Myosin molecular motors bind to and exert force upon actin filaments. Between 24 (1) and 37 (2) myosin subfamilies are currently recognized (3), the myriad members of which are specialized for tasks ranging from muscle contraction through to mechano-sensation and signaling, cell polarization, the sliding and tensioning of cytoskeletal elements, organelle transport, exocytosis and endocytosis, and vesicle transport. The molecular mechanism by which myosins step along actin filaments is accordingly a central problem, studies of which have been greatly facilitated in recent years by the discovery of processive myosins. Processive myosins can take many sequential steps along an actin filament, remaining attached throughout. One such is myosin-V (4), a twin-headed myosin that walks along actin filaments toward their fast-growing (blunt) ends, measuring out paces that correspond more or less exactly to the ≈36-nm helical repeat of the actin filament. The precisely metered stepping action is a function of the length of the neck (5), an extended regulatory domain of six calmodulin-binding IQ motifs that link each head to the coiled-coil tail. The favored ≈36-nm step allows the motor to pick out a linear pathway along a helical actin filament; however, recent work has revealed that off-axis binding can and does occur (6, 7). Like other molecular motors, myosin-V works directionally, with forward steps being favored over backward steps. Sustained directional stepping by any motor against a load requires an energy source, which, in the case of the myosins, is supplied by the hydrolytic conversion of ATP into ADP and Pi. Coupling the mechanical stepping cycle to ATP turnover in this way allows the myosin-V motor to step continuously against a load of up to ≈2–3 pN, consistent with its using one ATP per 36-nm step. Above ≈2–3 pN, the motor stalls, pausing with both heads attached to the actin filament (8–10) but unable to step forward because the work involved exceeds the energy available from ATP hydrolysis. Importantly, the evidence suggests that, in this stalled state, ATP turnover is halted, so that the myosin-V motor only consumes ATP when it is actively stepping. This invariant mapping of exactly one ATP molecule per step, and one step per ATP molecule, is called tight coupling. An obvious question is, what happens if one pulls backward on a walking myosin-V molecule with greater force than the stall force? Will the motor simply be torn from its track, or will it step back? And if it steps back, will it step in a controlled way, and will the tight requirement for one ATP molecule per step be maintained? In a recent issue of PNAS, Gebhardt et al. (11) reported answers to exactly these questions. Remarkably, they found that pulling backward on a walking myosin-V molecule causes the motor to reverse its mechanical action, again taking measured-out 36-nm steps, but without a requirement for ATP binding. Backstepping continues, at a rate that depends on the load, until the load diminishes to the point where the motor can once again set off in its natural progress direction, stepping in an ATP-dependent manner toward the barbed end of the actin filament.
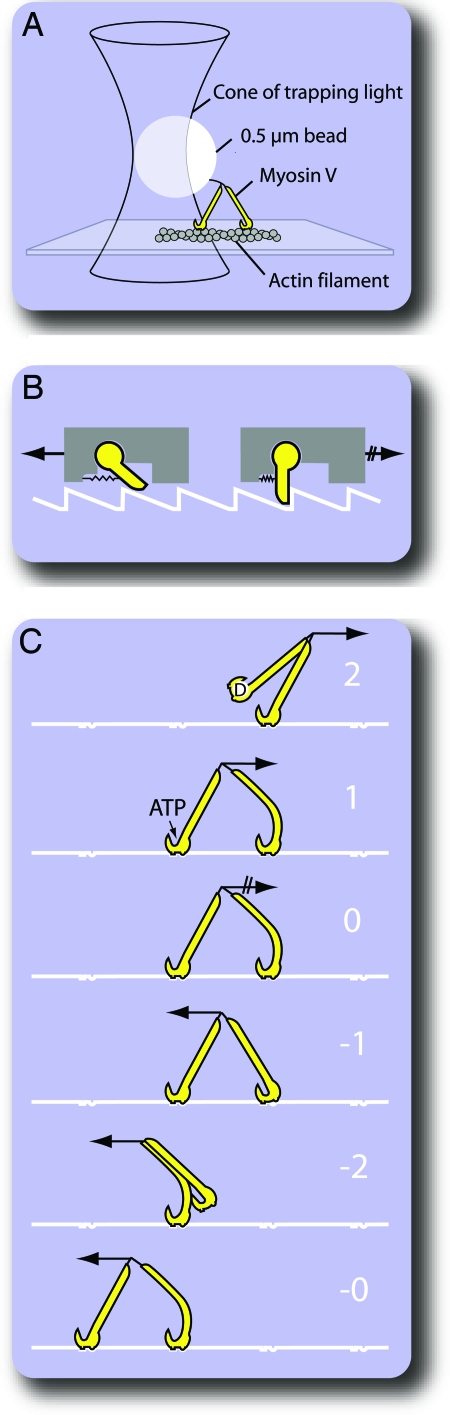
The Gebhardt et al. (11) experiments build on earlier work from the same laboratory (12) using classical single-bead optical trapping (13) to maneuver and track individual myosin-V molecules. In this type of experiment, single myosin-V molecules are attached to a ≈0.5-μm plastic bead. The bead is then trapped by a focused infrared laser beam and brought close to an immobilized actin filament (Fig. 1A). As the motor steps along the actin filament (pulling the bead toward the periphery of the trapping beam), the optical force (tending to return the bead to trap-center) increases. Eventually, the optical force is equal to the maximum force that the motor can develop, and it stalls. What Gebhardt et al. (11) have done is to ask what now happens if greater backward force is applied. To answer this question, they arranged a feedback so that the force experienced by the motor could be set to a particular level and maintained automatically; this feedback is achieved by having the apparatus sense when the motor has taken a step and then react by shifting the microscope stage slightly to compensate. Using this arrangement, they were able to apply a range of forward and backward forces to walking myosin-V molecules and track the mechanical responses of the motor.
Fig. 1.
Aspects of molecular walking under load. (A) Force-feedback single-bead optical trap. A single myosin-V molecule is attached to a plastic bead, and the bead is trapped by a focused infrared laser. Defined levels of forward or backward force are maintained by monitoring bead position and automatically stepping the stage to compensate for steps taken by the motor. (B) Ratchet-and-pawl. The gray block can be dragged easily to the left, because the pawl flips up and can slip along the toothed track. By contrast, the mechanism blocks movement to the right, because the pawl flips down and engages. Bicycle freewheels work this way. (C) A possible scheme for force-induced backstepping. A backstepping sequence (states 0, −1, −2, −0) can be driven by applying backward force, but a forward stepping sequence (states 0, 1, 2, 0) cannot. Forward stepping has an absolute requirement for ATP. The structure of the binding interface between the motor and its track means that backward force is able to unpeel the myosin head from its track, whereas forward force is not.
Pulling forward (in the progress direction) had little effect. The forward walking rate of the motor was dependent on the ATP concentration, as expected when each step requires an ATP to bind, but was essentially independent of the forward load. At low ATP concentration, there was no effect at all, and, at saturating ATP concentration, forward pulling produced maximally a 50% increase in stepping rate. Pulling backward, by contrast, induced backstepping, at a rate that depended on the load but not on the ATP concentration; indeed, backstepping occurred in the absence of ATP.
This result is remarkable. Its clear implication is that the myosin-V motor is to some extent a purely mechanical device, a kind of directional sliding clamp that grips the track more firmly when loaded in one direction than when loaded in the other. Ratchets have the property that they rectify applied force to allow motion in one direction only. Obvious analogues in the macroscopic world might be the freewheel on a bicycle or the clutch mechanism on a winch, wherein the sprung pawls slip along the track when moved in one direction but engage and grip in the other direction. Fig. 1B shows such a ratchet-and-pawl machine.
The Gebhardt et al. (11) data establish for the first time that a purely mechanical ratchet mechanism can exist at the molecular scale. They identify a mechanical pathway whereby applied force can distort the binding interface between the myosin-V motor and its track, thereby stabilizing or destabilizing binding in a direction-dependent way (Fig. 1C).
Importantly, however, myosin-V is much more than a passive mechanical ratchet. The turnover of ATP adds a large set of properties that overlays the basal mechanical ratchet, converting myosin-V from a machine that resists forward motion to a machine that steps actively in the direction that is forbidden by the underlying mechanical ratchet. How does this work, and do the Gebhardt et al. (11) results tell us anything about the ATP-driven part of the stepping mechanism?
The key to coordinated ATP-driven walking by myosin-V appears to be strain-dependent nucleotide exchange. This property has been called gating (14, 15). Kinetic studies on myosin-V in solution indicate that ADP release is rate limiting, and, crucially, that strain between the two heads of a walking myosin-V molecule can modulate the rate constant for ADP release and, thereby, the rate constant for detachment of a head from the actin filament (summarized in ref. 4). In the absence of external strain, backward strain on the leading head inhibits its ADP release by ≈100-fold compared with the trail head (16). Because ADP release controls ATP binding and thereby detachment, this strain dependence predicts that the trail head will be ≈100 times more likely to detach than the lead head. Applying extra, external loads to the lead head is likely to further exaggerate the differential.
In addition to strain-sensitive ADP release, the data also hint at strain-sensitive ATP binding. The finding that backstepping is not accelerated by ATP indicates that ATP binding is inhibited by backward load. This observation contrasts markedly with recent findings for kinesin, another molecular walking machine, for which backstepping rates do vary with ATP concentration (17). For myosin-V, the suggestion must be that not only is ADP release inhibited by backward load but also ATP binding. Perhaps the active site is closed by backward load, thereby inhibiting all nucleotide exchange. A stable apo (nucleotide-free) structure of the myosin-V head was recently determined (18) in which the nucleotide-binding site was closed off so as to block the docking of nucleotide.
What might be the biological significance of backward ratcheting by myosin-V? As discussed by Gebhardt et al. (11), ratcheting would be useful in a tug-of-war situation in which myosin and an oppositely directed, stronger motor were doing battle. The myosin-V could then maintain contact with its track while being driven passively backward at no energy cost and then rapidly reestablish forward progress once the oppositely directed motor relented. Further experiments are needed to test the behavior of individual motor molecules in these “social” situations.
Concerning the reversibility of molecular motors, the motors field now has examples of all three possible types of behavior. The rotary F1/F0 ATPase of mitochondria appears freely reversible. In its natural state, it synthesizes ATP while being driven by a proton pump; in vitro, the isolated motor spins the other way and hydrolyses ATP. Kinesin-1 ordinarily walks along microtubules toward their plus ends; by hauling backward on the molecule by using an optical trap, it can be made to walk backward, but the forced backward walking requires ATP binding, just as does the forward walking. Finally, we now have myosin-V, which can be forced backward without a requirement for ATP.
Conflict of interest statement: No conflicts declared.
See companion article on page 8680 in issue 23 of volume 103.
References
- 1.Foth B. J., Goedecke M. C., Soldati D. Proc. Natl. Acad. Sci. USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards T. A., Cavalier-Smith T. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 3.Goodson H. V., Dawson S. C. Proc. Natl. Acad. Sci. USA. 2006;103:3498–3499. doi: 10.1073/pnas.0600045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sellers J. R., Veigel C. Curr. Opin. Cell Biol. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Sakamoto T., Yildez A., Selvin P. R., Sellers J. R. Biochemistry. 2005;44:16203–16210. doi: 10.1021/bi0512086. [DOI] [PubMed] [Google Scholar]
- 6.Toprak E., Enderlein J., Syed S., McKinney S. A., Petschek R. G., Ha T., Goldman Y. E., Selvin P. R. Proc. Natl. Acad. Sci. USA. 2006;103:6495–6499. doi: 10.1073/pnas.0507134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syed S., Snyder G. E., Franzini-Armstrong C., Selvin P. R., Goldman Y. E. EMBO J. 2006;25:1795–1803. doi: 10.1038/sj.emboj.7601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker M. L., Burgess S. A., Sellers J. R., Wang F., Hammer J. A., III, Trinick J., Knight P. J. Nature. 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 9.Burgess S., Walker M., Wang F., Sellers J. R., White H. D., Knight P. J., Trinick J. J. Cell Biol. 2002;159:983–991. doi: 10.1083/jcb.200208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volkmann N., Liu H., Hazelwood L., Krementsova E. B., Lowey S., Trybus K. M., Hanein D. Mol. Cell. 2005;19:595–605. doi: 10.1016/j.molcel.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Gebhardt J. C. M., Clemen A. E.-M., Jaud J., Rief M. Proc. Natl. Acad. Sci. USA. 2006;103:8680–8685. doi: 10.1073/pnas.0510191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemen A. E., Vilfan M., Jaud J., Zhang J., Barmann M., Rief M. Biophys. J. 2005;88:4402–4410. doi: 10.1529/biophysj.104.053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Block S. M., Goldstein L. S., Schnapp B. J. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 14.Veigel C., Wang F., Bartoo M. L., Sellers J. R., Molloy J. E. Nat. Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 15.Veigel C., Schmitz S., Wang F., Sellers J. R. Nat. Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 16.Rosenfeld S. S., Sweeney H. L. J. Biol. Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 17.Carter N. J., Cross R. A. Curr. Opin. Cell Biol. 2006;18:61–67. doi: 10.1016/j.ceb.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Coureux P. D., Wells A. L., Menetrey J., Yengo C. M., Morris C. A., Sweeney H. L., Houdusse A. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]