Abstract
Members of the WNK family of serine/threonine kinases have been implicated as important modulators of salt homeostasis, regulating the balance between renal sodium reabsorption and potassium excretion. Gain-of-expression mutations in the WNK1 gene uncouple Na+ and K+ balance and cause a familial disorder of diminished renal potassium excretion, excessive sodium retention, and hypertension (pseudohypoaldosteronism type II or Gordon’s syndrome). Alternative splicing of the WNK1 gene produces a kidney-specific short form of WNK1 (KS-WNK1) and a more ubiquitous long form (L-WNK1), but it is not clear how either of these isoforms influence renal potassium excretion. Here we demonstrate that KS-WNK1 and L-WNK1 converge in a pathway to regulate the renal outer-medullary K+ channel, Kir1.1. Reconstitution studies in Xenopus oocytes reveal that L-WNK1 significantly inhibits Kir1.1 by reducing cell surface localization of the channel. A catalytically inactive L-WNK1 mutant has no inhibitory effect on Kir1.1, indicating that channel inhibition depends on kinase activity. KS-WNK1, lacking an intact kinase domain, does not directly alter Kir1.1. Instead, KS-WNK1 negatively regulates L-WNK1 to release Kir1.1 from inhibition. Acute dietary potassium loading increases the relative abundance of KS-WNK1 to L-WNK1 transcript and protein in the kidney, indicating that physiologic up-regulation of Kir1.1 activity involves a WNK1 isoform switch and KS-WNK1-mediated release from L-WNK1 inhibition. Thus, these observations provide evidence for the physiological regulation of Na+ and K+ balance by a kinase isoform switch mechanism.
Keywords: Gordon’s syndrome, hypertension, Kir1.1, renal outer-medullary K+ channel, WNK kinase
The WNK family of serine/threonine kinases has been shown to function as important regulators of salt homeostasis (1–9). Mutations in the WNK4 or WNK1 genes cause a familial disorder [pseudohypoaldosteronism type II, (PHAII)] of diminished renal potassium excretion, excessive sodium retention, and hypertension (10). Molecular reconstitution studies in Xenopus oocytes demonstrate that wild-type WNK4 inhibits the NaCl cotransporter (NCC) (encoded by SLC12A3) (3, 5), and secretory potassium channel, Kir1.1 (encoded by KCNJ1) (1), consistent with WNK4-dependent regulation of sodium reabsorption and potassium excretion in the distal nephron. Significantly, WNK4 modulation of NCC and Kir1.1 occurs by different mechanisms (1, 3), and PHAII missense mutations in WNK4 have opposite effects on the regulation of the two renal transport proteins. These actions of WNK4 explain the enhanced Na retention, as well as the hypertension, reduced K secretion, and hyperkalemia in the disease. As first postulated by Kahle et al. (1), the observations also raise the possibility that WNK4 may function physiologically to “switch” the distal nephron between high-Na-reabsorption and high-K-secretion states.
The physiological mediators of the WNK4 switch have not been identified, but the products of the WNK1 gene are excellent candidates. Alternative splicing of the WNK1 gene produces a kidney-specific short form of WNK1 (KS-WNK1) and a more ubiquitous long form (L-WNK1) (11, 12). Consistent with a switch mechanism, L-WNK1 blocks the inhibitory effect of WNK4 on the NCC transporter, restoring Na transport activity to near normal levels (5). In addition, both forms of WNK1 have been reported to stimulate the epithelial sodium channel (ENaC) (2, 4). Because PHAII-causing mutations in the WNK1 gene are large intronic deletions that increase WNK1 expression, the stimulatory effects of WNK1 isoforms on the two sodium transporters in the distal nephron explain the exaggerated sodium reabsorption in PHAII, but the action of WNK1 isoforms on potassium handling is unclear. Here we explore how WNK1 isoforms regulate the ROMK Kir1.1, a major route for potassium transport into the distal tubule lumen (13, 14) and the channel responsible for regulated potassium secretion by the kidney (15). These observations provide evidence for the physiological regulation of Na+ and K+ balance via a WNK1 kinase isoform switch mechanism.
Results
WNK1 Isoforms Differentially Affect Kir1.1 Activity.
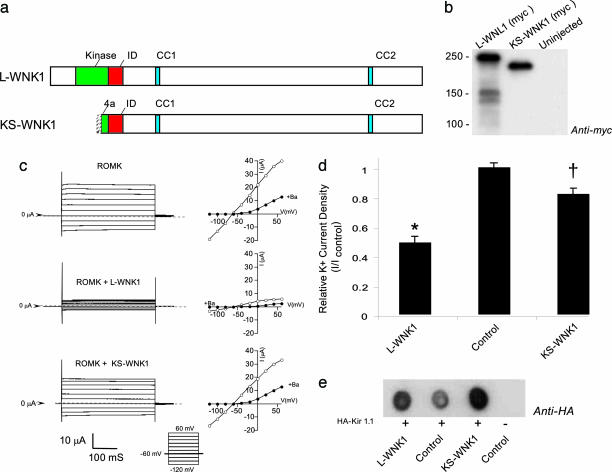
To test whether the WNK1 isoforms (Fig. 1a) regulate Kir1.1, Xenopus oocytes were injected with Myc-tagged L-WNK1 or KS-WNK1 along with hemagglutinin (HA)-tagged Kir1.1 (HA-Kir1.1) and potassium channel activity was measured under two-microelectrode voltage clamp (Fig. 1c) as described in ref. 16. Although similar levels of L-WNK1 and KS-WNK1 protein expression were achieved (Fig. 1b), the two WNK1 isoforms had remarkably different effects on Kir1.1 activity (Fig. 1 c and d). L-WNK1 significantly inhibited Kir1.1 (P < 0.01), contrasting with the significantly different (P < 0.01) and very modest (Fig. 1d) or absent (see Fig. 3b) effects of KS-WNK1. Evaluation of Kir1.1 abundance by Western dot blot revealed that the inhibitory effect of L-WNK1 cannot be explained by simple translational interference of Kir1.1 (Fig. 1e), suggesting a more direct mechanism.
Fig. 1.
WNK1 isoforms differentially affect Kir1.1 activity. The effects of L-WNK1 and KS-WNK1 isoforms on Kir1.1 activity were compared in Xenopus oocytes. Myc-tagged L-WNK1 or KS-WNK1 cRNAs were coinjected with HA-Kir1.1 at 10:1 molar excess of kinase over channel, and barium-sensitive potassium currents were recorded under a two-microelectrode voltage clamp in 5 mM potassium. (a) Domain structure of L-WNK1 and KS-WNK1. Both splice variants contain a kinase-inhibitory domain, ID (red), and two putative coiled-coil domains (blue), but KS-WNK1 lacks the full kinase domain (green). (b) L-WNK1 and KS-WNK1 (myc-tagged) protein expression in oocytes detected by immunoblot analysis after WNK1 isoform cRNA injection. (c) Whole-cell potassium-current tracings and current–voltage relationships of Kir1.1-expressing oocytes in the absence (Top) and presence of L-WNK1 (Middle) and KS-WNK1 (Bottom). ○, control; ●, +5 mM barium. (d) Kir1.1 current density of oocytes injected with KIR1.1 only (control), Kir1.1 plus L-WNK1, and Kir1.1 plus KS-WNK1 (mean ± SEM, n = 40 oocytes per condition for six frogs). L-WNK1 significantly reduces functional expression of Kir1.1 (∗, P < 0.01), contrasting the modest effects of KS-WNK1 (†, P < 0.05). (e) Kir1.1 protein abundance in the presence and absence of WNK1 isoforms. Shown is a dot blot evaluation of the total expression of HA-tagged Kir1.1 using anti-HA antibody, demonstrating that the inhibitory effect of L-WNK1 is not caused by a reduction in total expression of Kir1.1.
Fig. 3.
Dominant-negative regulation of L-WNK1 by KS-WNK1. (a) L-WNK1 inhibition of Kir1.1 requires kinase activity. Whole-cell potassium currents of oocytes injected with Kir1.1 cRNA only (control), Kir1.1 plus wild-type L-WNK1 cRNA, or Kir1.1 plus the kinase-defective mutant L-WNK1 (D368A) cRNA are shown relative to control (n = 20 oocytes per group for three frogs; ∗, P < 0.05). The D368A mutation blocks kinase activity and renders L-WNK incapable of inhibiting Kir1.1. (b) KS-WNK1 coexpression reduces L-WNK1 inhibition of Kir1.1. Coinjection of KS-WNK1 with L-WNK1 cRNA reduced the inhibition of Kir1.1 activity by L-WNK1, indicating that KS-WNK1 influences Kir1.1 function by a dominant-negative-type mechanism. P values are shown for the control vs. the groups with L-WNK1 (∗, P < 0.05 for L-WNK1 vs. all of the KS-WNK1 plus L-WNK1 groups). KS-WNK1 alone had no significant effect relative to control (n = 20 oocytes per group for four frogs). (c) KS-WNK1 coexpression does not significantly affect L-WNK1 abundance. Shown is a representative immunoblot of the myc-tagged L-WNK1 and KS-WNK1 of the oocytes studied in b. Oocytes were injected with L-WNK1 and increasing amounts of KS-WNK1 (molar ratios of KS-WNK1 to L-WNK1 of 0.5, 1.0, and 2.0). Quantification of all experiments showed that, although the amount of L-WNK1 tended to be lower when the molar ratio was 2.0, the difference was not significant (n = 4).
L-WNK1 Does Not Affect Single-Channel Properties but Reduces Surface Expression of Kir1.1.
We examined the mechanism by which L-WNK1 affects Kir1.1 by characterizing the single-channel properties of Kir1.1 channels expressed in the presence and absence of L-WNK1 in cell-attached, patch-clamp studies. L-WNK1 has no effect on the single-channel conductance (g) or channel-open probability (Po) of Kir1.1 (Fig. 2a). This finding indicates, albeit not quantitatively, that L-WNK1 alters the number of channels at the plasmalemma. Indeed, by using immunocytochemistry, we found that coexpression of L-WNK1 substantially reduces Kir1.1 labeling at the cell surface (Fig. 2b).
Fig. 2.
L-WNK1 does not affect single-channel properties but reduces surface expression of Kir1.1. (a) Expression of L-WNK1 has no effect on the single-channel conductance (g) or the channel-open probability (Po) of Kir1.1. (Left) Representative single-channel records in a cell-attached configuration at Vp = 80 mV in symmetrical 150 mM KCl. (Right) The results are summarized (n = 6 records per group). (b) Immunolabeling of HA-Kir1.1 at the oocyte surface consistently shows that coexpression of L-WNK1 reduces surface Kir1.1 levels (n = 6 oocytes from three frogs).
Dominant-Negative Regulation of L-WNK1 by KS-WNK1.
The absence of a remarkable effect of the KS-WNK1 on Kir1.1 suggests that L-WNK might act by a phosphorylation-dependent mechanism. To test this hypothesis directly, we examined the inhibitory effect of a well established, kinase-dead form of L-WNK that harbors a single point mutation, D368A, within the kinase domain catalytic core (17). We observed that L-WNK1 (D368A) has no significant effect on Kir1.1 (Fig. 3a), indicating that kinase activity is necessary for L-WNK1 to inhibit Kir1.1 function.
Because KS-WNK1 is abundantly and specifically expressed in the kidney distal nephron with L-WNK1 at the site where Kir1.1 occurs (11, 12), we reasoned that KS-WNK1 might indirectly influence Kir1.1 by regulating L-WNK1. Consequently, we examined whether KS-WNK1 expression could influence regulation of Kir1.1 by L-WNK1. When increasing amounts of KS-WNK1 are coexpressed with a fixed amount of L-WNK1, the ability of L-WNK1 to inhibit Kir1.1 is blocked (Fig. 3b). Assessment of L-WNK1 protein expression demonstrated that expression of KS-WNK1 has no significant effect on L-WNK1 levels (Fig. 3c), indicating that KS-WNK1 does not block L-WNK1 by translational interference. Recent findings showing the WNK1 isoforms physically interact (18) are consistent with a more direct and specific mechanism of dominant negative regulation.
K Diet Causes a WNK1 Isoform Switch in the Kidney.
Kir1.1 channel activity and potassium excretion is modulated in response to changes in dietary potassium by poorly understood mechanisms (19–21). To test the possible role of WNK1 isoforms, we examined the effect of acute changes in dietary K intake on WNK1 isoform expression in the kidney. Sprague–Dawley rats were fed matched diets that were either deficient in K (low-K diet) or high in K content (high-K diet) for 2 days and compared with control (control-K diet). These diets are well established to change Kir1.1 channel density and provoke strong K conservation (low-K diet) or strikingly increased renal K excretion (high-K diet) compared with animals on a control-K diet (22–25). Quantitative analysis of mRNA levels from the kidneys of these animals using isoform-specific primers and real-time PCR showed that KS-WNK1 (n = 3, P < 0.05) but not L-WNK1 was consistently increased by the high-K diet (Fig. 4a). Neither isoform was changed by the low-K diet. To test whether the response actually translates to a change in KS-WNK1 protein abundance, immunoblot analysis was performed with a C-terminal antibody (see Fig. 1a) that recognizes both WNK1 isoforms. Consistent with observations at the transcript level, the high-molecular-weight L-WNK1 protein was unaffected by different K diets, but the lower-molecular-weight protein (KS-WNK1) was significantly increased in animals on the high-K diet (Fig. 4b). The ability of the antibody to identify both isoforms was validated by immunoblots of oocytes injected with each isoform (Fig. 4c). Quantitative analysis of protein levels demonstrates a strong and specific increase in KS-WNK1 (n = 9, P < 0.02) in animals on the high-K diet (Fig. 4d). As with mRNA level, there is no change in KS-WNK1 for animals on the low-K diet. Additionally, a second antibody directed at the N-terminal that recognizes only the high-molecular-weight L-WNK1 in immunoblots (data not shown) confirmed that the protein abundance of L-WNK1 is not affected by variation in K diet.
Fig. 4.
K diet causes a WNK1 isoform switch in the kidney. (a) Quantitative analysis of mRNA levels in rat kidney using isoform-specific primers and real-time PCR showed that KS-WNK1 (n = 3, P < 0.05) but not L-WNK1 was consistently increased by the high-K diet. Neither isoform was changed by the low-K diet. (b) Western blots using an antibody that recognizes both isoforms show that the protein level of KS-WNK1 but not that of L-WNK1 was consistently increased by the high-K diet in rat kidney, and neither protein level was detectably changed by the low-K diet. Each lane was loaded with protein sample from a different rat. These observations indicate that L-WNK1 may function by a kinase-dependent mechanism to tonically inhibit Kir1.1; elevation of KS-WNK1 expression by the high-K diet would enhance Kir1.1 cell surface density and renal K excretion. (c) Western blots using the same antibody on oocyte-expressed isoforms show that the antibody recognizes L-WNK1 and the distinctly lower band of the of KS-WNK1. The antibody does not recognize any bands in uninjected oocytes. (d) Quantitative analysis of protein levels from blots of multiple animals on each diet show that KS-WNK1 (n = 9, P < 0.02) was consistently increased by the high-K diet. L-WNK1 was unchanged by the high-K diet, and neither isoform was changed by the low-K diet.
Discussion
These studies demonstrate that L-WNK1 acts to down-regulate Kir1.1 function by reducing surface expression. The reduced Kir1.1 surface expression explains how WNK1 gene-intronic mutations described in PHAII (10) could produce the striking hyperkalemia associated with this disease by overexpression of L-WNK1 in the kidney. As depicted in Fig. 5, overexpression of L-WNK1 in PHAII also would stimulate excessive Na reabsorption, leading to hypertension by its action on WNK4, causing excessive surface NCC (5, 6) and activation of ENaC via Sgk1 (4).
Fig. 5.
Role of the WNK1 kinase isoform switch in the regulation of distal nephron K and Na transport. Dietary potassium loading increases KS-WNK1 in distal nephron, which blocks the inhibitory tone of L-WNK1 and increases the number of K channels (Kir1.1) in the apical membrane by acting on vesicle trafficking and promoting K secretion. KS-WNK1 (2) and L-WNK1 (4) independently stimulate ENaC. This action depolarizes the apical membrane potential and provides enhanced driving force for K secretion.
The effect of L-WNK1 on Kir1.1 requires kinase activity. Remarkably, the kinase defective splice variant, KS-WNK1, blocks the inhibitory activity of L-WNK. Recent reports indicate that L-WNK1 assembles as a tetramer (26) and that KS-WNK1 can interact with L-WNK1 to inhibit kinase activity (18). Such a dominant-negative mechanism explains how KS-WNK1 modulates the effects of L-WNK1 on Kir1.1. It would be interesting to learn whether L-WNK1 inhibits by phosphorylation of Kir1.1 directly, phosphorylation of components involved in membrane traffic (27), or by both mechanisms.
Increased expression of KS-WNK1 in the kidney would release Kir1.1 from inhibition by L-WNK1 and cause a high surface expression of the potassium secretory channel necessary for efficient renal K excretion (Fig. 5). It remains to be determined whether the WNK1 isoform switch also regulates the flow-dependent potassium secretory pathway (28) governed by the BK channel (29). Nevertheless, our observation that KS-WNK1, but not the L-WNK1 isoform, is up-regulated in the kidney by a potassium-rich diet offers strong support for involvement of KS-WNK1 in the physiologic mechanism that maintains K balance by increasing renal K excretion via the ROMK channel when dietary K intake is elevated. In a recent paper using the HEK cell model, Lazrak et al. (30) report similar findings regarding the regulation of Kir1.1 by L-WNK1 and KS-WNK1, but we have somewhat different findings than they do for the effect of dietary potassium on WNK1 isoform abundance. Although we also found an increase in KS-WNK1 at the RNA level with the high-K diet, we did not detect the change in L-WNK1 RNA with reduced dietary K reported by Larak et al. (30). Importantly, our results at the transcript level correspond to changes at the protein level. Our studies assess changes in WNK1 isoform protein expression levels and reveal a substantial increase in KS-WNK1 with high-K diet, but we could not detect any changes in L-WNK1 abundance with the acute dietary treatments. Taken together, our findings suggest a central role for KS-WNK1 in the release of Kir1.1 from negative regulation by L-WNK1, involving changes in KS-WNK1 protein rather than L-WNK1.
The discovery is reminiscent of the negative regulation of Na reabsorption (31) by repressor factors, reinforcing the concept that the fidelity and flexibility of physiological responses can be enhanced by simultaneous action of negative and positive regulators. Importantly, the ability of KS-WNK1 to block L-WNK1 negative regulation is consistent with the idea that physiological repressors are exquisitely regulated. It would be interesting to learn whether c-Src (32), the other known negative regulator of ROMK, is organized in a signaling pathway with WNK1. In view of numerous findings indicating that WNK1 and WNK4 also regulate renal Na excretion (2–6, 10), our results also support the hypothesis that the WNK1 isoforms, as well as WNK4 (1), are involved in coordinating the balance between NaCl reabsorption and K secretion.
Importantly, the WNK1 isoform switch helps to explain the important and mysterious ability of the kidney to respond to changes in plasma K and aldosterone and achieve appropriate adjustment of either renal K or Na excretion without disturbing the plasma concentration of the other ion. The physiologic response to a high-K diet, as observed here, offers a salient and relevant example. Up-regulation of KS-WNK1 would block the inhibitory tone of L-WNK1 on Kir1.1 function and maximize K secretion in the late distal nephron segments (Fig. 5). At the same time, the action of the high-K diet to elevate aldosterone would stimulate ENaC. KS-WNK1 (2) and L-WNK1 (4) have been reported to independently stimulate ENaC. Interestingly, KS-WNK1 also has been found to be increased by aldosterone (2). Consequently, after dietary potassium loading, well established actions of elevated aldosterone and KS-WNK1 would be expected to activate ENaC, depolarize the apical membrane potential, and thereby provide enhanced driving force for K secretion. Determining how this effect might be achieved without decreasing overall renal Na excretion and disturbing Na balance has been problematic. Recent findings by Subramanya et al. (18), however, show that elevated KS-WNK1 may reduce NaCl reabsorption in the early distal tubule by blocking the action of L-WNK1 on WNK4 to reduce surface expression of NCC. We speculate that, in the case of the high-K diet, elevation of KS-WNK1 would sufficiently reduce Na reabsorption in the distal convoluted tubule to explain why overall renal Na reabsorption is unaffected by the high-K diet. In this way, the WNK1 kinase switch would promote renal K excretion without disrupting Na balance.
Methods
All studies were performed with modified rat ROMK1 (14) containing an external HA epitope tag as described in ref. 33. Melanie H. Cobb (University to Texas Southwestern Medical Center, Dallas) provided WNK1 cDNA. Site-directed mutagenesis was performed by using a PCR-based strategy with PfuTubo DNA polymerase (QuikChange; Stratagene). The sequence of all modified cDNAs was confirmed by dye termination DNA sequencing (University of Maryland School of Medicine Biopolymer Core).
cRNA Synthesis.
Complementary RNA was transcribed in vitro in the presence of capping analogue G(5′)ppp(5′) from linearized plasmids containing the cDNA of interest using SP6 RNA polymerase (mMessage Machine; Ambion, Austin, TX). cRNA was purified by phenolchloroform extraction and precipitated with ammonium acetate/isopropanol. Yield was quantified spectrophotometrically and confirmed by agarose gel electrophoresis.
Oocyte Isolation and Injection.
Oocytes from female Xenopus laevis (Xenopus Express, Homosassa, FL) were isolated and maintained by following standard procedures (34). Briefly, frogs were anesthetized with 0.15% 3-aminobenzoate, and a partial oophorectomy was performed through an abdominal incision. Oocyte aggregates were manually dissected from the ovarian lobes and then incubated in OR-2 medium (82.5 mM NaCl/2 mM KCl/1 mM MgCl2/5 mM Hepes, pH 7.5) containing collagenase (type 3; Worthington) for 2 h at room temperature to remove the follicular layer. After extensive washing with collagenase-free OR-2, oocytes were stored at 19°C in OR-3 medium (50% Leibovitz’s medium/10 mM Hepes, pH 7.4). Healthy-looking Dumont-stage V–VI oocytes were pneumatically injected 12–24 h later with 50 nl of diethyl pyrocarbonate-treated water containing 250 pg of ROMK cRNA in the absence and presence of a 10–20 M excess of WNK cRNAs, and then stored in OR-3 medium at 19°C for 4 days.
Electrophysiology.
Whole-cell currents in Xenopus oocytes were monitored by using a two-microelectrode voltage clamp as described in refs. 16 and 34. Briefly, oocytes were bathed in a 5 mM K solution (5 mM KCl/85 mM N-methyl-d-glucamine-Cl/1 mM MgCl2/1 mM CaCl2/5 mM Hepes, pH 7.4). Voltage-sensing and current-injecting microelectrodes had resistances of 0.5–1.5 MΩ when backfilled with 3 M KCl. Once a stable membrane potential was attained, oocytes were clamped to a holding potential of −60 mV, and currents were recorded during 500-ms voltage steps, ranging from −120 to +40 mV in 20-mV increments. Data were collected with an ITC16 analog-to-digital/digital-to-analog converter (Instrutech, Great Neck, NY), filtered at 1 kHz, and digitized online at 2 kHz with pulse software (HEKA Elektronik, Pfalz, Germany) for later analysis. ROMK currents were taken as the barium-sensitive current (2 mM Ba acetate) as we have done before (16). Values reported in the text are the barium-sensitive inward currents at 0 mV.
Single-channel properties were assessed in oocytes with the patch-clamp technique in the cell-attached mode as before (16). Single-channel currents were measured with an Axopatch 200B patch-clamp amplifier, digitized at a sampling rate of 47 KHz with a VR-10B digital data recorder (Instrutech), and stored on videotape. All single-channel recordings were performed under symmetrical [K+] conditions (150 mM KCl/1 mM CaCl2/5 mM Hepes, pH 7.4). Data were acquired and analyzed with the acquire and tac family of programs (Bruxton, Seattle). Data were replayed, filtered with an eight-pole Bessel filter (model 900; Frequency Devices, Haverhill, MA) at a cut-off frequency of 1 kHz, and sampled at not less than five times the filtering frequency. Patches that contained only one detectable channel were selected for analysis. A 50% threshold criterion was used to detect events. Open and closed dwell-time histograms (logarithmic time scale, 10 bins per decade) were constructed from 15- to 60-s records, and open probability was calculated as the time in the open state relative to the total recording time. The single-channel current magnitude was estimated by fitting Gaussian distributions to the current amplitude histograms, and single-channel conductance was taken as the inward slope conductance from current measurements at −120 to −40 mV.
Oocyte Fixation and Immunolocalizations.
Oocytes were fixed in 4% paraformaldehyde in OR-2 medium for 1 h followed by 60 min in a cryoprotectant of 10% EDTA in 0.1 M Tris. Cryostat sections (8 μm thick) were made and picked up on coverslips coated with gelatin. Sections were then treated with 0.5% SDS for 10 min to unmask antigenic sites. Surface HA-tagged ROMK1 was localized by using rat anti-HA antibody (Roche Molecular Biochemicals) detected by Alexa Fluor 488-labeled secondary antibody. To eliminate yolk autofluorescence, sections were then quenched by exposure to 0.3% OsO4 for 15 min on ice, and then the Alexa Fluor 488 label was detected by rabbit anti-Alexa Fluor 488 antibody (Molecular Probes) followed by goat anti-rabbit IgG with Alexa Fluor 488.
K Diet Studies.
Male Sprague–Dawley rats (175–210 g) were pair-fed K-deficient (Harlan Teklad 170550), matched control (Harlan Teklad 170555), or high-K (10% KCl, Harlan Teklad TD 76448) chow for 2 days.
All animal experiments were approved by the Institutional Animal Care and Use Committees of the University of Maryland School of Medicine. University of Maryland School of Medicine is fully accredited by the American Association for Accreditation of Laboratory Animal Care.
Immunoblotting.
Rat kidneys with papillary inner medulla excised were homogenized with a tissue homogenizer (Omni 1000 fitted with a microsawtooth generator) in ice-cold isolation solution containing 250 mM sucrose and 10 mM triethanolamine (Calbiochem) with 1 μg/ml leupeptin (Bachem) and 0.1 mg/ml phenylmethylsulfonyl fluoride (USB Corp.). The total protein concentration was measured by using a Pierce BCA protein assay reagent kit. The protein samples were solubilized in Laemmli sample buffer by heating at 70°C for 10 min and SDS/PAGE performed on 6% polyacrylamide gels. The proteins were transferred electrophoretically from unstained gels to nitrocellulose membranes. After being blocked with 5 g/dl nonfat dry milk for 30 min, the blots were probed with the respective antibodies overnight at 4°C. After washing, the nitrocellulose membranes were exposed to secondary antibody (donkey anti-rabbit IgG conjugated with horseradish peroxidase; Pierce no. 31458, diluted to 1:5,000) for 1 h at room temperature. Sites of antibody–antigen reaction were visualized with SuperSignal West Pico Chemiluminescence substrate (Pierce) before exposure to x-ray film (Kodak 165-1579 Scientific Imaging Film).
Antibodies.
Antibody to L-WNK1 were obtained from ADI (San Antonio, TX), which is to the C terminus and recognizes only L-WNK1. An antibody obtained from Novus Biologicals (Littleton, CO) is directed to a sequence in the N terminus that is shared between the full-length L-WNK1 isoform and the KS-WNK1 and thus recognizes both isoforms.
Data Presentation and Statistical Analysis.
All data are given as mean ± SEM. Analysis of variance was used with Dunnett’s multiple comparison as a posthoc test for significance.
Acknowledgments
We thank Dr. Melanie H. Cobb for providing the WNK1 cDNA and Donald Wade for preparing Fig. 5. This work was supported by grants from the National Institutes of Health (to J.B.W., A.R.S., D.H.E., and P.A.W.) and the American Heart Association (to J.B.W. and A.R.S.).
Abbreviations
- ENaC
epithelial Na channel
- KS-WNK1
kidney-specific WNK1
- L-WNK1
long-form WNK1
- NCC
NaCl cotransporter
- PHAII
pseudohypoaldosteronism type II
- ROMK
rat outer-medullary K channel
- HA
hemagglutinin.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Kahle K. T., Wilson F. H., Leng Q., Lalioti M. D., O’Connell A. D., Dong K., Rapson A. K., MacGregor G. G., Giebisch G., Hebert S. C., et al. Nat. Genet. 2003;35:372–376. doi: 10.1038/ng1271. [DOI] [PubMed] [Google Scholar]
- 2.Naray-Fejes-Toth A., Snyder P. M., Fejes-Toth G. Proc. Natl. Acad. Sci. USA. 2004;101:17434–17439. doi: 10.1073/pnas.0408146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson F. H., Kahle K. T., Sabath E., Lalioti M. D., Rapson A. K., Hoover R. S., Hebert S. C., Gamba G., Lifton R. P. Proc. Natl. Acad. Sci. USA. 2003;100:680–684. doi: 10.1073/pnas.242735399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu B. E., Stippec S., Chu P. Y., Lazrak A., Li X. J., Lee B. H., English J. M., Ortega B., Huang C. L., Cobb M. H. Proc. Natl. Acad. Sci. USA. 2005;102:10315–10320. doi: 10.1073/pnas.0504422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang C. L., Angell J., Mitchell R., Ellison D. H. J. Clin. Invest. 2003;111:1039–1045. doi: 10.1172/JCI17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang C. L., Zhu X., Wang Z., Subramanya A. R., Ellison D. H. J. Clin. Invest. 2005;115:1379–1387. doi: 10.1172/JCI22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahle K. T., Rinehart J., de los Heros P., Louvi A., Meade P., Vazquez N., Hebert S. C., Gamba G., Gimenez I., Lifton R. P. Proc. Natl. Acad. Sci. USA. 2005;102:16783–16788. doi: 10.1073/pnas.0508307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leng Q., Kahle K. T., Rinehart J., MacGregor G. G., Wilson F. H., Canessa C. M., Lifton R. P., Hebert S. C. J. Physiol. (London) 2005;571:275–286. doi: 10.1113/jphysiol.2005.102202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinehart J., Kahle K. T., de los Heros P., Vazquez N., Meade P., Wilson F. H., Hebert S. C., Gimenez I., Gamba G., Lifton R. P. Proc. Natl. Acad. Sci. USA. 2005;102:16777–16782. doi: 10.1073/pnas.0508303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson F. H., Disse-Nicodeme S., Choate K. A., Ishikawa K., Nelson-Williams C., Desitter I., Gunel M., Milford D. V., Lipkin G. W., Achard J. M., et al. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 11.Delaloy C., Lu J., Houot A. M., Disse-Nicodeme S., Gasc J. M., Corvol P., Jeunemaitre X. Mol. Cell. Biol. 2003;23:9208–9221. doi: 10.1128/MCB.23.24.9208-9221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Reilly M., Marshall E., Speirs H. J., Brown R. W. J. Am. Soc. Nephrol. 2003;14:2447–2456. doi: 10.1097/01.asn.0000089830.97681.3b. [DOI] [PubMed] [Google Scholar]
- 13.Palmer L. G., Choe H., Frindt G. Am. J. Physiol. 1997;273:F404–F410. doi: 10.1152/ajprenal.1997.273.3.F404. [DOI] [PubMed] [Google Scholar]
- 14.Ho K., Nichols C. G., Lederer W. J., Lytton J., Vassilev P. M., Kanazirska M. V., Hebert S. C. Nature. 1993;362:31–38. doi: 10.1038/362031a0. [DOI] [PubMed] [Google Scholar]
- 15.Giebisch G. Semin. Nephrol. 1999;19:458–471. [PubMed] [Google Scholar]
- 16.Flagg T. P., Tate M., Merot J., Welling P. A. J. Gen. Physiol. 1999;114:685–700. doi: 10.1085/jgp.114.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu B., English J. M., Wilsbacher J. L., Stippec S., Goldsmith E. J., Cobb M. H. J. Biol. Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 18.Subramanya A. R., Yang C. L., Zhu X., Ellison D. H. Am. J. Physiol. 2006;290:F619–F624. doi: 10.1152/ajprenal.00280.2005. [DOI] [PubMed] [Google Scholar]
- 19.Palmer L. G., Antonian L., Frindt G. J. Gen. Physiol. 1994;104:693–710. doi: 10.1085/jgp.104.4.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer L. G., Frindt G. Kidney Int. 2000;57:1324–1328. doi: 10.1046/j.1523-1755.2000.00970.x. [DOI] [PubMed] [Google Scholar]
- 21.Field M. J., Stanton B. A., Giebisch G. H. J. Clin. Invest. 1984;74:1792–1802. doi: 10.1172/JCI111598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mennitt P. A., Frindt G., Silver R. B., Palmer L. G. Am. J. Physiol. 2000;278:F916–F924. doi: 10.1152/ajprenal.2000.278.6.F916. [DOI] [PubMed] [Google Scholar]
- 23.Wei Y., Bloom P., Lin D., Gu R., Wang W. H. Am. J. Physiol. 2001;281:F206–F212. doi: 10.1152/ajprenal.2001.281.2.F206. [DOI] [PubMed] [Google Scholar]
- 24.Wang W. H., Schwab A., Giebisch G. Am. J. Physiol. 1990;259:F494–F502. doi: 10.1152/ajprenal.1990.259.3.F494. [DOI] [PubMed] [Google Scholar]
- 25.Palmer L. G., Frindt G. Am. J. Physiol. 1999;277:F805–F812. doi: 10.1152/ajprenal.1999.277.5.F805. [DOI] [PubMed] [Google Scholar]
- 26.Lenertz L. Y., Lee B. H., Min X., Xu B. E., Wedin K., Earnest S., Goldsmith E. J., Cobb M. H. J. Biol. Chem. 2005;280:26653–26658. doi: 10.1074/jbc.M502598200. [DOI] [PubMed] [Google Scholar]
- 27.Lee B. H., Min X., Heise C. J., Xu B. E., Chen S., Shu H., Luby-Phelps K., Goldsmith E. J., Cobb M. H. Mol. Cell. 2004;15:741–751. doi: 10.1016/j.molcel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Woda C. B., Bragin A., Kleyman T. R., Satlin L. M. Am. J. Physiol. 2001;280:F786–F793. doi: 10.1152/ajprenal.2001.280.5.F786. [DOI] [PubMed] [Google Scholar]
- 29.Pluznick J. L., Wei P., Grimm P. R., Sansom S. C. Am. J. Physiol. 2005;288:F846–F854. doi: 10.1152/ajprenal.00340.2004. [DOI] [PubMed] [Google Scholar]
- 30.Lazrak A., Liu Z., Huang C. L. Proc. Natl. Acad. Sci. USA. 2006;103:1615–1620. doi: 10.1073/pnas.0510609103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossier B. C. J. Clin. Invest. 2003;111:947–950. doi: 10.1172/JCI18232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W. H. Am. J. Physiol. 2006;290:F14–F19. doi: 10.1152/ajprenal.00093.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo D., Kim B. Y., Campo C., Nance L., King A., Maouyo D., Welling P. A. J. Biol. Chem. 2003;278:23066–23075. doi: 10.1074/jbc.M212301200. [DOI] [PubMed] [Google Scholar]
- 34.Welling P. A. Am. J. Physiol. 1997;273:F825–F836. doi: 10.1152/ajprenal.1997.273.5.F825. [DOI] [PubMed] [Google Scholar]