Abstract
In the cyanobacterium Synechococcus elongatus (PCC 7942) the kai genes A, B, and C and the sasA gene encode the functional protein core of the timing mechanism essential for circadian clock regulation of global gene expression. The Kai proteins comprise the central timing mechanism, and the sensor kinase SasA is a primary transducer of temporal information. We demonstrate that the circadian clock also regulates a chromosome compaction rhythm. This chromosome compaction rhythm is both circadian clock-controlled and kai-dependent. Although sasA is required for global gene expression rhythmicity, it is not required for these chromosome compaction rhythms. We also demonstrate direct control by the Kai proteins on the rate at which the SasA protein autophosphorylates. Thus, to generate and maintain circadian rhythms in gene expression, the Kai proteins keep relative time, communicate temporal information to SasA, and may control access to promoter elements by imparting rhythmic chromosome compaction.
Keywords: cyanobacteria, regulation
Circadian clocks have evolved within the cyanobacteria (an extremely diverse group of oxygenic photosynthesizing bacteria) and many, if not all, eukaryotes. These clocks effectively tune gene expression patterns, and thus metabolic activity and behavior, to distinct daily frequencies (1–3). In each of several well studied model systems, circadian gene expression patterns are generated and maintained by the combined functions of fairly small sets of proteins (1–3). Amazingly, in vitro combination of only three proteins, KaiA, KaiB, and KaiC, from the cyanobacterium Synechococcus elongatus results in a functional circadian timing mechanism (4). This phenomenon underscores recent data demonstrating that a transcription and translation feedback loop, once considered essential for circadian timing, is not required for rhythmic activity in this cyanobacterium (5). Interestingly, demonstration of this simple proteinaceous clock, and presumption of its straightforward transfer to newly formed daughter cells, explains the enigma of how a circadian (24-h) timing mechanism can function in cyanobacteria that have generation times of 8 h or less.
Despite those compelling data, questions concerning how this timing mechanism connects circadian clock function to global regulation of gene expression still loom (6). Existent data show functional interactions among the three Kai proteins and the SasA sensor kinase protein as essential for this global regulation (7). For example, the KaiC protein forms a homohexamer upon binding ATP and is an autokinase. It also interacts with double-stranded DNA molecules (8, 9). Overproduction of KaiC represses gene expression on a global scale (8, 9). In an sasA-null strain, except for kai gene expression patterns, all other tested genes are arrhythmically expressed (7). SasA protein thus appears to act as temporal output regulator from the clock. In addition, clock-regulated gene expression rhythms consist of at least two temporal classes (10–12). The major class includes kaiB gene expression and is illustrated in Fig. 1a. One minor class has a phase angle that is shifted (under constant illumination and relative to the major class) by 180°: so-called opposite phase expression (11–13). A lack of identifiable cis- or trans-acting elements that would explicitly determine gene expression pattern phase angles has goaded speculation that chromosome dynamics or DNA topology may be phase-determining (11, 12). Last, heterologous promoters from noncircadian clock-containing bacteria drive rhythmic gene expression patterns when operating in S. elongatus (1). Using these data as groundwork, we hypothesized that large-scale DNA topology or chromosome arrangement dynamics are circadian clock-controlled and that these dynamics could subsequently regulate global gene expression patterns. To begin testing this hypothesis, we treated samples from growing cultures with a fluorescent DNA-intercalating dye (DAPI) and visualized them using deconvolution fluorescence microscopy (14). We observed obvious and striking changes in chromosome compaction as a function of circadian time and demonstrated that this compaction rhythm depends on the kaiC gene. We concluded that the circadian clock in S. elongatus might control global gene expression, in large part, by regulating a chromosome compaction rhythm (15). Presumably, the degree of compaction would then prohibit or permit the transcription machinery access to particular promoter elements.
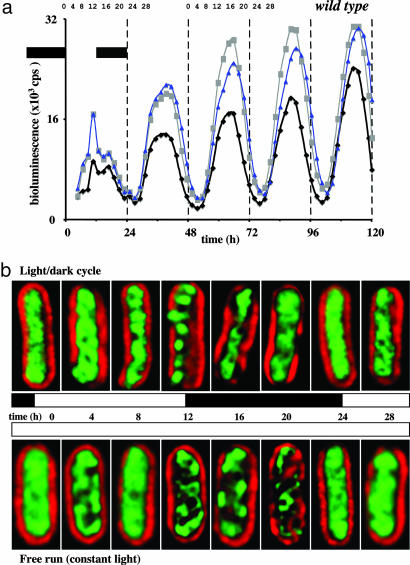
Fig. 1.
Gene expression and chromosome compaction rhythms in wild-type S. elongatus. (a) Bioluminescence (counts per second) recorded over time (hours) from a Φ(kaiB-luc+) reporter in an otherwise wild-type S. elongatus strain. Three independent data sets are graphed. The black bars indicate time without illumination. Numbers above the light/dark cycle (time 0–24) and the second free-running (constant condition) cycle (time 48–72) indicate sampling times for the cell images shown in b. (b) Deconvolved fluorescence microscopy images (red, autofluorescence from S. elongatus; green, DAPI-stained DNA) of wild-type cells sampled at the indicated times during the light/dark cycle (Upper) and the second free-running cycle (Lower). The chromosome arrangement image shown for each time point is representative of >99% (n = 300) of the cells from that sample. Each time course experiment was repeated six times with invariant results. For each of the cycles, note the slow arrangement of the DNA into distinct “nucleoids” and then the return to the time 0 diffuse state. Cells are ≈5 μm long.
Results and Discussion
Clock Control of Chromosome Compaction.
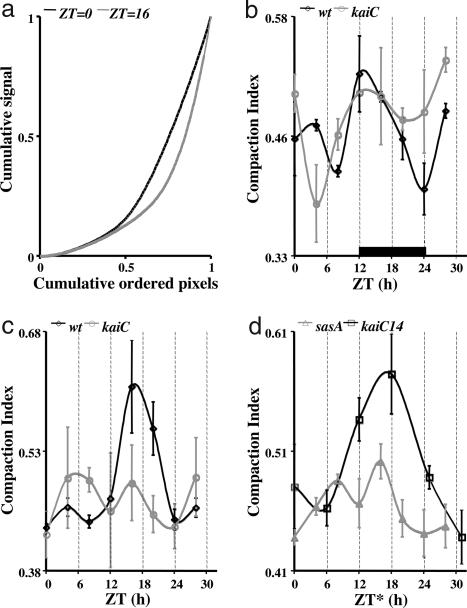
To examine clock control of chromosome compaction, we grew wild-type S. elongatus harboring a Φ(kaiB-luc+) translational fusion as reporter in liquid culture for several days, under constant illumination, before entraining it to a 12-h light/12-h dark diel cycle. Gene expression levels from this culture were monitored over time (Fig. 1a). Beginning near dawn (time 0) of the second complete diel cycle, culture samples were DAPI-stained and visualized every fourth hour for 28 h. As shown in Fig. 1b, chromosome compaction was a rhythmic process. Note the slow compaction (formation of distinct nucleoid regions) of chromosomal DNA from time 0 through time 12 and then the decompaction process from time 12 through time 24 (24 is equivalent to time 0 for the subsequent cycle). Quantification of the fluorescence images shown in Fig. 1b also supported our conclusion that the process was rhythmic (Fig. 2b). The compaction index (CI) (Fig. 2a and Materials and Methods) was low during the day and then peaked at Zeitgeber time (ZT) = 12 (Fig. 2b). Cells visualized at time 12 were sampled immediately before the lights went off. Remarkably, the chromosome(s) [S. elongatus typically harbors several identical copies of its chromosome (16)] had already fully compacted, presumably in anticipation of imminent darkness (Fig. 1b). Also, note that the chromosome was decompacted at times 0 and 24 and that these cells were sampled in darkness immediately before the lights came on.
Fig. 2.
Quantification of chromosome compaction. (a) Cumulative DAPI signal from the set of cell-interior pixels, as defined by the rhodamine signal threshold, plotted against cumulative pixels ranked by the strength of the DAPI signal in a wild-type strain. The black line represents a ZT = 0 sample, and the gray line represents a ZT = 16 sample. The area, A, under each curve was used to calculate a CI where CI = 1 − 2A (see Materials and Methods). Higher DAPI signal intensity values located in smaller areas are indicative of a compacted chromosome. Compacted states will impart a higher CI value than diffuse states. All data shown in b–d were analyzed by one-way ANOVA. For each data set (all experimental time points derived from a particular strain), we tested for heterogeneity of CI values among time points. Except for the kaiC strain under constant light (c), CI values were significantly heterogeneous among time points (b: wt, P = 0.00021; kaiC, P = 0.0012; c: wt, P = 0.000001; kaiC, P = 0.064; d: sasA, P = 0.00074; kaiC14, P = 0.00017). (b) CI versus time for a wild-type strain (black) and a kaiC strain (gray) under 12-h light/12-h dark conditions. Three data sets were analyzed for each strain. The black bar indicates time without illumination. (c) As in b, but data were collected under constant illumination. In the kaiC strain, five data sets were analyzed. For the wild-type strain, note that the CI values are low during the subjective day, are high during the subjective night, and return to low CI values at ZT = 24, corresponding to a diffuse chromosome during the subjective day and compacted chromosome during the subjective night. For the kaiC strain, no discernable rhythm in the CI values was observed. Large variations in CI values were apparent between experiments at each time point. (d) As in c, but data are from a kaiC14 strain (black) and an sasA strain (gray). The asterisk indicates that sample times for the kaiC14 strain were converted to circadian time (see kai-Dependent Chromosome Compaction and Fig. 4b). The time point ZT = 30 thus represents CT = 6 for that strain. For each strain, note the gradually increasing CI values that peak around the midpoint of the cycle and then decrease at ZT (or CT) = 24.
Other than obvious conjecture regarding the regularity of daily oscillations in light quality and quantity, in temperature variation, and in humidity levels, the selective pressures that have guided the evolutionary progression of a timing mechanism in cyanobacteria remain enigmatic (17). For obligate photosynthetic organisms like S. elongatus speculation about these pressures has included suggestion of a clock-based anticipatory timing strategy that allows the organism to ready itself for renewed interspecies competitions over energy acquisition and utilization just ahead of imminent sunrise. These compaction data support the idea of an anticipatory character underlying circadian clock function.
By definition, circadian clock-controlled processes do not require cyclic environmental input for continued circadian rhythmicity. We asked whether the chromosome compaction rhythm observed during the diel cycle was clock-controlled by assaying our culture as it grew under constant illumination. We sampled during the second free-running (constant conditions) cycle. The chromosome compaction rhythm continued and had a near 24-h periodicity (Figs. 1b and 2c). Under these constant conditions the rhythm was surprisingly similar to that seen during the diel cycle. Note that the period change in the expression rhythm [a slight lengthening under constant conditions (Fig. 1a) (18, 19)] resulted in our absolute sampling times during free run, occurring at points in the expression cycle different from those taken during the diel cycle (Fig. 1). The CI data also reflect the period shift as peak compaction occurred in the ZT = 16 sample (Fig. 2c). These data support our hypothesis that chromosome compaction is a circadian clock-controlled process because it remained rhythmic under constant conditions.
Some degree of compaction was noted in the 4-h sample of the experiment whose data are shown in Fig. 1b. This finding was typical for samples taken at that time. Based on our image data and the CI data (Fig. 2 b–d) we think that there may also be an expeditious chromosome compaction cycle that takes place during the cell division cycle. The clock runs independent of cell division but does gate (allow) cell division by only allowing it at specific circadian times (around the time we generally see this compaction) (20). We are investigating this phenomenon with a higher frequency sampling protocol.
kai-Dependent Chromosome Compaction.
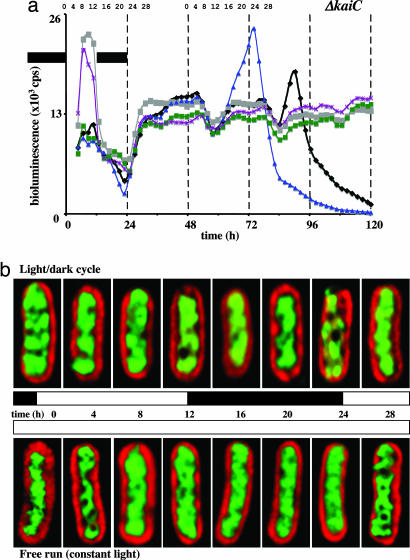
If chromosome compaction is clock-controlled, then it must be kai-dependent. To address this notion, we repeated the experiments above using a kaiC-null strain (Fig. 3). Gene expression patterns in this strain, again determined from a Φ(kaiB-luc+) reporter, were arrhythmic under constant illumination (Fig. 3a). Also, we did not observe chromosome compaction rhythms under constant illumination in this genetic background (Figs. 2c and 3b; n = 6). As we expected, the light/dark cycle did impart some rhythmicity to both gene expression patterns and the compaction process in this strain (Fig. 2 a and b). Throughout the constant light experiments, whose data are shown in Fig. 3b, the DAPI-stained chromosomes remain at basically the same compaction level. On occasion (these experiments were each repeated six times) some greater levels of chromosome compaction were apparent in our kaiC-null strain (Fig. 2 b and c). However, compaction during any given experiment was never rhythmic on a circadian time scale. This observation is especially pronounced when examining the standard deviation at each CI time point shown in Fig. 2c. Thus, we conclude that rhythmic chromosome compaction requires KaiC protein, but we suspect that KaiC is not directly responsible for all of the observed compaction. Experiments using a kaiA-null strain (arrhythmic gene expression patterns but producing the KaiC protein) gave the same results (data not shown). In fact, we could not distinguish phenotypic differences between kaiA- and kaiC-null strains using our fluorescence microscopy assay. Again, these data suggested that compaction required more than the simple presence of KaiC.
Fig. 3.
Gene expression and chromosome compaction in an S. elongatus kaiC strain. (a) As in Fig. 1a but with a kaiC-null strain. Five independent data sets are graphed. (b) As in Fig. 1b but with a kaiC-null strain. The chromosome arrangement shown for each time point is representative of >90% (n = 300) of the cells from that sample. Each time course experiment was repeated six times. For any given time point within each experiment, some variation in the extent of chromosome compaction was evident in this mutant strain. This variation was also reflected in the gene expression rhythms illustrated in a. However, we never saw rhythmic compaction patterns (n = 6). Note the partial compaction at several time points in each time course panel. Evidently, KaiC protein is not directly responsible for the entire chromosome compaction process. Cells are 4–5 μm long.
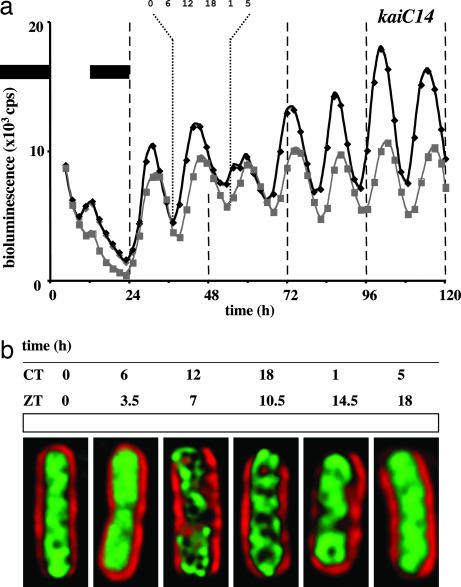
Many missense mutations in the kaiC gene result in period changes to S. elongatus gene expression rhythms (21). To further our argument for circadian clock control of chromosome compaction, we monitored this rhythmic behavior in a strain harboring the kaiC14 allele. This mutant allele results in a free-running gene expression rhythm with a period of 14 h (Fig. 4a). The chromosome compaction rhythm in this strain also had a near 14-h period (Figs. 2d and 4b). The image data are presented in circadian time (one 14-h cycle contains 24 circadian time hours) for easier comparison to the other data. The CI plotted as a function of circadian time also demonstrated the appropriate 14-h compaction rhythm (Fig. 2d). Clearly, chromosome compaction rhythms were in harmony with kaiB gene expression rhythms (Figs. 1–4).
Fig. 4.
Gene expression and chromosome compaction rhythms in an S. elongatus kaiC14 strain. (a) As in Fig. 1a but with a kaiC14 strain. Two independent data sets are graphed. (b) As in Fig. 1b but with a kaiC14 strain. Data are from the indicated free-running cycle. The chromosome arrangement shown for each time point is representative of >98% (n = 200) of the cells from that sample. Each time course experiment was repeated four times with invariant results. Note the arrangement of the DNA into distinct nucleoids and then the return to a diffuse state. For ease of comparison to the other image data, sample times were converted to circadian time (CT); the 14-h cycle was divided into 24 equal circadian hours. The ZT sample times are also provided. Cells are 5 μm long.
Is SasA Part of the Timing Process?
Our correlative data strongly suggested that kai-dependent compaction rhythms are the potential mechanism by which the circadian clock imparts rhythmicity to global gene expression. They also raised questions about the role of the sasA gene. Recall that only kai gene expression remains rhythmic in an sasA-null genetic background, implying that the circadian clock still functions in this mutant strain but that the timing information is not getting from the clock to output regulatory pathways (7). Also, there are data showing that the SasA protein is part of a subjective night Kai protein timing complex, interacts directly with the KaiC protein, and belongs to the large family of two-component-type sensory kinases. To examine the regulatory processes that these data suggested, we purified the Kai and SasA proteins and assayed for effects by these proteins on the rate at which SasA autophosphorylates (7, 22–25).
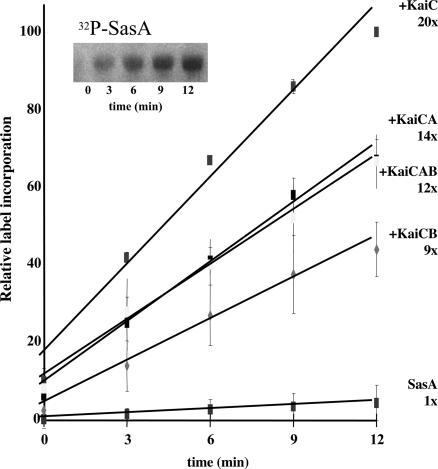
As expected, the SasA protein autophosphorylated using ATP as a phosphoryl group donor (Fig. 5). Moreover, the presence of KaiC protein increased the rate at which SasA autophosphorylated by 20-fold (Fig. 5). The other Kai proteins decreased the SasA protein maximum autophosphorylation rate, but only when KaiC was present (Fig. 5). It appears that the addition of KaiA, KaiB, or both KaiA and B alters the interaction between KaiC and SasA. Although these particular interactions are not well understood, we suspect that they reflect changes in the phosphorylation state of KaiC that in turn alter its interaction with the SasA protein (4, 24). However, the effect of KaiB may be due to a competitive interaction between SasA and KaiB for binding space on the KaiC protein. This notion is based on the sequence and structural similarities between the amino-terminal third of SasA and the entire KaiB protein and the fact that both proteins bind to KaiC (7, 26). The SasA protein did not alter the phosphorylation rate of KaiC (data not shown). This finding is consistent with a hypothesis that the direction of information flow is from the Kai protein complex to SasA. Given that the SasA protein is a two-component sensory kinase, we conclude that SasA mediates circadian-timed regulation of gene expression by having the Kai complex, via KaiC, regulate its rate of autophosphorylation. Cognate response regulators for SasA, which would likely demarcate circadian clock-based signal transduction pathways, have not been identified.
Fig. 5.
Influence of Kai proteins on the autophosphorylation rate of SasA. Assays were run under initial rate conditions as described (see Materials and Methods). The graph illustrates a time course of SasA autophosphorylation with ATP in the presence of (from high rate to low) KaiC; KaiC and KaiA; KaiC, KaiA, and KaiB; KaiC and KaiB; and no additional proteins. Without KaiC present, the other Kai proteins had no effect on SasA autophosphorylation. The SasA protein autophosphorylation rate was also unaffected by the addition of thioredoxin or BSA (data not shown). All time points are the average of four independent experiments, including the one represented by the gel image. Error bars indicate standard deviation from the mean (n = 4). Relative rates determined from the line slopes are indicated.
Chromosome Compaction Without SasA Function.
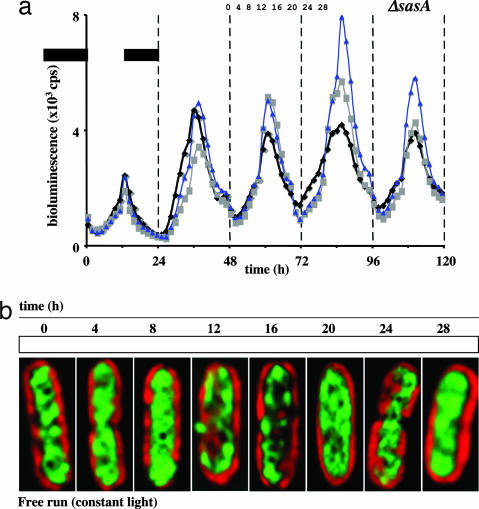
Next we examined chromosome compaction in an sasA-null strain. It remained rhythmic under constant illumination (Figs. 2d and 6). Like the wild-type strain, the sasA strain’s chromosome slowly compacted during the subjective day, with a CI value peak at ZT = 16, and then slowly decompacted throughout the subjective night, with low CI values at ZT = 0 and 24 (Figs. 2d and 6b). Unlike the wild-type strain, this mutant strain’s compaction rhythm was not robust. The amplitude of the compaction rhythm was relatively small (compare Fig. 2 c and d). Recall that gene expression is not rhythmic in this sasA strain (7). We saw some variability in the extent of chromosome compaction at each time point across several independent experiments. However, all experiments with this strain showed a compaction rhythm with the chromosome being most compact midway through a circadian cycle and clearly decompacted at the beginning and end. Consequently, even though chromosome compaction remained rhythmic in the sasA-null strain, the absence of regulatory information from the Kai complex to the SasA protein evidently renders gene expression arrhythmic. So we hypothesize that the circadian clock-controlled chromosome compaction rhythm may be necessary but not sufficient for rhythmic patterns of global gene expression.
Fig. 6.
Gene expression and chromosome compaction rhythms in an S. elongatus sasA strain. (a) As in Fig. 1a but with an sasA-null strain. Three independent data sets are graphed. (b) As in Fig. 1b but with an sasA-null strain. Data are from the indicated free-running cycle. The chromosome arrangement shown for each time point is representative of >98% (n = 200) of the cells from that sample. Each time course experiment was repeated four times with invariant results regarding rhythmic compaction. Again, note the arrangement of the DNA into more distinct nucleoids and then the return to a diffuse state. Recall that, with the exception of the kai genes, gene expression patterns in the sasA strain are arrhythmic (7). Cells are ≈4 μm long.
Functional Circadian Clock Model.
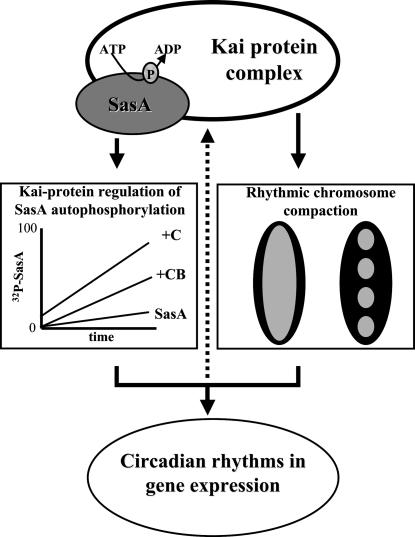
We offer a straightforward model for circadian clock regulation of global gene expression in S. elongatus. The Kai complex imparts a noncompacted, transcription-accessible chromosome during the first half of a circadian cycle and then a compact, less transcription-accessible chromosome during the latter half. The fundamental nature of the Kai circadian clock is therefore proposed to be this rhythmic breathing of the entire S. elongatus chromosome: compact, decompact, compact, decompact. Although the chromosome is absolutely accessible to transcription during part of the circadian cycle, if the proper regulatory elements, as controlled by the clock-output protein SasA, are not available then important temporal information is lost and gene expression patterns are arrhythmic. To generate and maintain circadian rhythms in gene expression, the Kai proteins keep relative time, communicate temporal information to SasA, and appear to control access to promoter elements by imparting a rhythm to chromosome compaction (Fig. 7). An experimental test of this model will be to eliminate the compaction rhythm, via mutation, in a strain with a functional circadian clock and then assay for circadian rhythms in gene transcription.
Fig. 7.
A schematic depiction of circadian clock control of global gene expression in S. elongatus (PCC 7942) is shown. The Kai protein complex keeps relative time (4). Temporal information flows directly from this complex to the SasA protein, regulating its rate of autophosphorylation. As a two-component-type sensor kinase, phosphorylated SasA then controls an undefined signal transduction pathway. Temporal information from the Kai complex also regulates rhythmic chromosome compaction. Together the signal transduction pathway and the chromosome compaction rhythm act to generate circadian rhythms in global gene expression. The dotted arrow represents putative feedback from the expressed gene products upon the Kai complex timing function (1).
Our model would also suggest that the particular phase angle of a gene expression rhythm could depend on the physical location of that gene on the chromosome. While chromosome compaction is expected to somehow sequester most promoter elements (because compaction is greatest during the subjective night while gene expression levels are low) it could also newly expose other promoter elements as a result of the presumed dramatic changes in chromosome topology. This speculation suggests a mechanism for the minor class of genes with increased expression during the subjective night when the chromosome is compacted (11, 12).
The use of chromosome superhelicity, compaction, and topology dynamics is a well known mechanism of both localized and global gene regulation in the enteric bacterium Escherichia coli (27). More apropos is the observation that the chloroplast in the unicellular green alga Chlamydamonas reinhardtii uses genome-wide fluctuations in DNA topology and superhelicity to regulate large-scale gene transcription patterns (28). These topological fluctuations continue under constant-light growth conditions, suggesting an endogenous control mechanism. The homology among modern cyanobacteria and chloroplasts is widely accepted, and thus temporally regulating global gene transcription patterns with large-scale, timed chromosome dynamics may be an ancient mechanism (29–33). Interestingly, recent data from other circadian biology model systems also suggest that the circadian clock may use rhythmic histone acetylation and deacetylation for chromatin remodeling and gene transcription regulation in both insects and mammals (34–38).
We have not yet coupled the large-scale, timed chromosome dynamics that we observed in S. elongatus to the smaller-scale, more direct regulation of gene expression patterns. However, our data suggest the fascinating possibility that a small, three-protein timing mechanism, the Kai clock, temporally regulates global gene expression patterns in part by organizing the structure of an entire bacterial chromosome.
Materials and Methods
Measurement of in Vivo Bioluminescence.
In vivo bioluminescence from a Φ(kaiB-luc+) reporter was measured with a TopCount scintillation counter as described in ref. 39.
Culture Growth and Cloning.
Except where indicated, all Synechococcus strains were cultured under constant illumination at 30°C in BG-11 medium (40). Under these conditions, the wild-type S. elongatus strain has a 25-h free-running period, the S. elongatus kaiC14 strain has a 14-h free-running period, and the S. elongatus sasA strain has an ≈26-h free-running period. Standard techniques were used for Synechococcus strain construction and isolation (39). E. coli strains for plasmid maintenance and protein overproduction were handled as previously described (24, 41). Standard cloning techniques were used (41). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA).
Fluorescence Microscopy.
S. elongatus cultures were grown at 30°C in completely baffled Erlenmeyer flasks to a cell density (OD750) of 0.10–0.15 under constant illumination (80 μmol of photons per m−2 s−1 from white fluorescent lights) and continuous shaking. Cultures grown to that density were then placed in the dark for at least 8 h to reset their circadian clocks. The S. elongatus clock requires ≈5 h of darkness to reset (42). Cultures were then grown for two 12-h light and 12-h dark cycles before being returned to continuous-light conditions. AMC 1297 (kaiC14) was grown in two 7-h light and 7-h dark cycles before being returned to continuous light. Samples (1 ml) were taken every 4 h (every 3.5 h for AMC 1297) starting at ZT = 0 and ending at ZT = 28 during the second light/dark cycle and the second continuous light (free-running) cycle. At each time point, cells were collected by centrifugation (16,000 × g for 1 min). Cells were washed with 0.5 ml of PBS (pH 7.2) and then resuspended in 100 μl of PBS. Next, 80 μl of the DNA stain DAPI (20 μg/ml in water) was added to the cell suspension. Cells were incubated in the dark at 30°C for 20 min, washed twice with 1 ml of H2O, and then resuspended in ≈100 μl of H2O. Stained cell samples (2.5 μl) were spread onto a microscope slide and covered with a slip. Cells were visualized by fluorescence microscopy. The time between taking the sample and visualizing the cells ranged from 30 to 35 min. The cell membrane autofluoresces and was visualized under the rhodamine channel (excitation at 555 ± 14 nm and emission at 617 ± 36 nm). The DAPI-stained DNA was visualized under the DAPI channel (excitation at 360 ± 20 nm and emission at 457 ± 25 nm).
Quantification of Chromosome Compaction.
Cell boundaries in the fluorescence-based images were delineated by changes in the intensity of the rhodamine fluorescence channel signal, which suddenly rises to many times the average background level over distances of just a few pixels. Cell interiors were then defined as the set of pixels with rhodamine channel signal intensities greater than approximately one-fourth of the maximum value seen in a given frame. In a few experiments, thresholds lower than this were needed to include the entire cell in the definition. The relative compaction of the chromosome was then estimated from the distribution of DAPI signals over the set of cell-interior pixels as defined by the rhodamine signal threshold. Pixels were ranked by the strength of the DAPI signal, and then the DAPI background (estimated as the signal at the 2% lower quantile of the distribution) was subtracted from each pixel above this point in the distribution; pixels below this point were set to 0. The area (A) under the resulting normalized cumulative distribution of the summed DAPI signal (Fig. 2a) was then used to calculate a relative CI where CI = 1 − 2A. If the signal were distributed evenly over the entire cell, then the cumulative distribution function would be a straight diagonal line from (0,0) to (1,1), and A would be equal to 0. Conversely, if the whole signal were concentrated into just a handful of pixels, then the cumulative distribution function would consist of a narrow spike, A would approach 0, and CI would approach 1. Between these extremes, different values of CI reflect the degree to which the DAPI signal is concentrated spatially within the cell.
Protein Purification.
The genes encoding KaiB and SasA were cloned into the plasmid pET32a+ vector (Novagen) and subsequently overexpressed in E. coli BL21 (DE3) (Novagen). Genes encoding KaiC and KaiA were cloned into plasmid pQE32 (Qiagen, Valencia, CA) and subsequently overexpressed in E. coli M15 (Qiagen). DNA from each of these clones was sequenced for authenticity (Gene Technologies Laboratory, Texas A&M University, College Station). The Kai proteins were purified as previously described (24). SasA protein was purified after adding isopropyl β-d-thiogalactoside (1 mM) to an aerated 1-liter culture that had reached an OD600 of 0.5. Cells were harvested 3.5 h later. Cell pellets were resuspended in aqueous 50 mM Tris·HCl at pH 7.5 with 300 mM KCl, 10 mM MgCl2, and 1 mM 2-mercaptoethanol. Cell suspensions were passed twice through a French press cell, and the lysates were clarified by centrifugation at 20,000 × g for 30 min. The tagged protein was purified from the supernatant fraction on a Ni-charged chelating column and dialyzed against the recommended enterokinase cleavage buffer (Novagen). After enterokinase cleavage (Novagen), the thioredoxin tag was separated from SasA protein by using a Ni-charged chelating column. This purification step proved unnecessary, and the autophosphorylation data presented are from SasA protein samples that include enterokinase and the free thioredoxin tag. All proteins were analyzed for purity by SDS/PAGE and dialyzed to an appropriate final buffer for the autophosphorylation assay.
Autophosphorylation Assays.
Autophosphorylation assays were run at 25°C. SasA protein was at 0.5 μM in aqueous buffer containing 25 mM Tris·HCl, 200 mM KCl, 1 mM 2-mercaptoethanol, and 5 mM MgCl2 (pH 7.5). To ensure initial velocity measurements, we did experiments over a range of constituent concentrations and established that 1 mM ATP, 1 μM KaiC, 3 μM KaiA, and 3 μM KaiB were saturating for SasA autophosphorylation rate determinations (24). The addition of BSA (New England Biolabs), thioredoxin (Novagen plasmid pET32a+), or CikA (43) protein had no effect on SasA autophosphorylation rates under any of our assay conditions (data not shown). Relevant proteins were mixed for at least 2 min before the assays were started by the addition of radiolabeled ATP. Time 0 samples were taken immediately after this latter addition. Samples, taken at the indicated times, were thermally denatured, and the proteins were separated by SDS/PAGE. Phosphorimages of dried gels were used to quantify the amount of 32PO3 incorporated into SasA. Numerical values representing the percentage of the highest level of incorporation for a given experiment were determined. These values were averaged for each reaction condition (protein components) and plotted as a linear function of time. Relative rates are the slopes of the calculated regression lines.
Acknowledgments
We are most grateful to Dr. Jon Seger, Dr. Fred Adler, and Mr. Brendan O’Fallon (University of Utah) for their development of our image data quantification method. We also thank Dr. S. Golden and Mrs. S. Canales (Texas A&M University), Dr. J. Ditty (University of St. Thomas, St. Paul), and Dr. M. Babst and Mr. M. Curtiss (University of Utah) for their technical, editorial, and general assistance. The PNAS peer review system also led to manuscript improvement. S.B.W. was supported by National Institutes of Health Grant GM19644 and the University of Utah. Protein purification and assay were initiated in S. Golden’s laboratory, and support to her by National Institutes of Health Grant R01 GM62419 and National Science Foundation Grants MCB-9982852 and 0235292 is also acknowledged.
Abbreviations
- CI
compaction index
- ZT
Zeitgeber time.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ditty J. L., Williams S. B., Golden S. S. Annu. Rev. Genet. 2003;37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- 2.Dunlap J. C., Loros J. J. J. Biol. Rhythms. 2004;19:414–424. doi: 10.1177/0748730404269116. [DOI] [PubMed] [Google Scholar]
- 3.Young M. W., Kay S. A. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., Kondo T. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 5.Tomita J., Nakajima M., Kondo T., Iwasaki H. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 6.Imai K., Nishiwaki T., Kondo T., Iwasaki H. J. Biol. Chem. 2004;279:36534–36539. doi: 10.1074/jbc.M405861200. [DOI] [PubMed] [Google Scholar]
- 7.Iwasaki H., Williams S. B., Kitayama Y., Ishiura M., Golden S. S., Kondo T. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 8.Mori T., Saveliev S. V., Xu Y., Stafford W. F., Cox M. M., Inman R. B., Johnson C. H. Proc. Natl. Acad. Sci. USA. 2002;99:17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakahira Y., Katayama M., Miyashita H., Kutsuna S., Iwasaki H., Oyama T., Kondo T. Proc. Natl. Acad. Sci. USA. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Tsinoremas N. F., Johnson C. H., Lebedeva N. V., Golden S. S., Ishiura M., Kondo T. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 11.Min H., Golden S. S. J. Bacteriol. 2000;182:6214–6221. doi: 10.1128/jb.182.21.6214-6221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Min H., Liu Y., Johnson C. H., Golden S. S. J. Biol. Rhythms. 2004;19:103–112. doi: 10.1177/0748730403262056. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y., Tsinoremas N. F., Golden S. S., Kondo T., Johnson C. H. Mol. Microbiol. 1996;20:1071–1081. doi: 10.1111/j.1365-2958.1996.tb02547.x. [DOI] [PubMed] [Google Scholar]
- 14.Oldenburg D. J., Bendich A. J. J. Mol. Biol. 2004;344:1311–1330. doi: 10.1016/j.jmb.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Gitai Z., Thanbichler M., Shapiro L. Trends Microbiol. 2005;13:221–228. doi: 10.1016/j.tim.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Mori T., Binder B., Johnson C. H. Proc. Natl. Acad. Sci. USA. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roenneberg T., Merrow M. J. Biol. Rhythms. 2002;17:495–505. doi: 10.1177/0748730402238231. [DOI] [PubMed] [Google Scholar]
- 18.Kondo T., Ishiura M. J. Bacteriol. 1994;176:1881–1885. doi: 10.1128/jb.176.7.1881-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo T., Tsinoremas N. F., Golden S. S., Johnson C. H., Kutsuna S., Ishiura M. Science. 1994;266:1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 20.Mori T., Johnson C. H. J. Bacteriol. 2001;183:2439–2444. doi: 10.1128/JB.183.8.2439-2444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C. R., Tanabe A., Golden S. S., Johnson C. H., Kondo T. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 22.Kageyama H., Kondo T., Iwasaki H. J. Biol. Chem. 2003;278:2388–2395. doi: 10.1074/jbc.M208899200. [DOI] [PubMed] [Google Scholar]
- 23.Williams S. B., Stewart V. Mol. Microbiol. 1997;26:911–925. doi: 10.1046/j.1365-2958.1997.6262002.x. [DOI] [PubMed] [Google Scholar]
- 24.Williams S. B., Vakonakis I., Golden S. S., LiWang A. C. Proc. Natl. Acad. Sci. USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolanin P. M., Thomason P. A., Stock J. B. Genome Biol. 2002;3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vakonakis I., Klewer D. A., Williams S. B., Golden S. S., LiWang A. C. J. Mol. Biol. 2004;342:9–17. doi: 10.1016/j.jmb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Hatfield G. W., Benham C. J. Annu. Rev. Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 28.Salvador M. L., Klein U., Bogorad L. Mol. Cell. Biol. 1998;18:7235–7242. doi: 10.1128/mcb.18.12.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson J. O., Roger A. J. Curr. Biol. 2002;12:115–119. doi: 10.1016/s0960-9822(01)00649-2. [DOI] [PubMed] [Google Scholar]
- 30.Cavalier-Smith T. Curr. Biol. 2002;12:R62–R64. doi: 10.1016/s0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- 31.Driessche T. V., Bonotto S. Arch. Int. Physiol. Biochim. 1968;76:205–206. [PubMed] [Google Scholar]
- 32.Durnford D. G., Deane J. A., Tan S., McFadden G. I., Gantt E., Green B. R. J. Mol. Evol. 1999;48:59–68. doi: 10.1007/pl00006445. [DOI] [PubMed] [Google Scholar]
- 33.Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., Leister D., Stoebe B., Hasegawa M., Penny D. Proc. Natl. Acad. Sci. USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown S. A., Ripperger J., Kadener S., Fleury-Olela F., Vilbois F., Rosbash M., Schibler U. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 35.Curtis A. M., Seo S. B., Westgate E. J., Rudic R. D., Smyth E. M., Chakravarti D., FitzGerald G. A., McNamara P. J. Biol. Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- 36.Naruse Y., Oh-hashi K., Iijima N., Naruse M., Yoshioka H., Tanaka M. Mol. Cell. Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ripperger J. A., Schibler U. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 38.Smolen P., Hardin P. E., Lo B. S., Baxter D. A., Byrne J. H. Biophys. J. 2004;86:2786–2802. doi: 10.1016/S0006-3495(04)74332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson C. R., Tsinoremas N. F., Shelton J., Lebedeva N. V., Yarrow J., Min H., Golden S. S. Methods Enzymol. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- 40.Bustos S. A., Golden S. S. J. Bacteriol. 1991;173:7525–7533. doi: 10.1128/jb.173.23.7525-7533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maloy S. R., Stewart V. J., Taylor R. K. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1996. Genetic Analysis of Pathogenic Bacteria: A Laboratory Manual. [Google Scholar]
- 42.Kiyohara Y. B., Katayama M., Kondo T. J. Bacteriol. 2005;187:2559–2564. doi: 10.1128/JB.187.8.2559-2564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mutsuda M., Michel K. P., Zhang X., Montgomery B. L., Golden S. S. J. Biol. Chem. 2003;278:19102–19110. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]