Abstract
The Sec23p/Sec24p complex functions as a component of the COPII coat in vesicle transport from the endoplasmic reticulum. Here we characterize Saccharomyces cerevisiae SEC24, which encodes a protein of 926 amino acids (YIL109C), and a close homologue, ISS1 (YNL049C), which is 55% identical to SEC24. SEC24 is essential for vesicular transport in vivo because depletion of Sec24p is lethal, causing exaggeration of the endoplasmic reticulum and a block in the maturation of carboxypeptidase Y. Overproduction of Sec24p suppressed the temperature sensitivity of sec23-2, and overproduction of both Sec24p and Sec23p suppressed the temperature sensitivity of sec16-2. SEC24 gene disruption could be complemented by overexpression of ISS1, indicating functional redundancy between the two homologous proteins. Deletion of ISS1 had no significant effect on growth or secretion; however, iss1Δ mutants were found to be synthetically lethal with mutations in the v-SNARE genes SEC22 and BET1. Moreover, overexpression of ISS1 could suppress mutations in SEC22. These genetic interactions suggest that Iss1p may be specialized for the packaging or the function of COPII v-SNAREs. Iss1p tagged with His6 at its C terminus copurified with Sec23p. Pure Sec23p/Iss1p could replace Sec23p/Sec24p in the packaging of a soluble cargo molecule (α-factor) and v-SNAREs (Sec22p and Bet1p) into COPII vesicles. Abundant proteins in the purified vesicles produced with Sec23p/Iss1p were indistinguishable from those in the regular COPII vesicles produced with Sec23p/Sec24p.
INTRODUCTION
Vesicular transport from the endoplasmic reticulum (ER) to the Golgi is the first step in the intracellular trafficking of proteins destined for the Golgi apparatus, lysosomes (vacuoles), plasma membrane, and extracellular space. Most, if not all, anterograde vesicular transport from the ER to the Golgi complex is carried out by vesicles coated with a protein complex known as COPII. In vitro assays have shown that the minimal components needed for formation of the COPII coat are three cytosolic proteins: Sar1p, the Sec23p/Sec24p complex, and the Sec13p/Sec31p complex (Barlowe et al., 1994). During vesicle formation, coat proteins are sequentially recruited to the ER: first Sar1p, then Sec23p/Sec24p, and finally Sec13p/Sec31p (Matsuoka et al., 1998).
Sar1p is a small GTPase (21 kDa) of the Ras superfamily (Nakano and Muramatsu, 1989; Oka et al., 1991). Sec12p, an integral membrane protein of the ER, facilitates exchange of GTP for GDP on Sar1p and is thought to recruit Sar1p to the ER (Barlowe and Schekman, 1993). The GTP-bound form of Sar1p is required for budding, and GTP must be hydrolyzed to GDP before vesicles can fuse with the Golgi (Barlowe et al., 1994). A nonhydrolyzable GTP analogue, 5′-guanylyl imidodiphosphate (GMP-PNP), satisfies the nucleotide requirement for vesicle formation, but the vesicles produced with GMP-PNP cannot fuse with the Golgi membrane. Presumably, GTP hydrolysis by Sar1p is a prerequisite for the dissociation of coat proteins from the vesicles to produce fusion-competent vesicles. Yoshihisa et al. (1993) found that Sec23p (85 kDa) stimulates the hydrolysis of GTP by Sar1p. Sec24p (105 kDa) was discovered as a subunit of a protein complex containing Sec23p (Hicke et al., 1992). Although the Sec24p subunit of this complex is required for in vitro vesicle formation, it has no significant effect on the GTPase-activation activity of Sec23p (Yoshihisa et al., 1993). Sec13p is a 33-kDa protein containing six WD repeat motifs that make up most of the protein (Salama et al., 1993; Saxena et al., 1996). Sec31p is a 150-kDa phosphoprotein that contains seven WD repeats near the N terminus (Salama et al., 1997). Phosphorylation of this protein is important for vesicle formation. Direct interactions among COPII components have been shown by two-hybrid analysis and in vitro binding assays: the N terminus of Sec24p binds to Sec23p (Gimeno et al., 1996); Sec13p and the N-terminal region of Sec31p interact with each other, possibly through their WD repeat motifs (Shaywitz et al., 1997); and Sec23p and Sec24p interact with a central region of Sec31p (Shaywitz et al., 1997). Sec16p, a 240-kDa peripheral ER membrane protein, has also been implicated in COPII coat assembly, because Sec16p is required for vesicle formation in vivo and Sec16p binds to Sec23p, Sec24p, and Sec31p (Espenshade et al., 1995; Gimeno et al., 1996; Shaywitz et al., 1997).
Cargo molecules are selectively packaged into COPII vesicles. Because some cargo molecules are known to be concentrated into COPII vesicles (Balch et al., 1994), an active uptake mechanism must exist for at least some proteins. Sar1p and Sec23p/Sec24p probably play a central role in cargo recruitment. When Sar1p and Sec23p/Sec24p are incubated with microsomes in the presence of GTP or GMP-PNP and in the absence of Sec13p/Sec31p, a prebudding complex forms that contains various cargo molecules, such as glycosylated pro-α-factor (gpαF), amino acid permeases, Emp24p, and SNARE molecules in yeast (Kuehn et al., 1998) and vesicular stomatitis virus glycoprotein in a mammalian system (Aridor et al., 1998). Importantly, resident ER proteins are excluded from these prebudding complexes, indicating that at this stage at least some cargo sorting has already taken place. The v-SNARE vesicle proteins Bet1p and Bos1p bind to Sec23p/Sec24p as well as to Sar1p, indicating that sorting may be achieved by direct interaction with these coat proteins (Springer and Schekman, 1998). Recent work in mammalian cells has identified two sorting motifs within the cytoplasmic domains of transmembrane cargo molecules: a di-acidic motif (Asp-X-Glu, where X represents any amino acid) on the cytoplasmic tail of vesicular stomatitis virus glycoprotein (Nishimura and Balch, 1997), and a double phenylalanine motif (Phe-Phe) on the cytoplasmic tail of p24 proteins (Fiedler et al., 1996; Dominguez et al., 1998) and ERGIC-53 (Kappeler et al., 1997). These motifs are important for the efficient exit of these cargo molecules out of the ER. Moreover, peptides containing the latter motif were shown to bind to the mammalian Sec23p/Sec24p complex (Kappeler et al., 1997; Dominguez et al., 1998).
Despite the fact that Sec24p has long been recognized to play an essential role in the budding of COPII-coated vesicles, the structure and function of this protein have not yet been described in detail. Here we report the characterization of Sec24p and a homologue, Iss1p. We demonstrate that Sec24p and Iss1p can function interchangeably in vesicle formation from the ER.
MATERIALS AND METHODS
Strains and Plasmids
Strains and plasmids used in this study are listed in Table 1 and Table 2, respectively, and their construction is described below
Table 1.
Strains used in this study
| Name | Genotype |
|---|---|
| RSY255 | MATα leu2-3,112 ura3-52 |
| RSY612 | MATα can1-100 leu2-3,112 his3-11,15 trp1-1 ura3-lade2-1 GAL2 |
| MATα can1-100 leu2-3,112 his3-11,15 trp1-1 ura3-lade2-1 GAL2 | |
| RSY620 | MATα leu2-3,112 ura3-52 ade2-1 trp1-1 his3-11,15PEP4∷TRP1 |
| RSY866 | RSY612 derivative, one of whose two SEC24 genes is disrupted by LEU2 |
| RSY875 | sec24∷LEU haploid carrying pTYY214 derived from RSY866 [pTYY214] |
| CKY496 | MATα sec24-1 ura3-52 leu2-3,112 |
| CKY499 | MATα iss1-Δ2∷TRP1 ura3-52 leu2-Δ1 his3-Δ200 lys2-801 ade2-101 trp1-Δ63 |
| CKY500 | MATa iss1-Δ2∷TRP1 ura3-52 leu2-Δ1 his3-Δ200 lys2-801 ade2-101 trp1-Δ63 |
| CKY39 | MATα sec12-4 ura3-52 his4-619 |
| CKY40 | MATa sec12-4 ura3-52 |
| CKY46 | MATa sec13-1 ura3-52 his4-619 |
| CKY51 | MATa sec16-2 ura3-52 |
| CKY55 | MATa sec17-1 ura3-52 his4-619 |
| CKY58 | MATα sec18-1 ura3-52 his4-619 |
| CKY59 | MATa sec18-1 ura3-52 his4-619 |
| CKY70 | MATα sec22-3 ura3-52 his4-619 |
| CKY71 | MATa sec22-3 ura3-52 |
| CKY72 | MATα sec22-1 ura3 |
| CKY78 | MATa sec23-1 ura3-52 leu2-3,112 |
| CKY85 | MATa bet1-1 ura3-52 |
| GPY60 | MATα ura3-52 trp1-289 his4-579 leu2-3,112 prb1 pep4∷URA3 gal2 |
| YPH501 | MATa ura3-52 lys2-801amber ade2-101ochee trp1-Δ63 his3-Δ200 leu2-Δ1 |
| MATα ura3-52 lys2-801amber ade2-101ochee trp1-Δ63 his3-Δ200 leu2-Δ1 | |
| TKY5 | MATa SEC24 (pTKY6) derived from RSY866 (pTKY6) |
| TKY6 | MATα SEC24 (pTKY6) derived from RSY866 (pTKY6) |
| TKY7 | MATa sec24∷LEU2 (pTKY6) derived from RSY866 (pTKY6) |
| TKY8 | MATα sec24∷LEU2 (pTKY6) derived from RSY866 (pTKY6) |
| TKY10 | MATα ISS1 ura3-52 lys2-801amber ade2-101ochee trp1-Δ63 his3-Δ200 leu2-Δ1 |
| TKY12 | MATα iss1-Δ1∷HIS3 ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 |
| TKY22 | Haploid (sec24∷LEU2 [pTKY11]) derived from RSY866 (pTKY11) |
| TKY23 | Haploid (SEC24 [pTKY11]) derived from RSY866 (pTKY11) |
Table 2.
Plasmids used in this study
| Name | Description | Source |
|---|---|---|
| pTYB121 | pBS II KS(+) carrying partial SEC24 (0.45 kb) amplified by PCR | This study |
| pTYB131 | pBS II SK(+) carrying 4.2-kb XhoI–HindIII fragment containing SEC24 | This study |
| pTYY111–114 | Positive clones from YEp24 library in SEC24 screening | This study |
| pTYY211–213 | Positive clones from YCp50 library in SEC24 screening | This study |
| pTYY115 | YEp352 (2μ URA3) with 4.2-kb fragment containing SEC24 | This study |
| pTYY116 | YEp352 (2μ URA3) with 4.2-kb fragment containing SEC24 and 3.5-kb fragment containing SEC23 | This study |
| pTYY122 | YEp352 (2μ URA3) with 3.5-kb fragment containing SEC23 | This study |
| pTYY214 | pBM743 (CEN URA3 GAL1 promoter) with SEC24 | This study |
| pTYY303 | pTYB131 derivative with SEC24 disrupted by LEU2 | This study |
| pRH200 | pCT3 (CEN URA3) with 7.0-kb ClaI–XbaI fragment containing ISS1 | This study |
| pTKB1 | pGEM-4Z with 7.0-kb ClaI–XbaI fragment containing ISS1 | This study |
| pTKB2 | pGEM-4Z with 7.0-kb ClaI–XbaI fragment containing His6-tagged version of ISS1 | This study |
| pTKY4 | YEp352 (2μ URA3) with 7.0-kb HindIII–XbaI fragment containing ISS1 | This study |
| pTKY6 | YEp352 (2μ URA3) with 7.0-kb HindIII–XbaI fragment containing His6-tagged version of ISS1 | This study |
| pTKY7 | pGAL425GAL1 (2μ LEU2 GAL1 promoter and CYC1 terminator) with 3.5-kb PstI fragment containing His6-tagged version of ISS1 | This study |
| pTKY9 | pGAL426GAL1 (2μ URA3 GAL1 promoter and CYC1 terminator) with 2.9-kb NcoI–HindIII fragment containing SEC23 | This study |
| pTKY11 | pGAL426GAL1 (2μ URA3 GAL1 promoter and CYC1 terminator) with 3.5-kb fragment containing His6-tagged version of ISS1 | This study |
Media
YPD is a complex medium with 1% Bacto yeast extract (Difco, Detroit, MI) and 2% Bacto peptone (Difco) supplemented with glucose (2%, unless noted otherwise). YPGal is the same as YPD except that it contains galactose (2%, unless noted otherwise) instead of glucose. MVCA medium consists of 0.67% yeast nitrogen base without amino acids (Difco), 0.5% vitamin assay casamino acid (Difco), and 5% carbon source. MV-lowS medium contains 0.67% yeast nitrogen base without amino acids and ammonium sulfate (Difco), 0.1 mM (NH4)2SO4, and 5% carbon source. MV-noS medium lacks (NH4)2SO4 from MV-lowS medium. Nutrients corresponding to auxotrophic markers were supplemented to the MVCA, MV-lowS, and MV-noS media. Synthetic complete (SC) medium contains 0.67% yeast nitrogen base without amino acids and 2% glucose as a carbon source (unless noted otherwise) as well as various nutrients (Sherman, 1991). SC dropout medium lacks one or two nutrients from SC medium.
Purification and Amino Acid Sequencing of Sec24p
The Sec23p/Sec24p complex was purified as described (Hicke et al., 1992), precipitated with 5% trichloroacetate (TCA), denatured in SDS-PAGE sample buffer, and separated by 7.5% SDS-PAGE. After transfer to a nitrocellulose membrane, proteins were stained with Ponceau S. The protein band of Sec24p (105 kDa) was excised, destained with Tris-buffered saline, and washed thoroughly with distilled water. Sec24p bound to nitrocellulose was digested with trypsin, and the released peptides were purified with C18 reverse-phase HPLC. Several peaks were recovered and sequenced: P1, IWQIFQ; P2, SVQ(D/F)ILATYK; P3, VGLLATTINTLLQNL; and P4, VTAQLLSCQDSTY.
Cloning of SEC24
Three sets of sense and antisense degenerate oligonucleotides were synthesized based on the amino acid sequences of tryptic peptides P1 (1 and rev-1) and P2 (2A, rev-2A, 2B, and rev-2B): 1, 5′-AT(A/T/C)TGGCA (A/G)AT(A/T/C)TT(T/C)CT-3′; rev-1, 5′-TG(A/G)AA(A/T/G)AT (T/C)TGCCA(A/G/T)AT-3′; 2A, 5′-AT(A/T/C)TT(A/G)GCNACNTA (T/C)AA-3′; rev-2A, 5′-TT(A/G)TANGTNGC(T/C)AA(A/T/G)AT-3′; 2B, 5′-AT(A/T/C)CTNGCNACNTA(T/C)AA-3′; rev-2B, 5′-TT(A/G)TANGTNGCNAG(A/T/G)AT-3′.
PCR was conducted with the primer pairs 1/rev-2A or rev-2B and rev-1/2A or 2B with the use of genomic DNA from RSY255 as a template. Thirty reaction cycles (each cycle was 0.5 min at 93°C, 1.5 min at 50°C, and 3 min at 72°C) were carried out, followed by a 5-min incubation at 72°C. A PCR product of 0.45 kilobase (kb) was obtained with primers 1 and rev-2A. This product was also detected in a similar reaction at a higher annealing temperature (53°C). This fragment was subcloned into an SmaI site of pBluescript II KS(+) (Stratagene, La Jolla, CA), giving pTYB121, and sequenced. An ORF was shown to span the entire 453-base pair (bp) insert. This insert was isolated, labeled with [α-32P]dCTP by means of the random primer DNA-labeling system (Amersham, Arlington Heights, IL), and used as a probe to screen yeast genomic libraries in YEp24 and YCp50. Screening was carried out according to the protocol provided by Amersham. We isolated four positive clones from YEp24 libraries (pTYY111–pTYY114) and three from YCp50 libraries (pTYY211–pTYY213). All seven clones shared a 0.5-kb EcoRI fragment, a 0.9-kb PstI fragment, and a 3.0-kb NcoI fragment that hybridized with the probe DNA. The EcoRI fragments from pTYY113 (3.3 and 0.5 kb) were subcloned into an EcoRI site of pBluescript II KS(+), and the NcoI fragment from pTYY212 (3.0 kb), converted to blunt ends by the Klenow enzyme, was introduced into a SmaI site of pBluescript II KS(+). These plasmids were used for sequencing.
Construction of Plasmids for Purification of YNL049C-encoded Protein (Iss1p)
A sequence coding for a stretch of six histidine residues was introduced in front of the termination codon of YNL049C (ISS1) as follows. The following four PCR primers were synthesized: TKPr1, 5′-CAGTAACCTCACTTAACCTATG-3′; TKPr2, 5′-TGTTAGTGATGGTGATGGTGATGTCTGTTGATACTAGTCTTCATAC-3′; TKPr3, 5′-ACAGACATCACCATCACCATCACTAACAATCAGTCTTTCTTTAATCTT-3′; and TKPr4, 5′-ATATGGCCATTTATCACGAATAC-3′.
His6 residues are encoded by the underlined sequence of TKPr3 and the sequence complementary to the underlined sequence of TKPr2. TKPr1 anneals to the antisense strand of the ISS1 ORF, ∼650 nucleotides upstream from the termination codon. A part of TKPr2, TCTGTTGATACTAGTCTTCATAC, anneals to the sense strand of the 3′ end of ISS1 ORF. A part of TKPr3, TAACAATCAGTCTTTCTTTAATCTT, anneals to the 3′-flanking region and termination codon of ISS1. TKPr4 anneals to the 3′-flanking region of ISS1, ∼900 nucleotides downstream from the termination codon. TKPr2 and TKPr3 anneal to each other. The plasmid pRH200 carries the ISS1 gene on a ClaI–XbaI genomic DNA fragment. The first-stage PCR was carried out with TKPr1 and TKPr2 and with TKPr3 and TKPr4 with the use of pRH200 as a template. We obtained an ∼650-bp fragment with TKPr1 and TKPr2 and an ∼900-bp fragment with TKPr3 and TKPr4. We used these two fragments as templates in the second-stage PCR, in which TKPr1 and TKPr4 were used as primers. This second-stage PCR yielded an ∼1.6-kb DNA encoding the C terminus of Iss1p that included His6 residues as well as a termination codon and the 3′-flanking region of ISS1.
The ClaI–XbaI fragment of pRH200 containing ISS1 was ligated with pGEM4Z (Promega, Madison, WI) digested with AccI and XbaI to generate pTKB1. The 1.4-kb AccI–SnaI region of pTKB1 corresponding to the 3′ end and 3′-flanking region of ISS1 was replaced by the 1.4-kb AccI–SnaI fragment of the above PCR product encoding the His6-tagged C terminus of Iss1p. This plasmid was named pTKB2. We sequenced the region of pTKB2 derived from the PCR product to ensure that no mutation had occurred as a result of PCR. XbaI–HindIII fragments of pTKB1 and pTKB2 were introduced into the XbaI–HindIII site of YEp352 (2μ URA3) to obtain pTKY4 and pTKY6, respectively. The PstI fragment (3.5 kb) of pTKY6 encoding His6-tagged Iss1p was introduced into the PstI site of p425GAL1 (2μ LEU2) to express the ISS1 coding sequence from the GAL1 promoter (Mumberg et al., 1994). This plasmid, pTKY7, was used for Iss1p purification.
The NcoI (blunted)–HindIII fragment (2.9 kb) of pTYY121 (YEp351 containing SEC23) was introduced into p426GAL1 (2μ URA3) digested with SpeI (blunted) and HindIII so that SEC23 could be expressed under control of the GAL1 promoter (Mumberg et al., 1994). This plasmid was named pTKY9.
Purification of His6-tagged Iss1p
RSY620 was transformed with pTKY7 and pTKY9. The cells were grown in SC-Ura-Leu (2% glucose) to early stationary phase and used to inoculate 6 l of SC-Ura-Leu (2% raffinose) at an initial OD600 of 0.005. The cells were grown at 30°C for 1 d until an OD600 of ∼1.2 was reached. At this time, 1/100 volume of 20% galactose was added (final concentration of 0.2%) and incubation continued for 5 h, to an OD600 of ∼2.7, for overproduction of Iss1p and Sec23p. The cells were harvested and washed twice with distilled water. About 25 g of cells (wet weight) were obtained from a 6-l culture. The cells were stored at −80°C until use.
The frozen cells were suspended with HSLB (0.75 M potassium acetate, 50 mM HEPES, 0.1 mM EGTA, 20% [wt/vol] glycerol; final pH was adjusted to 7.0 with 5 M KOH) to a final volume of 70 ml. Immediately before cell disruption, protease inhibitors and reducing agent were added to the following final concentrations: 1.4 mM 2-mercaptoethanol, 1 μM leupeptin, 1 μM pepstatin A, 1 mM ε-aminocaproic acid, and 0.5 mM PMSF. Cells were disrupted in a bead-beater chamber filled with a half-volume of glass beads (0.5 mm diam, Biospec Products, Bartlesville, OK) by five 1-min periods of agitation with 2-min intervals for chilling. The lysate was recovered, and the glass beads were washed once with 20 ml of HSLB supplemented with the protease inhibitors and 2-mercaptoethanol. The total lysate (80 ml) was centrifuged at 13,000 × g for 10 min, and the resulting supernatant was centrifuged at 40,000 rpm (∼186,000 × g) for 75 min (45Ti rotor, Beckman, Palo Alto, CA) to obtain a high-speed supernatant fraction.
The supernatant (46 ml) was loaded onto a 10-ml nickel-nitriloacetic acid (Ni-NTA) agarose column (Qiagen, Valencia, CA) equilibrated with HSLB with the protease inhibitors and 2-mercaptoethanol. The column was washed successively with 90 ml of B-II [0.75 M potassium acetate, 50 mM 2-(N-morpholino)ethanesulfonic acid, 0.1 mM EGTA, 20% (wt/vol) glycerol, 40 mM imidazole (pH 6.3 adjusted with 5 M KOH)], 20 ml of B-III [0.75 M potassium acetate, 50 mM HEPES, 0.1 mM EGTA, 0.25 M sorbitol, 20% (wt/vol) glycerol, 40 mM imidazole (pH 7.0)], 35 ml of B-IV100 (same as B-III except 100 mM imidazole), and finally 35 ml of B-IV200 (same as B-III except 200 mM imidazole). B-II, B-III, and B-IV100 contained the protease inhibitors. Sec23p/Iss1p was eluted from the column with B-IV500 (same as B-III except 500 mM imidazole). In a typical preparation, 1.5 mg of Sec23p/Iss1p was obtained from 25 g of cells (wet weight). Fractions that contained Sec23p/Iss1p were frozen in liquid nitrogen and stored at −80°C.
Disruption of SEC24 and ISS1
pTYB131 is a pBluescript II SK(+) derivative harboring the 4.2-kb XhoI–HindIII fragment from pTYY113 with the entire SEC24 gene. The BglII–SalI fragment (1.6 kb) of pTYB131 was replaced by the 2.5-kb BglII–SalI fragment of YEp13 containing the LEU2 gene to yield pTYY303. A 3.7-kb BamHI–HindIII fragment from pTYY303 containing a partial SEC24 disrupted by LEU2 was introduced into the diploid strain RSY612 to disrupt one of the chromosomal copies of SEC24. The resulting heterozygous disruption was named RSY866. We confirmed the disruption by Southern blot analysis. Tetrad analysis was performed as described (Sherman et al., 1983).
We deleted one of the chromosomal copies of ISS1 in the diploid strain YPH501 (Sikorski and Hieter, 1989) as follows. Two PCR primers (TKPr12, 5′-CCTTCTTCCATTAATGATCGACAGCTGCAGTGAATAGCAGATTGTACTGAGAGTGCACC-3′; and TKPr13, 5′-GGTTAATAAAGATAAAGATTAAAGAAAGACTGATTGGCATAT-GATCCGTCGAGTTCAA-3′) were used to amplify the HIS3 gene on pRS313 (Sikorski and Hieter, 1989). The underlined sequences of TKPr12 and TKPr13 anneal to the 5′ and 3′ regions of HIS3, respectively. YPH501 was transformed with the amplified DNA fragment, and His+ transformants were selected. The transformants were sporulated and dissected to obtain a haploid cell with a disruption of the ISS1 gene (iss1-Δ1::HIS3).
A second disruption (iss1-Δ2::TRP1) that replaced amino acids 116–622 of ISS1 with the TRP1 marker was made by one-step disruption of the chromosomal ISS1 gene. The disruption plasmid (pRH247) was constructed as follows. A SpeI fragment of pRH200 was cloned into pRS306, creating pRH217. After deletion of the EcoRI site from the polylinker, the 1.5-kb BglII–EcoRI fragment of pRH217 was replaced with a 1-kb fragment containing the TRP1 marker, creating pRH247.
A trp1 diploid, CKY19, was transformed with the 2.3-kb SpeI fragment of pRH247, yielding CKY498. Tetrad analysis of CKY498 gave 2:2 segregation of TRP1. Integration of TRP1 at the ISS1 locus was confirmed by Southern blotting
Construction of Yeast Strains for the Galactose Shut-Off Experiment
The SEC24 ORF was fused to the GAL1 promotor as follows. A 5′-terminal region of SEC24 was amplified and mutated by PCR to introduce an XbaI site in front of the initiation codon. No misincorporation was found by sequencing. The amplified fragment was subcloned into the HincII site of pBluescript II SK(+) to obtain pTYB133. We replaced the BamHI–SacI region of pTYB133 with a 3.0-kb BamHI–SacI fragment from pTYB131 to obtain the complete ORF (pTYB134). A 3.1-kb XbaI–HindIII fragment of pTYB134 was isolated and introduced downstream of the GAL1 promoter on pBM743 followed by a multicloning site. The resulting plasmid, pTYY214, was introduced into RSY866. After sporulation of the transformant, a haploid segregant was obtained in which SEC24 was expressed under the control of the GAL1 promoter. RSY875 is leu2-3,-112 his3-12,15 trp1-1 ura3-1 ade2-1 GAL2 sec24::LEU2 (pTYY214 [URA3 Gal1p–SEC24]).
Multicopy Suppression Analysis
The XhoI–HindIII fragment (4.2 kb) from pTYB131containing SEC24 was ligated into YEp352 (2μ URA3) digested with SalI and HindIII to obtain pTYY115. The HindIII fragment containing SEC23 was introduced into the HindIII site of YEp352 and pTYY115, giving pTYY122 and pTYY116, respectively. Various temperature-sensitive sec mutant strains were transformed with these plasmids.
Construction of ISS1-overexpressing Yeast Strains with SEC24 Disrupted
RSY866 (MATa/α SEC24/sec24::LEU2) was transformed with pTKY4 (2μ URA3 ISS1). The resultant strain was sporulated, and the asci were dissected. Four strains derived from the same tetrad were named TKY1 (MATa SEC24 [pTKY4 (URA3 ISS1)]), TKY2 (MATα SEC24 [pTKY4 (URA3 ISS1)]), TKY3 (MATa sec24::LEU2 [pTKY4 (URA3 ISS1)]), and TKY4 (MATα sec24::LEU2 [pTKY4 (URA3 ISS1)]).
Similar strains were constructed with pTKY6 (2μ URA3 His6-tagged version of ISS1) instead of pTKY4 and named TKY5 (MATa SEC24 [pTKY6]), TKY6 (MATα SEC24 [pTKY6]), TKY7 (MATa sec24::LEU2 [pTKY6]), and TKY8 (MATα sec24::LEU2 [pTKY6]).
We constructed the following plasmid and yeast strain to regulate the ISS1 expression level in the sec24-disrupted background. The BamHI–HindIII fragment (3.5 kb) of pTKY7 containing ISS1 was introduced into p426GAL1 (2μ URA3) digested with BamHI and HindIII. The resultant plasmid (pTKY11) was introduced into the diploid strain RSY866, in which one of the SEC24 genes had been disrupted by LEU2. The transformant was sporulated and dissected on a YPGal plate to allow the expression of ISS1. We obtained a haploid Leu+ and Ura+ strain, TKY22 (sec24::LEU2 [pTKY11 (URA3 Gal1p–ISS1)]), and a Ura+ strain, TKY23 (SEC24 [pTKY11 (URA3 Gal1p–ISS1)]).
Pulse-Chase Experiment
For pulse-chase analysis of carboxypeptidase Y (CPY) during Sec24p depletion, RSY875 was grown in MV-lowS (galactose) with appropriate nutrients and then transferred to fresh MV-lowS (galactose) or MV-lowS (glucose) with the supplements. After 9, 12, and 15 h of incubation, 3.0-OD600-unit cells were collected, washed, and transferred to 5 ml of MV-noS (galactose) or MV-noS (glucose). Cells were labeled with 9.3 MBq Trans35S-label (ICN, Costa Mesa, CA) for 10 min and then chased for 60 min. Aliquots (1.2 OD600 units of cells) were withdrawn before and after the chase, and lysates were prepared with glass beads as described (Rothblatt and Schekman, 1989). Radioactive proteins immunoprecipitated with anti-CPY antibody were separated on SDS-PAGE and detected with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
We conducted a pulse-chase experiment for the iss1 null strain and the ISS1-overexpressing, sec24-disrupted strain as follows. The cells were grown in SC dropout medium to late log phase and then transferred to fresh SC dropout medium (initial OD600 = 0.15). After a 2.5-h incubation at 30°C, the cells were collected, washed three times with SC-Met dropout medium, and suspended in SC-Met dropout medium (OD600 = 0.3). After a 15-min incubation at 30°C, 35S-Promix (Amersham) was added (1.5 MBq for 0.3-OD600-unit cells). After a 7-min incubation at 30°C, methionine and cysteine were added (final concentration of each amino acid was 0.6 mg/ml) and incubation was continued at 30°C. Cells (0.3 OD600 unit) were taken from the solution 0, 5, 15, 30, and 60 min after the addition of methionine and cysteine. The cell suspension was mixed with an equal volume of 10 mM NaN3/10 mM NaF on ice, collected by centrifugation, and washed once with 10 mM NaN3/10 mM NaF. The cells were resuspended in lysis buffer (1% SDS, 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM PMSF) (100 μl for 0.3-OD600-unit cells) and disrupted with glass beads. Radioactive proteins immunoprecipitated with anti-CPY antibody or anti-Gas1p antibody were separated by SDS-PAGE and detected with a PhosphorImager (Molecular Dynamics).
Electron Microscopy
RSY875 grown in YPGal (5%) to OD600 = 0.15–0.6 was collected and suspended in sterile distilled water to OD600 = 6. Fifty milliliters of YPD (5%) and YPGal (5%) were inoculated with 0.1 ml of the RSY875 suspension. After a 9-h incubation at 30°C, the cells were fixed with glutaraldehyde followed by potassium permanganate, as described by Kaiser and Schekman (1990). Briefly, 50% glutaraldehyde was added to cultures (final concentration, 1%) for 10 min. Then the cells were centrifuged, washed, and resuspended in 4% KMnO4 for 2–4 h at 4°C. Fixed cells were collected, washed several times in water, and incubated in 2% uranyl acetate for 12–16 h at 4°C. After several rinses in water, the samples were dehydrated in an ethanol series and embedded in Spurr's medium. Thin sections were stained with lead citrate and viewed in a JEOL100 electron microscope (JEOL, Tokyo, Japan).
α-Factor Halo Assay
A Δsst2 strain, whose growth is arrested in the presence of α-factor, was grown in YPD at 30°C to exponential phase and suspended in YPD containing 1% agar to a final OD600 of 3 × 10−4. A YPD plate was overlaid with this suspension. To examine α-factor secretion, strains were grown to stationary phase in SC dropout medium and spotted (0.01 OD600 unit/spot) on the Δsst2-covered plate. The plate was incubated at 30°C for 2 d.
In Vitro ER Vesicle-budding Assay
GPY60 was grown at 30°C in YPD to exponential phase, and microsomes were prepared as described (Wuestehube and Schekman, 1992). Purified microsomes were adjusted to 40 OD280 (∼8 mg protein/ml) in B88 (20 mM HEPES, pH 6.8, 250 mM sorbitol, 150 mM potassium acetate, 5 mM magnesium acetate). The preparation was frozen in liquid nitrogen and stored at −80°C.
The microsome-based α-factor packaging assay was carried out as follows based on the method described (Baker et al., 1988; Rexach and Schekman, 1991; Kuehn et al., 1996). [35S]Prepro-α-factor was posttranslationally translocated into microsomes in the presence of 1× ATP regeneration mix (Baker et al., 1988) at 10°C for 30 min. Microsomes (400 μg of protein) containing [35S]gpαF were washed once with 1 ml of B88, resuspended in 50 μl of B88, mixed with 50 μl of B88 containing 4.2 M urea (final concentration, 2.1 M), and incubated at 0°C for 10 min. After addition of 1 ml of B88, microsomes were collected by centrifugation and washed twice with 1 ml of B88. Budding reactions were carried out in 50 μl of B88 containing 20 μg of urea-washed microsomes, 1× ATP regeneration mix, 0.1 mM GMP-PNP, and appropriate amounts of Sar1p, Sec13p/Sec31p, Sec23p/Sec24p, and Sec23p/Iss1p, whose concentrations are described in RESULTS. The mixture was incubated at 20°C for 30 min, unless noted otherwise, and chilled on ice for 5 min. Portions of the total reaction and the medium-speed supernatant (MSS) (12,000 × g, 4 min) were collected. The amount of trypsin-resistant, concanavalin A–precipitable [35S]gpαF in the MSS was divided by the amount in the total fraction to determine the percentage of α-factor packaged into the vesicles.
The large-scale budding reaction was carried out as follows to isolate the vesicles derived from the ER. For each reaction, microsomes containing 2 mg of proteins were used. Microsomes were incubated with 1× ATP regeneration mix in 1 ml of B88 for 30 min at 10°C. After being washed once with 1 ml of B88, they were incubated with 2.1 M urea in 300 μl of B88 for 10 min at 0°C. After addition of 1 ml of B88, microsomes were collected by centrifugation and washed twice with 1 ml of B88. A budding reaction was carried out at 20°C for 30 or 60 min in 1 ml of B88 containing 2 mg of microsomes, 1× ATP regeneration mix, 0.2 mM GMP-PNP, 65 μg of Sar1p, 120 μg of Sec13p/Sec31p, and 35 μg of either Sec23p/Sec24p or Sec23p/Iss1p. After 5 min on ice, a 50-μl aliquot reaction mixture was taken as total, and the remaining solution was centrifuged (14,000 × g, 4 min) to obtain a MSS fraction. A sucrose density gradient consisting of 0.3 ml of B88 containing 70% (wt/wt) sucrose and 2.5 ml of B88 containing 15% (wt/wt) sucrose was overlaid with 750 μl of the MSS. After centrifugation at 50,000 rpm (∼250,000 × g) for 2 h (SW55, Beckman), the interface (∼0.5 ml) between 15 and 70% sucrose was collected, and its sucrose concentration was adjusted to 55% (wt/wt) with the use of B88 containing 70% (wt/wt) sucrose. The final volume was ∼0.8 ml, and 0.55 ml of this solution was placed on the bottom of a sucrose density gradient consisting of B88 containing 52.5, 50, 45, 40, 35, and 25% (wt/wt) sucrose (from the bottom to the top). The volume of each of the bottom three layers was 0.5 ml, and the volume of each of the top three layers was 1 ml. This gradient was centrifuged at 50,000 rpm (∼250,000 × g) for 20 h (SW55, Beckman), and fractions (0.4 ml × 13) were collected from the top with a density gradient fractionator (ISCO, Lincoln, NE). Proteins in these fractions were concentrated by TCA precipitation.
Other Methods
Sarlp, Sec13p/Sec31p, and Sec23p/Sec24p were purified as described previously (Barlowe et al., 1994; Yeung et al., 1995; Salama et al., 1997). DNA manipulation was done according to Sambrook et al. (1989). Yeast cells were transformed by the lithium acetate method (Ito et al., 1983). Protein concentrations were determined with the Bio-Rad (Richmond, CA) protein assay kit with the use of BSA as a standard. Silver staining was carried out as described (Bloom et al., 1987). Western blot analysis was performed with a nitrocellulose membrane, and ECL (Amersham) was used for detection.
RESULTS
Cloning and Sequence Analysis of SEC24
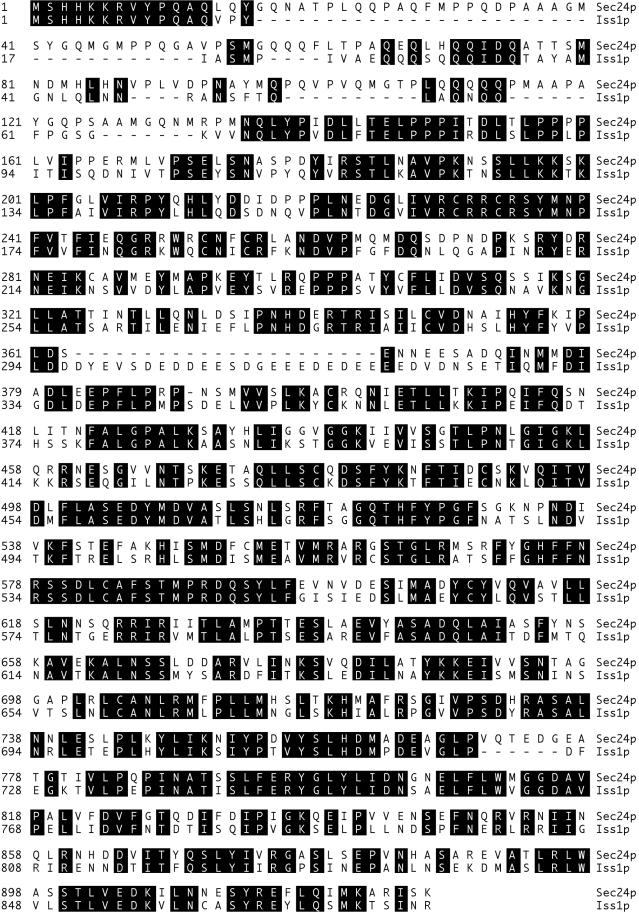
We purified the Sec23p/Sec24p complex from crude yeast cytosol and obtained peptide sequence information from tryptic fragments of Sec24p. A part of the SEC24 gene was PCR amplified with the use of degenerate primers designed according to the peptide sequence information. Finally, a DNA fragment containing the entire SEC24 gene was obtained by screening a S. cerevisiae genomic DNA library with the use of the PCR fragment as a hybridization probe. The identified ORF (YIL109C) encoded 926 amino acid residues (Figure 1), including four sequences corresponding to those obtained by sequencing of the tryptic fragments. The predicted molecular mass of 103,614 Da agrees with the estimated mass of purified Sec24p (∼105 kDa). The coding sequence does not appear to contain a hydrophobic signal sequence or transmembrane domain, consistent with the observation that Sec23p/Sec24p is a cytosolic protein (Hicke et al., 1992). As a final verification of identity, we constructed a hybrid of the trpE gene fused to the ORF YIL109C and showed that this fusion protein expressed in Escherichia coli could be recognized by anti-Sec24p (our unpublished data). These data confirm that SEC24 is YIL109C.
Figure 1.
Sequence alignment of Sec24p (YIL109C) with Iss1p (YNL049C). The CLUSTAL method was used for alignment.
We searched a nonredundant sequence database for similarity with the Sec24p sequence with the use of BLAST2 (http://www.ncbi.nlm.nih.gov/BLAST/) and found that human (DDBJ accession number D38555), Caenorhabditis elegans (SPTREMBL accession numbers Q19371 and Q23368), and Arabidopsis thaliana (EMBL accession number AL022537 [PID accession number e1287285]) have homologous proteins (our unpublished data).
Interestingly, a second ORF of S. cerevisiae, YNL049C, showed striking similarity with SEC24. YNL049C was first isolated in a two-hybrid interaction assay with the use of the central region of Sec16p as a bait construct and was named ISS1 (interactor with SEC sixteen) (our unpublished data). The protein sequence of Iss1p is 62% identical to that of Sec24p (Figure 1). This sequence similarity extends throughout the length of the protein and defines two variable regions in the N-terminal part of the proteins, a glutamine-rich domain (amino acids 17–143 in Sec24p, amino acids 17–67 in Iss1p) and a charged domain (amino acids 362–372 in Sec24p, amino acids 295–327 in Iss1p).
Sec24p Depletion is Lethal and Causes a Defect in CPY Maturation and Exaggeration of the ER
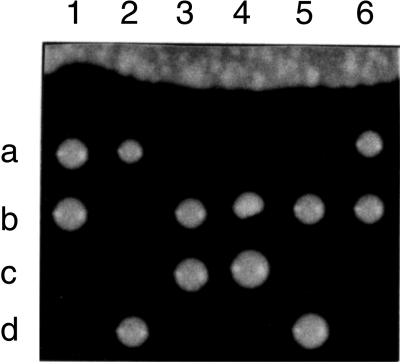
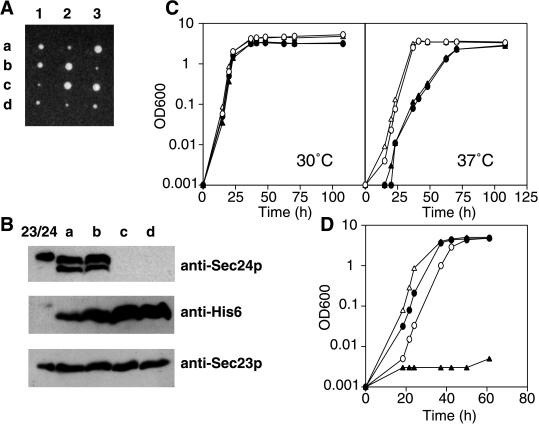
The SEC24 gene was disrupted with LEU2 in a diploid strain. After sporulation, most tetrads contained two viable and two dead spores (Figure 2). The viable spores were all Leu−, indicating that they retained wild-type SEC24. This was confirmed by Southern blot analysis (our unpublished data). SEC24 is thus essential for cell viability.
Figure 2.
Tetrad analysis of a SEC24/sec24::LEU2 diploid (RSY866). All viable spores are Leu−.
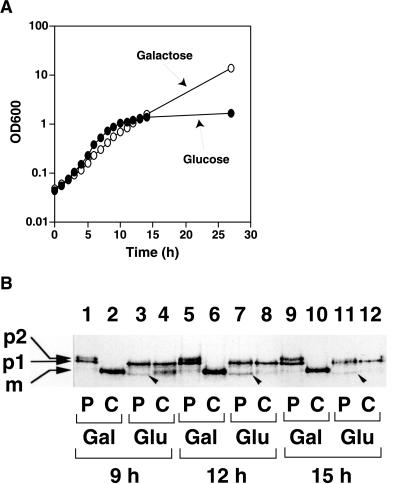
To investigate the phenotype of a strain depleted of Sec24p, we constructed haploid strain RSY875 that has a disrupted chromosomal SEC24 locus covered by a plasmid-borne copy of SEC24 expressed from the GAL1 promoter. When RSY875 cells were grown to early exponential phase in MVCA (galactose) and then transferred to MVCA (glucose) to repress expression of Sec24p, cell growth slowed after 8 h and ceased after ∼14 h (Figure 3A).
Figure 3.
Secretion defects caused by depletion of Sec24p. (A) RSY875, carrying SEC24 expressed from the GAL1 promoter, was grown to log phase. After washing, the culture was divided into two portions and transferred to either fresh MVCA (galactose) or MVCA (glucose) at 30°C, and growth was monitored. To keep exponential growth, cultures were diluted 10-fold after 10 h. (B) A culture of RSY875 was transferred to MV-lowS (galactose) (lanes 1, 2, 5, 6, 9, and 10) or MV-lowS (glucose) (lanes 3, 4, 7, 8, 11, and 12). After 9, 12, or 15 h of cultivation at 30°C, 3.0-OD600-unit cells were pulse labeled with [35S]methionine for 10 min (lanes marked with P) and then chased for 60 min with an excess of unlabeled methionine (lanes marked with C). CPY was immunoprecipitated and analyzed by SDS-PAGE and autoradiography. p1, p2, and m represent p1 precursor (67 kDa), p2 precursor (69 kDa), and mature form (61 kDa) of CPY, respectively. Bands marked with arrowheads (60 kDa) may represent untranslocated precursors.
Using the cells grown in the glucose or galactose medium for 9, 12, and 15 h, we conducted a pulse-chase experiment to monitor the maturation of CPY as an assay for the function of the early part of the secretory pathway. Newly synthesized pro-CPY is translocated into the ER, where it is glycosylated to become the p1 form (67 kDa), and then transported to the Golgi to be modified to the p2 form (69 kDa). Mature CPY (61 kDa) is produced by proteolytic cleavage after pro-CPY enters the vacuole. In RSY875 cells grown on galactose medium, the p1 form of pulse-labeled CPY was processed to the mature form within 60 min of the chase period (Figure 3B, lanes 2, 6, and 10). In contrast, cells that had grown in glucose for 9 h processed only a portion of CPY to the mature form, and after 12 or 15 h of growth in glucose medium most of the CPY remained as the p1 form (Figure 3B, lanes 4, 8, and 12). Thus, depletion of cellular Sec24p causes a block in ER-to-Golgi transport. In the glucose-grown cells, some CPY in a 60-kDa form was detected during the pulse period (Figure 3B, lanes 3, 7, and 11, arrowheads), and this form appeared to be converted to the p1 form after the chase. This 60-kDa form of CPY likely corresponds to prepro-CPY, an ER membrane translocation precursor. An accumulation of prepro-CPY may be a consequence of a prolonged ER-to-Golgi transport defect.
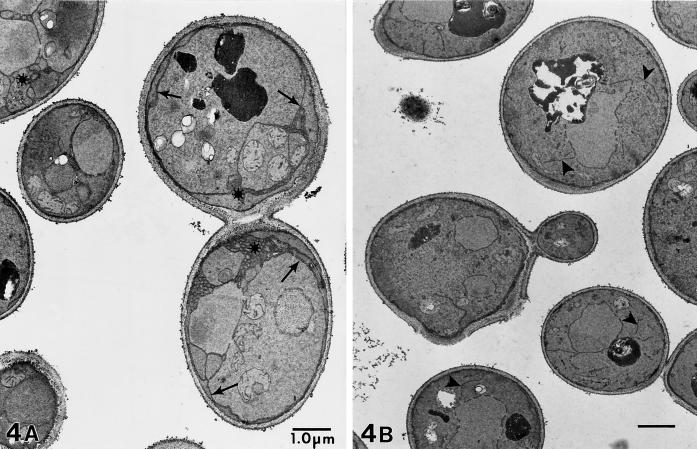
We examined the morphological consequences of Sec24p depletion by electron microscopy of fixed cells. RSY875 was grown in YPGal and then transferred to either YPGal or YPD for 9 h. Cells were fixed with glutaraldehyde followed by 4% KMnO4, a fixation and stain that gives contrast to membrane-derived structures (Kaiser and Schekman, 1990). Cells depleted of Sec24p by growth on glucose showed extensive ER proliferation: long sheets of ER could be seen in the cytoplasm and concentrated at the cell periphery. In a few cases, the ER sheets or tubules formed mesh-like structures (Figure 4A). The cells that contained Sec24p because of growth in galactose appeared to have normal ER morphology (Figure 4B). A control strain with a wild-type copy of SEC24 on its chromosome showed normal morphology in both glucose and galactose media (our unpublished data). The morphological phenotype of Sec24p depletion is similar to that seen in the class I ER-to-Golgi sec mutants such as sec12 (Kaiser and Schekman, 1990) and is in agreement with the in vitro observation that Sec24p is required for COPII vesicle formation (Hicke et al., 1992).
Figure 4.
Electron microscopy of RSY875 cells grown for 9 h in YPGlu (A) or YPGal (B). (A) Glucose-grown Sec24p-depleted cells show amplification of ER membranes (arrows) and occasional tubules or meshwork structures (asterisks). (B) Cells in YPGal have normal ER morphology (arrowheads). Bars, 1 μm.
Genetic Interaction between SEC24 and Other ER-to-Golgi SEC Genes
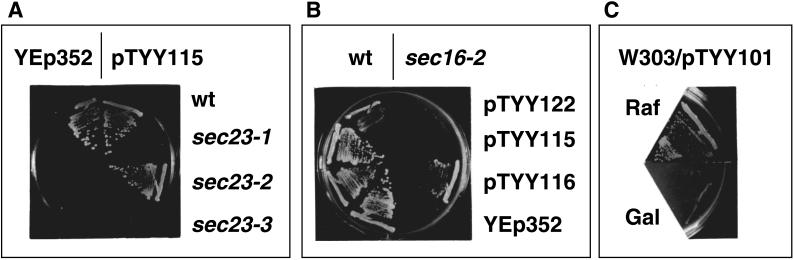
We next investigated the effect of Sec24p overproduction on the early sec mutants. Either pTYY122 (SEC23 on YEp352 [2μ URA3]), pTYY115 (SEC24 on YEp352), pTYY116 (SEC23 and SEC24 on YEp352), or vector (YEp352) was introduced into a Sec+ strain as well as into the following sec mutants: sec12-1, sec13-1, sec16-2, sec17-1, sec18-1, sec19-1, sec20-1, sec22-3, sec23-1, sec23-2, sec23-3, and sec23-4. The growth of these transformants at 23 and 37°C was compared on plates. Although the temperature-sensitive phenotypes of sec12-1, sec13-1, sec17-1, sec18-1, sec20-1, sec22-3, sec23-1, sec23-3, and sec23-4 were not suppressed by these plasmids (our unpublished data), sec23-2 and sec16-2 were. Overproduction of Sec24p suppressed sec23-2 (Figure 5A), and overproduction of both Sec23p and Sec24p in the same cell suppressed sec16-2 (Figure 5B). These genetic interactions are consistent with the binding interactions among Sec24p, Sec23p, and Sec16p that have been detected in vitro (Hicke et al., 1992; Gimeno et al., 1996).
Figure 5.
Effect of Sec24p and/or Sec23p overproduction on yeast growth. (A) The growth of wild-type (wt), sec23-1, sec23-2, and sec23-3 strains with either pTYY115 (2 μm SEC24) or YEp352 (2 μm without SEC24) at 37°C. pTYY115 suppressed temperature-sensitive sec23-2. (B) Wild-type and sec16-2 strains with either pTYY122 (2 μm SEC23), pTYY115 (2 μm SEC24), pTYY116 (2 μm SEC23 SEC24), or YEp352 were streaked on a MVCA (glucose) plate and incubated at 37°C for 3 d. Only pTYY116 suppressed sec16-2. (C) RSY612 (pTYY101 [2 μm Gal1p–SEC23]) precultured on MVCA (raffinose) was streaked on either MVCA (raffinose; Raf) or MVCA (galactose; Gal). This strain did not grow on galactose plates, whereas RSY612 with vector plasmid pCG109 grew on both raffinose and galactose plates (our unpublished data).
Overproduction of Sec23p was toxic to both sec12-1 and sec13-1 (our unpublished data). Even the wild-type cells could not tolerate the overproduction of Sec23p by a multicopy plasmid with SEC23 under the control of the GAL1 promoter (Figure 5C). These phenomena are consistent with the fact that excess Sec23p monomer inhibits vesicle formation in vitro (Yoshihisa et al., 1993). The introduction of SAR1 on multicopy plasmid only partially mitigated the growth inhibition by overexpression of SEC23 (our unpublished data).
A temperature-sensitive allele of SEC24, sec24-1, was isolated from a collection of random temperature-sensitive S. cerevisiae mutants by screening for secretion defects at 37°C. The sec24-1 mutant is unable to grow at temperatures >27°C and accumulates the ER (p1) form of the secretory marker protein CPY (Chitouras, Frand, and Kaiser, unpublished observations). We crossed sec24-1 to different sec mutant strains to test for possible synthetic lethal interactions. Because Sec24p is required for COPII vesicle formation in vitro, we expected to find strong synthetic lethal interactions between sec24-1 mutations and other vesicle-formation mutations (sec12-4, sec13-1, sec16-2, and sec23-1), which was the case (Table 3). In addition, we detected significant synthetic lethal interactions between sec24-1 and mutations in v-SNARE genes (sec22-3 and bet1-1) and other mutants defective in vesicle fusion (sec17-1 and sec18-1) (Table 3). Such interactions with docking or fusion mutations were not observed for sec12, sec13, sec16, or sec23 mutations (Kaiser and Schekman, 1990), suggesting that Sec24p may have a unique function among the COPII genes in the packaging of SNARE proteins, or in the docking or fusion of ER-derived vesicles with the Golgi apparatus (Peng et al., 1999).
Table 3.
Genetic interactions of iss1 and sec24 mutants
| Genotype | Incubation temperature
|
||
|---|---|---|---|
| 28°C | 30°C | 33°C | |
| sec24-1 | ++ | − | − |
| sec24-1 iss1-Δ1∷TRP1 | ++ | − | − |
| SNAREs | |||
| sec22-3 | ++ | ± | − |
| sec22-3 iss1-Δ1∷TRP1 | Double mutants not viable at 25°C | ||
| sec22-3 sec24-1 | Double mutants not viable at 25°C | ||
| bet1-1 | +++ | +++ | +++ |
| bet1-1 iss1-Δ1∷TRP1 | +++ | − | − |
| bet1-1 sec24-1 | Double mutants not viable at 25°C | ||
| Vesicle fusion | |||
| sec17-1 | +++ | ++ | ± |
| sec17-1 iss1-Δ1∷TRP1 | +++ | ++ | ± |
| sec17-1 sec24-1 | − | − | − |
| sec18-1 | +++ | − | − |
| sec18-1 iss1-Δ1∷TRP1 | +++ | − | − |
| sec18-1 sec24-1 | Double mutants not viable at 25°C | ||
| Vesicle formation | |||
| sec12-4 | +++ | − | − |
| sec12-4 iss1-Δ1∷TRP1 | +++ | − | − |
| sec12-4 sec24-1 | Double mutants not viable at 25°C | ||
| sec13-1 | +++ | + | − |
| sec13-1 iss1-Δ1∷TRP1 | +++ | + | − |
| sec13-1 sec24-1 | Double mutants not viable at 25°C | ||
| sec16-2 | ++ | − | − |
| sec16-2 iss1-Δ1∷TRP1 | ++ | − | − |
| sec16-2 sec24-1 | Double mutants not viable at 25°C | ||
| sec23-1 | +++ | − | − |
| sec23-1 iss1-Δ1∷TRP1 | +++ | − | − |
| sec23-1 sec24-1 | Double mutants not viable at 25°C | ||
Growth of single colonies on YPD after 24–48 h. +++, growth comparable to wild type; −, no growth.
ISS1 Is Not Essential for Growth or Transport of CPY and Gas1p
Next, we examined the function of the SEC24 homologue ISS1. We used HIS3 to disrupt one copy of the ISS1 gene in a diploid strain. On sporulation, the heterozygous diploid yielded four spores from most tetrads. Two of the spores were His+, and the other two were His−. The presence of the iss1-Δ1::HIS3 disruption in the His+ strain was verified by PCR with the use of primers to the 5′-noncoding region of ISS1 and HIS3 coding sequences. The iss1-Δ1::HIS3 haploids grew normally at 24, 30, and 37°C. Therefore, ISS1 is not essential for cell growth.
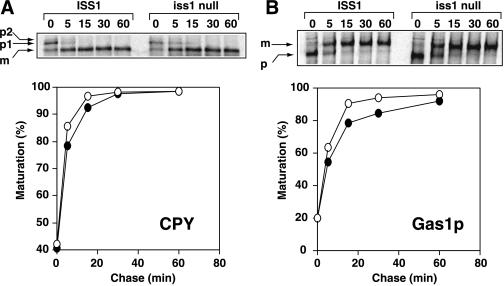
The rate of maturation of CPY and Gas1p in the iss1-Δ1::HIS3 strain was examined by a pulse-chase experiment. Gas1p is first modified in the ER to form a 105-kDa GPI-anchored precursor with N- and O-linked core oligosaccharides. Then it is modified to a 125-kDa mature form in the Golgi and delivered to the plasma membrane. After a 7-min pulse, TKY10 (ISS1) and TKY12 (iss1-Δ1::HIS3) were chased for up to 60 min. The maturation rate of CPY in TKY12 was almost the same as that in TKY10 (Figure 6A). Gas1p maturation was slightly slower in TKY12 than in TKY10, but the effect was moderate (Figure 6B). These data indicate that ISS1 is not essential for the transport of these proteins.
Figure 6.
Effect of ISS1 deletion on maturation of CPY (A) and Gas1p (B). After pulse-chase analysis of CPY or Gas1p, the intensities of radioactive bands were quantified with a PhosphorImager (Molecular Dynamics). Maturation was calculated as follows: CPY maturation (%) = m/(p1 + m) × 100; Gas1p maturation (%) = m/(p + m) × 100, where p indicates precursor and m indicates mature form. (○) ISS1 strain; (●) iss1 null strain.
Suppression of SEC24 Mutations by ISS1
Because Iss1p is similar to Sec24p in both sequence and protein–protein interactions, we tested whether Iss1p could substitute for Sec24p during ER-to-Golgi transport. An initial indication of functional overlap came from the observation that the expression of ISS1 from a high-copy-number (2μ) plasmid could restore growth of sec24-1 at temperatures up to 36°C (our unpublished data). We further tested the ability of ISS1 overexpression to suppress a chromosomal deletion of SEC24. RSY866, a diploid strain heterozygous for a SEC24 gene disruption (sec24::LEU2), was transformed with pTKY4 (2μ plasmid with ISS1 gene). Sporulation of this diploid often yielded tetrads with four viable spores. In the tetrads with four viable spores, two of the spore clones were Leu+ and the other two were Leu−. The absence of the SEC24 gene in Leu+ spore clones was confirmed by Western blot analysis for Sec24p. Thus, sec24::LEU2 can be suppressed by increased dosage of the ISS1 gene. We could not obtain a sec24::LEU2 haploid strain with the use of pRH200 (CEN ISS1) instead of pTKY4 (2 μm ISS1) (our unpublished data).
We also found that ISS1 tagged with a His6-encoding sequence can suppress SEC24 disruption (Figure 7A), indicating that the His6-tagged version of Iss1p is functional in vivo. Immunoblot analysis showed that all four strains derived from one tetrad produced Iss1p-His6 as well as Sec23p and that Sec24p was absent from the two Leu+ spores (Figure 7B). A strain with the chromosomal disruption sec24::LEU2 suppressed by pTKY4 (2 μm ISS1) grew more slowly than the wild-type strain at 37°C, but the growth was indistinguishable from that of the wild-type strain at 24 and 30°C (Figure 7C).
Figure 7.
Complementation of sec24 disruption by ISS1. (A) Tetrad analysis of RSY866 (SEC24/sec24::LEU2 [pTKY6 (2 μm His6-tagged version of ISS1)]). Four viable spores were obtained. Two smaller colonies in each set grew on SC-Leu, indicating the disruption of SEC24 (our unpublished data). (B) Western blot analysis of TKY5 (MATa SEC24 [pTKY6]) (a), TKY6 (MATα SEC24 [pTKY6]) (b), TKY7 (MATa sec24:: LEU2 [pTKY6]) (c), and TKY8 (MATα sec24:: LEU2 [pTKY6]) (d). Sec23p/Sec24p-His6 (0.15 μg) (Yeung et al., 1995) was loaded onto the lane indicated by 23/24. The cells were grown in SC-Ura to stationary phase and disrupted with glass beads in HSLB containing protease inhibitors and 2-mercaptoethanol. After centrifugation at 14,000 × g for 10 min, protein in the supernatant was concentrated by TCA precipitation. The precipitated protein (100 μg) was analyzed by immunoblotting with the use of either anti-Sec24p, anti-His6, or anti-Sec23p antibody. The slower mobility of Sec24p in lane 23/24 is probably due to the presence of His6 tag, which was attached to Sec24p in the cells analyzed here. In lanes a and b, a degradation product of Sec24p is also seen. (C) Growth curve of TKY5, TKY6, TKY7, and TKY8 in SC-Ura at 30 and 37°C. (○) TKY5; (▵) TKY6; (●) TKY7; (▴) TKY8. (D) TKY22 (sec24:: LEU2 [pTKY11 (Gal1p–ISS1)]) and TKY23 (SEC24 [pTKY11 (Gal1p–ISS1)]) were cultured in SC-Ura (2% galactose) and then transferred to either SC-Ura (2% galactose) or SC-Ura (2% glucose). Growth at 30°C was monitored. (○) TKY23 in galactose medium; (▵) TKY23 in glucose medium; (●) TKY22 in galactose medium; (▴) TKY22 in glucose medium.
The apparent indispensability of Iss1p overexpression in cells with the chromosomal disruption sec24::LEU2 was confirmed by a galactose shut-off experiment. TKY22 (sec24::LEU2 [pTKY11 (GAL1p–ISS1)]) grew well when Iss1p expression was induced by growth in the galactose medium, but the growth of this strain was severely inhibited when Iss1p expression was repressed by growth on glucose medium (Figure 7D).
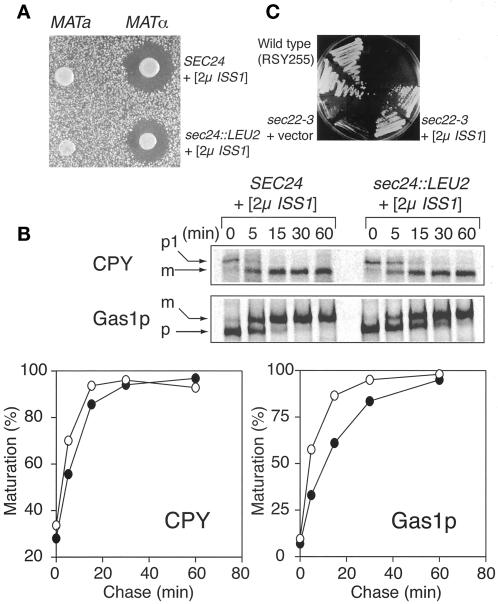
We next examined whether protein secretion was fully restored in strains with the chromosomal disruption sec24::LEU2 suppressed by pTKY4 (2 μm ISS1). A convenient test for secretion of α-factor is to assay the extent of growth inhibition of a MATa sst2Δ strain by a halo assay. Clear halos of equal diameter formed around both a MATα sec24::LEU2 (pTKY4 [2μ ISS1]) strain and the MATα control strain, showing that the secretion of α-factor was normal (Figure 8A).
Figure 8.
ISS1 overexpression suppresses sec24::LEU2 and sec22-3. (A) Halo assay for α-factor secretion comparing MATα control strain TKY6 (MATα SEC24 [pTKY6 (2μ ISS1)]) with TKY8 (MATα sec24::LEU2 [pTY6 (2μ ISS1)]). (B) Pulse-chase analysis of CPY and Gas1p transport in TKY6 (SEC24 [pTKY6 (2μ ISS1)]) and TKY8 (sec24::LEU2 [pTKY6 (2μ ISS1)]). Cells were labeled for 7 min and chased for the times indicated. Maturation was calculated as for Figure 6. (○) TKY6; (●) TKY8. (C) Suppression of sec22-3 by 2μ ISS1. CKY70 (sec22-3) carrying either ISS1 on a 2μ plasmid or a control plasmid was grown for 3 d at 30°C. Duplicate transformants are shown.
We also examined the maturation kinetics of CPY and Gas1p in the sec24::LEU2 (pTKY4 [2μ ISS1]) strain by a pulse-chase experiment. Maturation of CPY occurred in the sec24::LEU2 (pTKY4 [2μ ISS1]) cells at almost the same rate as in SEC24 control cells (Figure 8B). Maturation of Gas1p occurred more slowly, but after a 60-min chase, >90% of Gas1p was converted to the mature form (Figure 8B). These data indicate that transport of CPY and Gas1p from the ER to the Golgi can be achieved in the Sec24p-depleted cells if there is sufficient Iss1p.
Genetic Interactions between ISS1 and v-SNARE Mutants
To learn more about the function of Iss1p in the secretory pathway, we tested for synthetic lethal interactions between iss1-Δ2::TRP1 and a panel of secretion mutants (Table 3). Synthetic lethal interactions have been helpful for detecting the step in the secretory pathway at which a gene product acts, because synthetic lethal interactions usually occur only between two genes that affect the same step of the pathway (Newman et al., 1987; Salminen and Novick, 1987; Rothblatt et al., 1989; Kaiser and Schekman, 1990; Gimeno et al., 1996). Surprisingly, iss1-Δ2::TRP1 did not affect mutants defective in vesicle formation (sec12-4, sec13-1, sec16-2, and sec23-1) or vesicle fusion (sec17-1 and sec18-1) but showed strong synthetic lethal interactions with mutants defective in v-SNARE proteins required for ER-to-Golgi transport (sec22-3 and bet1-1) (Table 3).
We also examined the corresponding effects of ISS1 overexpression on v-SNARE mutations. ISS1 on either a low- or high-copy-number plasmid was transformed into a variety of sec and bet mutant strains, and the transformants were tested for their temperature sensitivity. ISS1 overexpression suppressed the growth defect of both sec22 alleles tested (Figure 8C) but had no effect on the growth of any of the other mutants, with the exception of sec24-1. The ability of ISS1 overexpression to suppress sec22 is consistent with the synthetic lethality of iss1 sec22 double mutants. Thus, both ISS1 and SEC24 are implicated in vesicle docking or fusion by their genetic interactions.
Purification of Iss1p as a Complex with Sec23p
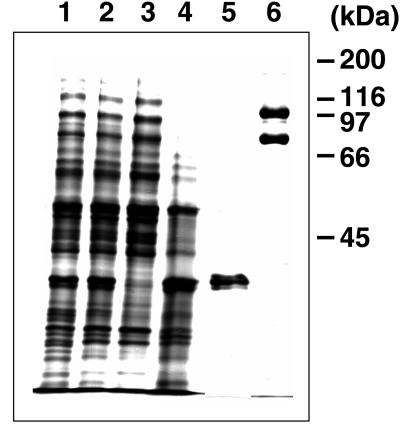
Iss1p was purified from RSY620 harboring pTKY7, which expresses a His6-tagged version of the ISS1 gene under the control of the GAL1 promoter, and pTKY9, which expresses SEC23 under the control of the GAL1 promoter. A high-speed supernatant fraction prepared from the lysate was loaded onto a Ni-NTA agarose column and eluted with a stepwise gradient of imidazole-containing buffers. The peak fractions, eluted with 500 mM imidazole, contained two proteins (Figure 9, lane 6), one of which (100 kDa) was identified as His6-tagged Iss1p and the other of which (85 kDa) was identified as Sec23p by Western blotting with either His6 antibody or Sec23p antibody (our unpublished data). The coelution of Iss1p with Sec23p indicated that these proteins assemble into a complex (when a control extract from cells expressing Iss1p without the His6 tag was used, Sec23p did not bind to the column). An extract from 25 g (wet weight) of cells yielded 1.5 mg of the Sec23p/Iss1p complex. We found that glycerol was necessary to stabilize purified Sec23p/Iss1p; in buffers that lacked glycerol, the purified complex lost ∼80% of its activity in 5 d at 4°C, whereas >80% of its activity remained under the same conditions in the presence of 20% glycerol.
Figure 9.
Purification of Sec23p/Iss1p from RSY620 (pTKY7 [2 μm GAL1p–ISS1 (His6-tagged)], pTKY9 [2μ GAL1p–SEC23]). Proteins in the cell extract and eluted from the Ni-NTA agarose column were separated on SDS-PAGE (10%) and stained with Coomassie brilliant blue R-250. Lane 1, cell lysate (5 μg); lane 2, high-speed supernatant (5 μg); lane 3, flow through from the column (5 μg); lane 4, B-II eluate (5 μg); lane 5, B-III eluate (2 μg); lane 6, B-IV (500 mM imidazole) eluate (1 μg).
Sec23p/Iss1p Drives Formation of COPII-like Vesicles from the ER
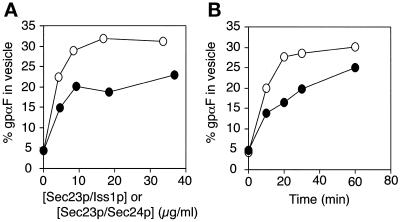
The α-factor halo assay (Figure 8A) with sec24-disrupted cells suggested that Iss1p could substitute for Sec24p in α-factor trafficking in vivo and suggested that Iss1p might also substitute for Sec24p in the incorporation of gpαF into COPII vesicles in vitro (Barlowe et al., 1994; Bednarek et al., 1995). We examined this activity with the use of purified Sec23p/Iss1p and found that 20–25% of gpαF was captured in vesicles budded from ER membranes incubated with Sar1p, Sec13p/Sec31p, and Sec23p/Iss1p (without Sec23p/Sec24p) (Figure 10). The packaging of gpαF into vesicles required added GTP or GMP-PNP; in the absence of these nucleotides, <5% of gpαF was released into the vesicle fraction (our unpublished data). The amount of gpαF packaged increased with increasing concentrations of Sec23p/Iss1p (Figure 10A) and increasing incubation times (Figure 10B). Together, these results indicate that Sec23p/Iss1p, like Sec23p/Sec24p, drives the formation of gpαF-containing vesicles from the ER.
Figure 10.
Packaging of gpαF precursor into vesicles produced from microsomes. (A) Microsomes (containing 23 μg of proteins) were incubated for 30 min at 20°C with 64 μg/ml Sar1p, 144 μg/ml Sec13p/Sec31p, and indicated concentrations of either Sec23p/Iss1p (●) or Sec23p/Sec24p (○) in the presence of 0.1 mM GMP-PNP in 50 μl of B88. The percentage of gpαF packaged into the vesicles was determined as described in MATERIALS AND METHODS. (B) Experiment carried out under the same conditions as described for A except that the fixed concentration of Sec23p/Iss1p (9.2 μg/ml) or Sec23p/Sec24p (8.4 μg/ml) was used and that the reaction was conducted for the times indicated. (○) Reaction with Sec23p/Sec24p; (●) reaction with Sec23p/Iss1p.
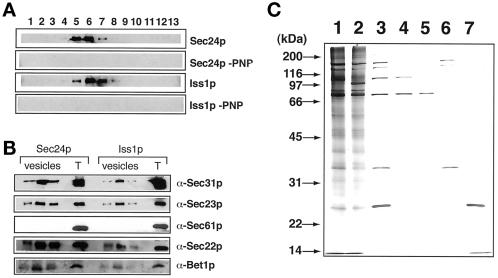
To compare the cargo molecules in the vesicles produced with Sec24p and Iss1p, we carried out the budding reaction on a large scale in the presence of Sec23p complexes containing Sec24p or Iss1p and isolated the vesicles on a sucrose density gradient. As shown in Figure 11A, Sec23p was present in the fractions whose sucrose concentration was ∼38% when the budding reaction was carried out in the presence of Sec23p/Iss1p or Sec23p/Sec24p. Sec23p was not found in these fractions when the budding reaction was carried out in the absence of GMP-PNP (Figure 11A) or without Sec23p/Iss1p and Sec23p/Sec24p (our unpublished data). Thus, Sec23p/Iss1p produces vesicles from the ER in the presence of Sar1p, Sec13p/Sec31p, and GMP-PNP, and the densities of these vesicles are similar to those of regular COPII vesicles produced by Sec23p/Sec24p. Like COPII vesicles, the vesicles produced with Iss1p contained SNARE molecules (Sec22p and Bet1p) and did not contain the ER-resident protein Sec61p (Figure 11B). Silver staining of the proteins showed that the vesicles produced with Iss1p contained Iss1p (100-kDa band) as well as Sec31p, Sec23p, Sec13p, and Sar1p. The vesicles produced with Sec24p contained Sec24p (105-kDa band) instead of Iss1p. Other abundant proteins in the Iss1p-coated vesicles were indistinguishable from those in the Sec24p-coated regular COPII vesicles (Figure 11C).
Figure 11.
Characterization of COPII-like vesicles produced with Iss1p. (A) Budding reactions were carried out in the presence of Sec23p/Sec24p or Sec23p/Iss1p with or without GMP-PNP as described in MATERIALS AND METHODS. Vesicles (0.55 ml) obtained from each budding reaction were subjected to flotation in a sucrose density gradient (55–25% stepwise gradient). Proteins in each fraction were precipitated, and one-fifth of each precipitate was analyzed by immunoblotting with Sec23p antiserum. Sucrose concentrations of the fractions containing Sec23p were as follows. Fractions obtained with Sec24p: fraction 5, 36.1%; fraction 6, 38.4%; fraction 7, 40.5%. Fractions obtained with Iss1p: fraction 5, 36.0%; fraction 6, 38.2%; fraction 7, 40.3%. Sec24p-PNP and Iss1p-PNP indicate results obtained in the absence of GMP-PNP. (B) Vesicle fractions were obtained by flotation in a sucrose density gradient. Three successive fractions constituting the Sec23p peak were analyzed by immunoblotting of antisera specific for Sec31p, Sec61p, Sec22p, and Bet1p. Fractions were prepared as described for A. Lane T contains total budding reaction mixture (1 μl). (C) Silver staining of the vesicle fractions obtained with Sec24p (lane 1) and Iss1p (lane 2). Lane 3, COPII mix (50 ng of Sar1p, 50 ng of Sec23p/Sec24p, and 50 ng of Sec13p/Sec31p); lane 4, Sec23p/Sec24p (50 ng); lane 5, Sec23p/Iss1p (50 ng); lane 6, Sec13p/Sec31p (50 ng); lane 7, Sar1p (50 ng).
DISCUSSION
Sec24p Plays an Essential Role in COPII Vesicle Formation In Vivo
Here we have shown that Sec24p is indispensable for protein transport from the ER in vivo; deletion of the SEC24 gene is lethal, and depletion of Sec24p causes exaggeration of the ER as well as a defect in CPY exit from the ER (Figures 2–4). A homology search revealed that humans, C. elegans, and A. thaliana have SEC24 homologues. Thus, Sec24p probably performs a similar role in vesicle formation from the ER in all eukaryotes.
Gimeno et al. (1996) showed that Sec24p and Sec23p bind to adjacent regions of Sec16p: Sec23p binds to a C-terminal region and Sec24p binds to a more central region of Sec16p. The finding that the temperature sensitivity of sec16-2 is suppressed by the combined overproduction of Sec24p and Sec23p suggests that interaction between these proteins is important for budding events in vivo, although the requirement of Sec16p is not absolute in vitro (Matsuoka et al., 1998). It is also known that Sec31p interacts with Sec24p (Shaywitz et al., 1997). Therefore, it is probable that Sec24p serves as an essential structural unit of coat structure.
GTP hydrolysis by Sar1p renders the COPII coat unstable (Barlowe et al., 1994), and the GTPase activity of Sar1p is activated by Sec23p (Yoshihisa et al., 1993). Accordingly, the timing of GTP hydrolysis by Sar1p as activated by Sec23p may be regulated in some way to prevent premature release of the coat subunits before budding is complete. It is interesting to speculate that Sec24p may participate in the regulation of Sar1p GTPase activation. Although previous experiments indicated that Sec24p does not affect the GTPase-activation activity of Sec23p in solution (Yoshihisa et al., 1993), in the context of an intact coat, e.g., one that includes Sec16p (Espenshade et al., 1995), Sec24p may impede the interaction of Sec23p with Sar1p until vesicle fission has been completed. Because overproduction of Sec23p impairs cell growth (Figure 5C) and excess Sec23p inhibits the budding reaction in vitro (Yoshihisa et al., 1993), it may be that an appropriate balance of Sec24p and Sec23p is important for efficient budding.
Iss1p Replaces the Essential Role of Sec24p
The S. cerevisiae genomic sequence revealed a gene (ISS1) with striking similarity to SEC24 (Figure 1). We found that deletion of ISS1 does not have a significant effect on yeast growth or the maturation kinetics of CPY and Gas1p (Figure 6). We found that on overexpression, Iss1p can fulfill the essential function of Sec24p (Figure 7). Strains with a chromosomal disruption of SEC24 that were suppressed by overexpression of ISS1 grew slowly at 37°C, indicating that Iss1p itself or interactions between Iss1p and other proteins may be unstable at high temperature. Because disruption of SEC24 in an otherwise wild-type strain is lethal, the endogenous level of Iss1p expressed from the chromosomal gene must not be sufficient to carry out the essential function of Sec24p (Figures 2 and 3).
Genetic Interactions of SEC24 and ISS1
Tests for genetic suppression and synthetic lethality revealed a possible connection between the functions of SEC24 and ISS1 and SNARE proteins involved in COPII vesicle function. First, the loss-of-function mutants iss1 and sec24 were found to exacerbate mutations in the SNARE genes SEC22 and BET1 (Table 3). Second, overexpression of ISS1 partially suppressed mutations in SEC22 (Figure 8C). These genetic interactions of sec24 mutants with vesicle-docking mutants were particularly surprising, because none of the other mutants in COPII components shows interactions with vesicle-docking mutants (Kaiser and Schekman, 1990).
Four mechanisms can be postulated by which Sec24p and Iss1p could affect vesicle docking or fusion. First, Sec24p and Iss1p may be required for loading of the v-SNAREs Sec22p, Bet1p, and, possibly, Bos1p into vesicles during vesicle formation. Consistent with this idea is the observation that Bet1p and Bos1p are concentrated into prebudding complexes by the COPII proteins Sar1p, Sec23p, and Sec24p (Springer and Schekman 1998). It is conceivable that Sec24p and Iss1p similarly direct incorporation of docking factors into COPII-coated vesicles by binding to their cytosolic domains. To date, attempts to coprecipitate v-SNARE proteins with either Sec24p or Iss1p have been unsuccessful. A second possibility is that Sec24p and Iss1p may be required for the formation of retrograde transport vesicles that recycle integral membrane docking factors to the ER. If formation of these vesicles is blocked, Sec22p and other docking factors will be depleted from the ER, ultimately causing a defect in vesicle docking. Interestingly, mutants in coatomer components that block recycling (sec21, sec26, and sec27) have multiple genetic interactions with docking mutants similar to iss1 and sec24 mutants (Newman and Ferro-Novick, 1987; Duden et al., 1994). If Iss1p and Sec24p in fact participate in retrograde transport, it would be expected that mutants in these proteins cause missorting of recycled cargo proteins. However, we have been unable to detect a sorting defect in iss1 mutants. In particular, deletion of iss1 does not cause missorting of Kar2p, an ER protein that undergoes recycling, and does not affect the recycling of a KKXX-containing fusion protein (our unpublished data). A third possibility is that Sec24p and Iss1p participate in the fusion reaction itself. Recently, Peng et al. (1999) detected strong and specific interaction between Sec24p and Sed5p, a t-SNARE that marks the docking site for COPII vesicles targeted to the cis-Golgi compartment. It is possible that Sec24p and Iss1p participate in the formation of complexes between v-SNAREs and t-SNAREs. Finally, a role for Sec24p and Iss1p in vesicle docking or fusion could reflect a requirement for these proteins in regulated disassembly of the COPII coat, possibly stimulating GTP hydrolysis on Sar1p. More detailed biochemical studies will be required to resolve these possibilities.
Cargo Recruitment by Sec24p-coated Vesicles and Iss1p-coated Vesicles
Recently, it was shown that Sar1p and Sec23p/Sec24p cooperate to recruit cargo molecules into ER-derived vesicles (Aridor et al., 1998; Kuehn et al., 1998; Springer and Schekman, 1998). This suggests a direct interaction between these coat proteins and cargo molecules or adaptor molecules that bind to the cargo molecules. Because Iss1p and Sec24p differ, we considered the possibility that Sec23p/Iss1p serves to recruit a subset of cargo molecules different from those recruited by Sec23p/Sec24p. We found that purified Sec23p/Iss1p replaces Sec23p/Sec24p to drive vesicle formation from the ER in vitro (Figures 10 and 11). However, we did not see a clear difference in cargo composition between Sec24p- and Iss1p-coated vesicles (Figures 10 and 11). Therefore, at least the abundant cargo proteins as well as α-factor and SNAREs (Sec22p and Bet1p) can be recruited into the vesicles by both Sec24p and Iss1p in collaboration with the other COPII components. We cannot exclude the possibility that Iss1p recruits a different subset of cargo molecules whose amounts are too small to be detected by silver staining and that are not essential to cell viability under the conditions we used.
Sec24p and Iss1p may display quantitative rather than qualitative differences in the recognition of cargo molecules. In the titration experiments and kinetic studies measuring the packaging of α-factor precursor, we found that purified Sec23p/Iss1p was consistently less active than Sec23p/Sec24p (Figure 10). Of course, Sec23p/Iss1p may simply be inherently less active than Sec23p/Sec24p as a coat promoter. However, in sec24 null mutant cells suppressed by overexpression of ISS1, we found a measurable difference in the rates of maturation of CPY and Gas1p compared with wild-type cells (Figure 8). This result is most consistent with qualitative differences in cargo or cargo receptor recognition by Iss1p and Sec24p.
Pagano et al. (1999) and Roberg et al. (1999) reported the cloning and characterization of a more distant (23% identical) homologue of Sec24p. Pagano et al. found that this homologue (which they call Sec24C), when deleted, reduces the secretion of a subset of prominent proteins detected in the cell culture supernatant. Pagano et al. also report the cloning and deletion of ISS1 (which they call Sec24B). Deletion of this gene has no effect on the spectrum of proteins secreted into the culture supernatant. Roberg et al. (1999) discovered SEC24C by a screen for mutations lethal in a sec13-1 mutant strain. They call this gene LST1 and demonstrate that lst1 null cells are sensitive to low pH because of a deficit in transport of the major plasma membrane ATPase, Pma1p, out of the ER. Roberg et al. document a complex of Lst1p and Sec23p and suggest that this dimer may perform a specialized function in the packaging of Pma1p into COPII vesicles.
Together, our data and the recent reports suggest that Sec24p and its homologues may define the selectivity of cargo protein sorting. The signal that interacts with Sec24p, Iss1p (Sec24B), and Lst1p (Sec24C), and the binding pocket within these coat subunits, remain to be defined.
ACKNOWLEDGMENTS
We thank B. Lesch for the preparation of purified COPII components, A. Frand for isolation of the sec24-1 allele, A. Spang and S. Springer for comments on the manuscript, and members of the Schekman and Kaiser laboratories for helpful discussion. T.K. was supported by fellowships from the Human Frontier Science Program and the Ministry of Education, Science, Sports, and Culture of Japan. This work was supported by grants from the National Institutes of Health, the Human Frontier Science Program, and the Howard Hughes Research Foundation.
Abbreviations used:
- CPY
carboxypeptidase Y
- ER
endoplasmic reticulum
- GMP-PNP
5′-guanylyl imidodiphosphate
- gpαF
glycosylated pro-α-factor
- MSS
medium-speed supernatant
- Ni-NTA
nickel-nitriloacetic acid
- SC
synthetic complete
- TCA
trichloroacetate
REFERENCES
- Aridor M, Weissman J, Bannykh S, Nuoffer C, Balch WE. Cargo selection by the COPII budding machinery during export from the ER. J Cell Biol. 1998;141:61–70. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D, Hicke L, Rexach M, Schleyer M, Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- Balch W, McCaffery J, Plutner H, Farquhar M. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell. 1994;76:841–852. doi: 10.1016/0092-8674(94)90359-x. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Bednarek S, Ravazzola M, Hosobuchi M, Amherdt M, Perrelet A, Schekman R, Orci L. COPI- and COPII-coated vesicles bud directly from the endoplasmic reticulum. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Bloom H, Beier H, Gross HS. Improved silver staining of plant proteins, RNA and DNA, in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- Dominguez M, Dejgaard K, Füllekrug J, Dahan S, Fazel A, Paccaud J, Thomas D, Bergeron J, Nilsson T. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COPI and II coatomer. J Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R, Hosobuchi M, Hamamoto S, Winey M, Byers B, Schekman R. Yeast beta- and beta′-coat proteins (COP) J Biol Chem. 1994;269:24486–24495. [PubMed] [Google Scholar]
- Espenshade P, Gimeno R, Holzmacher E, Teung P, Kaiser C. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler K, Veit M, Stamnes MA, Rothman JE. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- Gimeno R, Espenshade P, Kaiser C. COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Sec16p. Mol Biol Cell. 1996;7:1815–1823. doi: 10.1091/mbc.7.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Yoshihisa T, Schekman R. Sec23p and a novel 105-kDa protein function as a multimeric complex to promote vesicle budding and protein transport from the endoplasmic reticulum. Mol Biol Cell. 1992;3:667–676. doi: 10.1091/mbc.3.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kappeler F, Klopfenstein DRC, Foguet M, Paccaud J-P, Hauri H-P. The recycling of ERGIC-53 in the early secretory pathway: ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COPII. J Biol Chem. 1997;272:31801–31808. doi: 10.1074/jbc.272.50.31801. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Schekman R, Ljungdahl PO. Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro. J Cell Biol. 1996;135:585–595. doi: 10.1083/jcb.135.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Muramatsu M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Ferro-Novick S. Characterization of new mutants in the early part of the yeast secretory pathway isolated by a [3H]mannose suicide selection. J Cell Biol. 1987;105:1587–1594. doi: 10.1083/jcb.105.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Balch WE. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- Oka T, Nishikawa S, Nakano A. Reconstitution of GTP-binding Sar1 protein function in ER to Golgi transport. J Cell Biol. 1991;114:671–679. doi: 10.1083/jcb.114.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano A, Letourneur F, Garcia-Estefania D, Carpentier J-L, Orci L, Paccaud J-P. Sec24 proteins and sorting at the endoplasmic reticulum. J Biol Chem. 1999;274:7833–7849. doi: 10.1074/jbc.274.12.7833. [DOI] [PubMed] [Google Scholar]
- Peng R, Grabowski R, DeAntoni A, Gallwitz D. Specific interaction of the yeast cis-Golgi syntaxin Sed5p and the coat protein complex II component Sec24p of endoplasmic reticulum-derived transport vesicles. Proc Natl Acad Sci USA. 1999;96:3751–3756. doi: 10.1073/pnas.96.7.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Schekman R. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J Cell Biol. 1991;114:219–229. doi: 10.1083/jcb.114.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg K, Crotwell M, Espenshade P, Gimeno R, Kaiser C. LST1 is a SEC24 homologue used for selective export of the plasma membrane ATPase from the endoplasmic reticulum. J Cell Biol. 1999;145:659–672. doi: 10.1083/jcb.145.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothblatt J, Schekman R. A hitchhiker's guide to analysis of the secretory pathway in yeast. Methods Cell Biol. 1989;32:3–36. doi: 10.1016/s0091-679x(08)61165-6. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, Chuang JS, Schekman R. SEC31 encodes an essential component of the COPII coat required for transport vesicle budding from the endoplasmic reticulum. Mol Biol Cell. 1997;8:205–217. doi: 10.1091/mbc.8.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, Yeung T, Schekman RW. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J. 1993;12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Novick PJ. A ras-like protein is required for a post Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Saxena K, Gaitatzes C, Walsh M, Eck M, Neer E, Smith T. Analysis of the physical properties and molecular modeling of Sec13: a WD repeat protein involved in vesicular traffic. Biochemistry. 1996;35:15215–15221. doi: 10.1021/bi961616x. [DOI] [PubMed] [Google Scholar]
- Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hickes JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer S, Schekman R. Nucleation of COPII vesicular coat complex by endoplasmic reticulum to Golgi vesicle SNAREs. Science. 1998;281:698–700. doi: 10.1126/science.281.5377.698. [DOI] [PubMed] [Google Scholar]
- Wuestehube L, Schekman R. Reconstitution of transport from the endoplasmic reticulum to the Golgi complex using an ER-enriched membrane fraction from yeast. Methods Enzymol. 1992;219:124–136. doi: 10.1016/0076-6879(92)19015-x. [DOI] [PubMed] [Google Scholar]
- Yeung T, Yoshihisa T, Schekman R. Purification of Sec23p-Sec24p complex. Methods Enzymol. 1995;257:145–151. doi: 10.1016/s0076-6879(95)57020-9. [DOI] [PubMed] [Google Scholar]
- Yoshihisa T, Barlow C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]