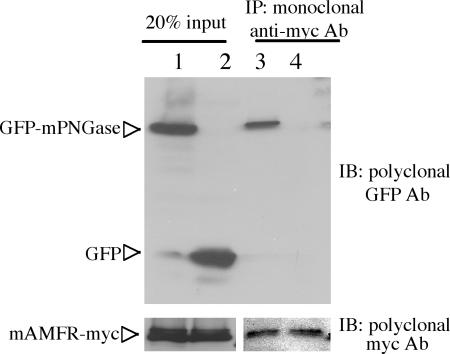
Coimmunoprecipitation using mouse monoclonal anti-myc Ab was performed in the cell lysate with transiently expressed mAMFR-Myc and GFP-mPNGase or GFP. We found that GFP-mPNGase, but not GFP itself, coimmunoprecipitated with mAMFR-myc when anti-myc Ab was used for the immunoprecipitation (Fig. 2). These results clearly establish that in vivo GFP-mPNGase and mAMFR-myc form a complex in COS1 cells.
Fig. 2.
GFP-mPNGase was coimmunoprecipitated with mAMFR-myc in COS1 cells. COS1 cells were cotransfected with plasmids encoding GFP and mAMFR-myc (lanes 2 and 4) and GFP-mPNGase and mAMFR-myc (lanes 1 and 3). Cell lysates were subject to immunoprecipitation (IP) with monoclonal anti-Myc Ab (lanes 3 and 4) in RIPA buffer. The monoclonal anti-myc Ab and bound proteins were recovered with agarose beads containing coupled protein A. The cell lysate (lanes 1 and 2) and the proteins coupled to the protein G beads (lanes 3 and 4) were subjected to SDS/PAGE and immunoblotted (IB) with polyclonal anti-GFP (Upper) or anti-myc (Lower).
The PUB Domain of mPNGase Is Essential for the Interaction with mp97.
Earlier, no direct interaction between GST-mAMFR(c) and His6-mPNGase could be detected by using GST pull-down experiments (18). Based on this finding we hypothesized that there was a mediator involved in formation of the complex containing GFP-mPNGase and mAMFR-myc in mammalian cells. A possible candidate was p97, which was recently reported to directly interact with mAMFR(c) (7, 19, 20) and mPNGase (18, 21). However, it was still not known whether mp97 mediates the formation of a ternary complex containing mPNGase, mp97, and mAMFR.
Because a direct interaction of mPNGase and mp97 was detected, we asked what region of mPNGase interacted with mp97. First, a set of His6-tagged mPNGase constructs (18) containing residues 1–111, 112–450, 451–651, or 1–651 were incubated with His6-mp97. Native gel analysis revealed that the fragment 1–111 and full-length mPNGase formed a complex with His6-mp97. The fragments of 112–450 and 451–651 did not form such a complex (data not shown). These results indicate that the first 111 residues at the N terminus of mPNGase are sufficient for the interaction with mp97.
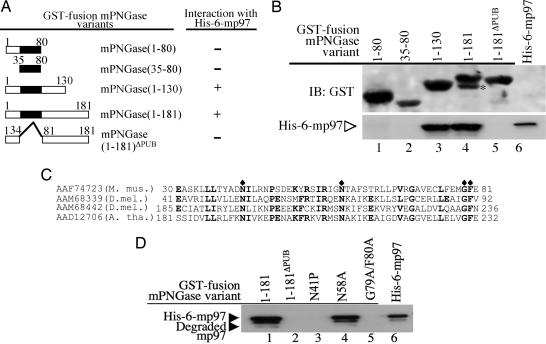
Next, a GST pull-down experiment was performed to investigate the possible role of the PUB domain in the interaction between mPNGase and mp97, because the 111 residues at the N terminus contain an intact PUB domain (residues 35–85) (22). A set of constructs with a GST tag at the N terminus of the following mPNGase fragments was prepared: residues 1–80, 35–80, 1–130, 1–181, and 1–181ΔPUB (Fig. 3A). GST pull-down experiments were performed by using the GST-fusion mPNGase variants and His6-mp97 as described in ref. 18. As shown in Fig. 3B, fragments of GST-mPNGase containing residues 1–130 (Fig. 3B, lane 3) and 1–181 (Fig. 3B, lane 4) interacted with His6-mp97, but residues 1–80 (Fig. 3B, lane 1), 35–80 (Fig. 3B, lane 2) and 1–181ΔPUB (Fig. 3B, lane 5) did not interact with His6-mp97. Two variants containing either residues 35–130 or 15–95 also failed to interact with His6-mp97 (data not shown). These results indicate that the PUB domain is necessary for the interaction between mPNGase and His6-mp97. However, it is clear that the interaction also requires flanking sequences at both sides of the PUB domain.
Fig. 3.
The PUB domain of mPNGase is responsible for the interaction with His6-mp97. (A) mPNGase constructs and the results of interactions between the indicated mPNGase variants and His6-mp97. (B) The PUB domain of mPNGase is responsible for the interaction with His6-mp97. GST fusion protein containing mPNGase variants was immobilized on glutathione beads and incubated with purified His6-mp97. Bound proteins were subjected to SDS/PAGE and blotted with monoclonal anti-GST (Upper) or anti-His Ab (Lower). The asterisk in lane 4 indicates degraded GST-mPNGase (1–181). (C) Sequence alignments, with conserved amino acids in bold. Amino acids chosen for mutation are labeled with a diamond. M. mus., Mus musculus; D.mel., Drosophila melanogaster; A. tha., Arabidopsis thalania. (D) The N41P and G79A/F80A mutants abolish the interaction between mPNGase and mp97. Pull-down experiments were as described in B.
To further determine whether the PUB domain is essential for the interaction between mPNGase and mp97, we screened for mutations in the mPNGase PUB domain that would alter their binding to mp97. Based on the conservation found upon sequence alignment of the PUB domains of various proteins (Fig. 3C), we constructed three variants of the PUB domain of mPNGase. Among them, two contain a mutation in one of the conserved residues (N41P and N58A), and one contains a mutation in two of the conserved residues (G79A/F80A). As shown above, GST-mPNGase(1–181) interacts with mp97. Accordingly, the three mutations were introduced in GST-mPNGase(1–181). GST pull-down experiments were performed to examine the interaction between the PUB domain variants and mp97. It was found that two mutations (N41P and G79A/F80A) completely abolished the interaction between mPNGase and mp97 (Fig. 3D, lanes 3 and 5), but the other mutation, N58A, did not affect the interaction (Fig. 3D, lane 4). These observations strongly support the idea that the PUB domain, with its flanking sequences, is critical for the interaction between mPNGase and mp97.
Different Regions in mp97 Interact with mPNGase, mAMFR, and mY33K.
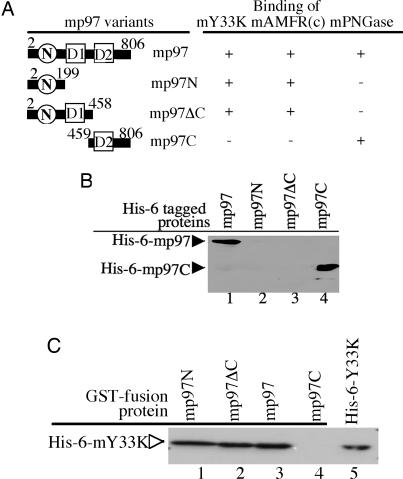
Because mp97 interacts with mPNGase and mAMFR, we asked whether there were different regions in mp97 responsible for the interaction with these two proteins. Different strategies were used to answer this question. First, GST pull-down experiments were performed to determine what region of mp97 interacts with mPNGase. A set of constructs of mp97 was prepared (Fig. 4A), His6-mp97N (2–199), His6-mp97ΔC (2–458), His6-mp97C (459–806), and full-length mp97. The GST-mPNGase(1–181) protein was used to perform the pull-down experiments. As shown in Fig. 4B, the full-length His6-mp97 (Fig. 4B, lane 1) and His6-mp97C (Fig. 4B, lane 4) were pulled down with GST-mPNGase(1–181). However, neither His6-mp97N (Fig. 4B, lane 2) nor His6-mp97ΔC (Fig. 4B, lane 3) were pulled down with GST-mPNGase(1–181). These results indicate that the C terminus of mp97 interacts with mPNGase.
Fig. 4.
Different regions on mp97 mediate binding to mPNGase, mAMFR, and mY33K. (A) Schematic diagram of the different regions of mp97. GST-mPNGase (1–181) (B) or GST-mY33K (C) bound on glutathione beads was incubated with His6-tagged mp97 variants. Bound proteins were separated by SDS/PAGE and blotted with monoclonal anti-His Ab. (B) The C terminus of mp97 associates with mPNGase. (C) The N terminus of mp97 interacts with mY33K.
As described in an earlier study (20), the 49-aa fragment at the C terminus of mAMFR is sufficient for interaction with mp97. We prepared a mAMFR variant with the 49-aa fragment (AMFRc49) in an intein vector. AMFRc49 was incubated with the mp97 variants mentioned above, and the mixtures were loaded onto a native gel. We found that AMFRc49 could form complexes with His6-mp97N, His6-mp97ΔC, and full-length mp97 but not with His6-mp97C (data not shown). These results clearly demonstrate that the N-terminal region of mp97 is sufficient for interaction with mAMFR.
Earlier, mY33K, a UBA and UBX domain-containing protein, was found to interact with mPNGase in the yeast two-hybrid system (17). Further studies were performed in the yeast two-hybrid system by using truncated proteins mY33KΔUBX (mY33K with its UBX domain deleted), mY33KΔUBA (with UBA domain deleted), and mPNGaseΔPUB (with PUB domain deleted) to determine which domains are necessary for the interaction between mY33K and mPNGase. As shown in Table 2, deletion of either the UBX domain on mY33K or the PUB domain on mPNGase interrupted the interaction between these two proteins. These results indicate that the UBX domain on mY33K and the PUB domain in mPNGase are essential for the interaction between these two proteins.
Table 2.
β-Galactosidase activity assay in yeast two-hybrid system shows that the UBX domain in mY33K and the PUB domain in mPNGase are responsible for the interaction between mY33K and mPNGase
| mY33K mutants | mPNGase | mPNGaseΔPUB |
|---|---|---|
| mY33K | + | − |
| mY33KΔUBX | − | − |
| mY33KΔUBA | + | − |
However, a direct interaction between GST-mY33K and His6-mPNGase was not detected by using GST pull-down experiments (data not shown). It has been suggested that UBX domains may act as general p97/CDC48p-binding modules (23). Therefore, the possibility of an interaction between GST-mp97 variants and His6-mY33K was examined by using GST pull-down experiments, which revealed a direct interaction between GST-mp97 and His6-mY33K (Fig. 4C, lane 3). Furthermore, the N-terminal region of mp97 is responsible for the interaction between the two proteins (Fig. 4C, lane 1). These results confirm and extend the previous observations (21) that mY33K directly interacts with mp97.
Combining the observations reported above, it is clear that the C-terminal regions of mAMFR and mY33K interact with the N-terminal region of mp97, and mPNGase associates with the C terminus of mp97 (for summary, see Fig. 4A).
mp97 Mediates the in Vitro Formation of Three Ternary Complexes and a Quintenary Complex.
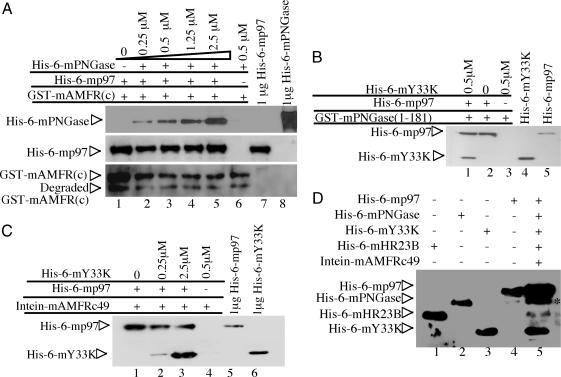
To detect whether the three proteins mAMFR, mP97, and mPNGase form a complex in vitro, a previously designed competition experiment (18) was used to detect the formation of this ternary complex. As shown in lane 6 of Fig. 5A, without His6-mp97, His6-mPNGase was not pulled down with GST-mAMFR(c) (Fig. 5A, lane 6). However, when His6-mp97 was added in this reaction system, both His6-mp97 and His6-mPNGase were pulled down with GST-mAMFR(c) (Fig. 5A, lanes 2–5). The amount of His6-mPNGase pulled down with GST-mAMFR(c) was increased when the input of His6-mPNGase was increased from 1- to 10-fold. This result indicates that mp97 mediates the formation of the ternary mAMFR(c)–mp97–mPNGase complex.
Fig. 5.
mp97 mediates formation of three ternary complexes, mAMFR–mp97–mPNGase, mPNGase–mp97–mY33K, and mAMFR–mp97–mY33K and a quintenary complex containing mAMFR, mp97, mY33K, mPNGase, and mHR23B. (A) Formation of the ternary mAMFR(c)–mp97–mPNGase complex. GST-mAMFR(c) (0.25 μM) bound on glutathione beads was incubated with 0.25 μM His6-mp97 in the presence of a 0- to 10-fold molar excess of His6-mPNGase. Bound proteins were subjected to SDS/PAGE and blotted with anti-mPNGase (Top), anti-mp97 (Middle), or anti-GST (Bottom) Ab. (B) Formation of the ternary of mPNGase–mp97–mY33K complex. GSH-agarose beads containing 0.25 μM GST-mPNGase(1–181) were incubated with 0.25 μM His6-mp97 and then incubated with or without 0.5 μM His6-mY33K. (C) Formation of the ternary mAMFR–mp97–mY33K complex. Chitin beads containing 0.25 μM intein-mAMFRc49 were incubated with 0.25 μM His6-mp97 and then incubated with 0, 0.25, or 2.5 μM His6-mY33K. Bound proteins were subjected to SDS/PAGE and blotted with a monoclonal anti-His Ab (C and D). (D) Formation of a quintenary complex. Chitin beads containing 0.25 μM intein-mAMFRc49 were incubated with 2.5 μM His6-mp97 and then harvested and successively incubated with 2.5 μM His6-mY33K, His6-mPNGase, and His6-mHR23B. The asterisk in lane 5 indicates His6-mPNGase and degraded His6-mp97.
Next, we investigated the formation of the ternary mPNGase–mp97–mY33K complex using GST-mPNGase(1–181). As shown in Fig. 5B, His6-mY33k alone was not pulled down with GST-mPNGase(1–181) (Fig. 5B, lane 3). However, when His6-mp97 was added to this system, His6-mY33k was pulled down with GST-mPNGase(1–181) (Fig. 5B, lane 1). This observation clearly indicates that mp97 also bridges mPNGase and mY33K.
Because both mAMFR(c) and mY33K can associate with the N terminus of mp97, we asked whether mAMFRc49 and mY33K competed with each other in binding to mp97. As shown in Fig. 5C, without His6-mp97, His6-mY33K was not pulled down (Fig. 5C, lane 4). In the presence of His6-mp97, His6-mY33K was pulled down (Fig. 5C, lanes 2 and 3), indicating the formation of a third ternary complex, composed of mAMFRc49, mp97, and mY33K.
Based on the ternary mp97–mPNGase–mHR23B complex (18), we asked whether a quintenary complex containing mAMFRc49, mY33K, mp97, mPNGase, and mHR23B might exist. Intein-mAMFRc49 bound on chitin beads was used to pull down His6-mp97, His6-mY33K, His6-mPNGase, and His6-mHR23B. As shown in Fig. 5D, all four proteins were pulled down with intein-mAMFRc49 (Fig. 5D, lane 5), which indicates the formation of a quintenary complex containing mAMFR, mp97, mY33k, mPNGase, and mHR23B.
Discussion
For certain misfolded glycoproteins, the action of PNGase does not appear to be required before proteasomal degradation (see review in ref. 2). Recently, however, it has been demonstrated that, in yeast, the glycoprotein ricin A must be deglycosylated before degradation by the proteasome and that the Png1–Rad23 complex directly couples the activities of deglycosylation and proteasomal degradation (16).
The object of this study in mammalian cells has been to investigate the possible existence of an assemblage of proteins that mediates retrotranslocation, ubiquitination, and deglycosylation of misfolded glycoproteins. Initially, we found that mp97 linked mPNGase to the ER membrane-associated E3 ligase mAMFR. The ternary complex produced by this interaction, mAMFR–mp97–mPNGase, may couple two important activities in the ER-associated degradation pathway, ubiquitination and deglycosylation. The formation of a ternary complex might explain the interaction between mAMFR(c) and mPNGase, detected in the yeast two-hybrid system, which lacks the mp97 protein. However, in yeast, there is a highly conserved homologue of mp97, Cdc48, which may function as the organizer of the mAMFR(c)–Cdc48p–mPNGase complex.
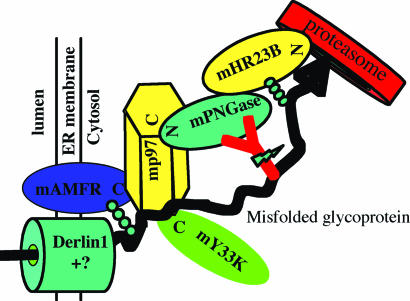
p97, one of the most abundant proteins in cells (≈1% of the cytosolic protein), plays an essential role in the ER-associated degradation pathway. Recently it has been reported that p97 recruits E3 ligases to the possible retrotranslocon component Derlin 1 and couples the activities of retrotranslocation, ubiquitination, and extraction of misfolded proteins from the ER membrane (7, 8). With the observations of the formation of three ternary complexes and the quintenary complex containing five proteins, mAMFR, mY33K, mp97, mPNGase, and mHR23B in this study, we propose a possible model for the retrotranslocation, ubiquitination, deglycosylation, and degradation of ER-associated degradation substrate (Fig. 6). It is possible that mp97 may recruit both the E3 ligase and mPNGase to the site of retrotranslocation at the surface of the ER membrane and allow the activities of ubiquitination and deglycosylation to proceed in close proximity to the emerging misfolded protein. This idea is consistent with the observation that the deglycosylation of TCRα is not inhibited by the PNGase inhibitor Z-VAD-fmk (13), suggesting that at least some PNGase is not accessible to the inhibitor and that dislocation and deglycosylation may be temporally coupled in this case. Sequentially, the deglycosylated substrate will be transferred through mHR23B to the proteasome for degradation. In our model, the components involved in retrotranslocation, ubiquitination, and deglycosylation form a complex through the mediator mp97, which might ensure a more efficient degradation pathway.
Fig. 6.
Model depicting the events of retrotranslocation, ubiquitination, deglycosylation, and degradation of a glycosylated ERAD substrate. Direct interactions among the five proteins mAMFR, mp97, mY33K, mPNGase, and mHR23B are depicted as determined in this study. Interactions among Derlin-1, p97, mAMFR (7, 8), mHR23B, and proteasome were reported in ref. 24. The arrangement of the glycoprotein being degraded is hypothetical.
Despite the identification of these different complexes, our results do not allow us to propose the precise order of events during and after retrotranslocation. Although the function of mY33K is still unclear, the study of a yeast ER membrane protein, Ubx2, with similar UBA and UBX domains may shed light on its function (5, 6). Ubx2 is speculated to transfer the ubiquitinated substrates from ER-associated ubiquitin ligases to the Cdc48 complex (5, 6). It will be interesting to see whether mY33K has a similar function in mammals.
While studying the formation of the ternary mAMFR(c)–mp97–mPNGase complex, we noted that the amount of His6-mPNGase brought down with GST-mAMFR(c) was very low and could be detected only by using the more efficient anti-mPNGase Ab rather than the anti-His Ab (see Fig. 5A). Furthermore, in the quintenary complex, the amount of His6-mPNGase and His6-mHR23B brought down with intein-mAMFRc49 was very low (see Fig. 5D). However, during the analysis of the formation of the ternary mHR23B–mPNGase–mp97 complex, His6-mp97 bound to GST-mHR23B–mPNGase was readily detected by using an anti-His Ab (18), and, when a 10-fold molar excess of His6-mPNGase and His6-mp97 were added sequentially in the ternary GST-mHR23B–mp97–mPNGase complex, the amount of pull-down His6-mPNGase is much lower than His6-mp97, even though it is His6-mPNGase that mediates the interaction between mHR23B and mp97 (G.L. and W.J.L., unpublished data). Given the hexametric structure of p97 (25), one possible explanation for this observation could be that six mp97 molecules bind to one mPNGase molecule rather than a one-to-one interaction. This issue will have to be addressed in future studies.
In this study, we found that the PUB domain of mPNGase is critical for the interaction between mPNGase and mp97. We have found that the PUB domain (residues 35–80), as defined in the literature (22), was not sufficient for this interaction. We propose that flanking sequences are necessary on both sides of the PUB domain for its proper function. Our results that variants lacking sequences extensions on both sides fail to bind to mp97 strongly support this idea, consistent with the observation that the mutation at the C-terminal edge (G79A/F80A) completely disrupted the interaction between mPNGase and mp97. Interestingly, secondary structure prediction with the psipred program revealed that residue 35 is in the middle of an α-helix, and residue 80 is at the edge of another α-helix. Thus, it seems possible that the three-dimensional structure of the PUB domain was destroyed when truncations were carried out at these sites. It is interesting that the PUB domain does not exist in yeast PNGase; it is restricted to higher organisms such as insects and vertebrates. This fact may reflect an evolutionary addition of the PUB domain to the N terminus of PNGase to facilitate the assembly of a more complicated ERAD pathway in mammals that involves multiple regulatory protein–protein interactions.
Materials and Methods
Abs and Chemicals.
Purified polyclonal antiserum against mPNGase was kindly provided by Tadashi Suzuki (University of Osaka, Osaka). Monoclonal antibodies against GST and poly- and monoclonal His and Myc antibodies were purchased from Santa Cruz Biotechnology. A monoclonal anti-p97 Ab was purchased from Fitzgerald Industries International (Concord, MA). Glutathione (GSH)-agarose beads and the Alexa Fluor 568 goat anti-mouse Ab were purchased from Molecular Probes.
Construction of Plasmids.
The pQE9 plasmid containing full-length mp97 was kindly provided by Hemmo H. Meyer (Swiss Federal School of Technology, Lausanne, Switzerland) and used for preparation of all mp97 variants. Plasmids pGEX-5x-1 (Amersham Pharmacia) and pET28a (Novagen) were used to construct the GST-fusion proteins and His-tagged proteins. The pACTII-mY33K, pGEX-5x-1-mAMFR(c), pET28a-mPNGase, pET28a-mHR23B, and pGEX-5x-1-mHR23B plasmids were described in ref. 18. pACTII-mY33K, pEGFP-mPNGase, and pACTII-mAMFR(c) were described in ref. 17. The plasmid PACTII-mY33K was used as template to construct the plasmids pBTM116-mY33K, pET28a-mY33K, and GEX-5x-1-mY33K. pBTM116-mY33K was used to prepare the deletion mutants mY33KΔUBA and mY33KΔUBX. pET28a-mPNGase was used to construct mPNGase variants.
Immunofluorescence and Microscopy.
COS1 cells were cultured in DMEM supplemented with 10% FBS, 10 mM l-glutamine and 100 μg/ml each penicillin and streptomycin, which were purchased from GIBCO/BRL. For immunofluorescence, COS1 cells were cultured on coverslips in six-well dishes and transfected with plasmids by using Lipofectamine 2000 (Invitrogen). Cells were fixed in 3% paraformaldehyde for 20 min and permeabilized with 0.5% Triton X-100 on ice. The staining of mAMFR-myc was done by using monoclonal mouse anti-Myc Ab and Alexa Fluor-586-conjugated goat anti-mouse IgG.
Immunoprecipitation and Immunoblotting.
The indicated plasmids were transiently transfected alone or together into COS1 cells. Cells were harvested 20–24 h after transfection and lysed in RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). Protein G beads (20 μl) were added to preclear the lysates before immunoprecipitation. Precleared lysates were incubated with 1 μg of Ab overnight at 4°C. Then, 20 μl of protein G beads were added, and the mixture was incubated at 4°C for 2 h. The beads were washed five times in buffer containing 50 mM Tris·HCl (pH7.5), 0.1 M NaCl, and 1% Triton X-100 before processing by Western Blot.
GST Pull-Down Experiments.
The expression and purification of proteins were carried out as described in ref. 18. Competition experiments between GST-AMFR(c) and His6-mp97 were carried out as follows: GSH-agarose beads containing 0.25 μM bound GST-AMFR(c) were incubated with 0.25 μM His6-mp97 in the presence of a 0- to 10-fold molar excess of His6-mPNGase. The other tests for complex formation were performed step by step, namely adding the first component, followed by harvesting the first complex and then adding the second component, and so on.
Acknowledgments
We thank Dr. Tadashi Suzuki for anti-mPNGase antiserum, Dr. Hemmo H. Meyer for the pQE9-p97 plasmid, Shivanjali Joshi (Stony Brook University) for providing the plasmid of pcDNA4.1-mAMFR-myc, Dr. Hangil Park for support, and members of the Lennarz laboratory for critical comments on the manuscript. This work was supported by National Institutes of Health Grants GM 33184 (to W.J.L.) and DK54835 (to H.S.).
Abbreviations
- AMFR
autocrine motility factor receptor
- ER
endoplasmic reticulum
- mAMFR
mouse AMFR
- PNGase
peptide N-glycanase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.McCracken A. A., Brodsky J. L. J. Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romisch K. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 3.Ahner A., Brodsky J. L. Trends Cell Biol. 2004;14:474–478. doi: 10.1016/j.tcb.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Woodman P. G. J. Cell Sci. 2003;116:4283–4290. doi: 10.1242/jcs.00817. [DOI] [PubMed] [Google Scholar]
- 5.Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. Nat. Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 6.Schuberth C., Buchberger A. Nat. Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- 7.Lilley B. N., Ploegh H. L. Proc. Natl. Acad. Sci. USA. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. Proc. Natl. Acad. Sci. USA. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T., Park H., Lennarz W. J. FASEB J. 2002;16:635–641. doi: 10.1096/fj.01-0889rev. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch C., Blom D., Ploegh H. L. EMBO J. 2003;22:1036–1046. doi: 10.1093/emboj/cdg107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch C., Misaghi S., Blom D., Pacold M. E., Ploegh H. L. EMBO Rep. 2004;5:201–206. doi: 10.1038/sj.embor.7400066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi S., Katiyar S., Lennarz W. J. FEBS Lett. 2005;579:823–826. doi: 10.1016/j.febslet.2004.12.060. [DOI] [PubMed] [Google Scholar]
- 13.Misaghi S., Pacold M. E., Blom D., Ploegh H. L., Korbel G. A. Chem. Biol. 2004;11:1677–1687. doi: 10.1016/j.chembiol.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T., Park H., Hollingsworth N. M., Sternglanz R., Lennarz W. J. J. Cell Biol. 2000;149:1039–1052. doi: 10.1083/jcb.149.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blom D., Hirsch C., Stern P., Tortorella D., Ploegh H. L. EMBO J. 2004;23:650–658. doi: 10.1038/sj.emboj.7600090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. J. Cell Biol. 2006;172:211–219. doi: 10.1083/jcb.200507149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park H., Suzuki T., Lennarz W. J. Proc. Natl. Acad. Sci. USA. 2001;98:11163–11168. doi: 10.1073/pnas.201393498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G., Zhou X., Zhao G., Schindelin H., Lennarz W. J. Proc. Natl. Acad. Sci. USA. 2005;102:15809–15814. doi: 10.1073/pnas.0507155102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grelle G., Kostka S., Otto A., Kersten B., Genser K. F., Muller E. C., Walter S., Boddrich A., Stelzl U., Hanig C., et al. Mol. Cell. Proteomics. 2006;5:234–244. doi: 10.1074/mcp.M500198-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. J. Biol. Chem. 2004;279:45676–45684. doi: 10.1074/jbc.M409034200. [DOI] [PubMed] [Google Scholar]
- 21.McNeill H., Knebel A., Arthur J. S., Cuenda A., Cohen P. Biochem. J. 2004;384:391–400. doi: 10.1042/BJ20041498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki T., Park H., Till E. A., Lennarz W. J. Biochem. Biophys. Res. Commun. 2001;287:1083–1087. doi: 10.1006/bbrc.2001.5688. [DOI] [PubMed] [Google Scholar]
- 23.Dreveny I., Kondo H., Uchiyama K., Shaw A., Zhang X., Freemont P. S. EMBO J. 2004;23:1030–1039. doi: 10.1038/sj.emboj.7600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katiyar S., Li G., Lannarz W. J. Proc. Natl. Acad. Sci. USA. 2004;101:13774–13779. doi: 10.1073/pnas.0405663101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huyton T., Pye V. E., Briggs L. C., Flynn T. C., Beuron F., Kondo H., Ma J., Zhang X., Freemont P. S. J. Struct. Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]