Abstract
The phosphoinositide 3-kinase (PI3K)/Akt pathway controls a vast array of normal physiological processes and is frequently aberrantly activated in cancer, thus identifying PI3K/Akt-signaling components as promising drug targets in oncology. However, implementation of rational cancer therapies for this pathway needs robust and simple tools to stratify patients according to PI3K pathway activation and to validate and measure the impact of targeted inhibition on primary cancer tissues. Herein we present a technique for the quantification of the PI3K/Akt-signaling pathway based on the mass spectrometric measurement of PI3K-dependent protein kinase activity in cell lysates. The concept of this application of MS is to exploit enzymatic activity to amplify the signal of the enzyme under study analogous to the PCR used to amplify nucleic acid sequences. We show that this approach allows quantitative analysis of a cell-signaling pathway with high sensitivity, precision of quantification, and specificity. Due to its special analytical capabilities and potential for multiplexing, this approach could contribute significantly to cell-signaling studies and to the development and implementation of personalized cancer therapies.
Keywords: quantitative analysis, signaling pathway, personalized therapies, cancer stem cells, patient stratification
Signal transduction pathways downstream of receptor tyrosine kinases control a large array of biological processes, including energy metabolism, cell proliferation, survival, cell motility, and immune responses. One of these pathways, the phosphoinositide 3-kinase (PI3K)/Akt axis, is one of the most frequently deregulated pathways in cancer (1–4) by virtue of oncogenic mutation, by overexpression of its signaling components (5–8), by inactivation of the PTEN tumor-suppressor protein, which opposes PI3K action (9–12), or by being at the crossroads of upstream oncogenic signals (reviewed in ref. 4).
Because of the importance of this pathway in the control of several biological and pathological processes (13–15), robust methods to quantify its activation are needed. PI3K catalyzes the phosphorylation of the lipid phosphatidylinositol-4,5-bisphosphate to form phosphatidylinositol-3,4,5-trisphosphate, a second messenger that is needed for the activation of downstream Ser/Thr kinase protein kinases, such as Akt [also known as protein kinase B (PKB)] and phosphoinositide-dependent kinase-1. The assessment of lipid production by various approaches has turned out to be cumbersome, and it is thus far not possible to directly measure production of phosphatidylinositol-3,4,5-trisphosphate in tissue samples. Activation of the PI3K pathway is therefore commonly assessed by examination of signaling components downstream of PI3K. These components include Akt/PKB and kinases farther downstream of the Akt/PKB cascade, such as p70-S6K. Most often, this examination is performed by indirect assessment using immunochemical methods that measure the phosphorylation status of Akt on Ser-473 or Thr-308 or of S6, a ribosomal protein substrate of p70-S6K, on Ser-235/236. A drawback of this strategy is its limited dynamic range and its subjective and semiquantitative nature, making it impossible to compare data from different laboratories and difficult to implement in routine analysis of tumor samples.
Quantification of phosphoproteins or their phosphorylated sites by MS has proven useful for studying the mechanisms of signal transduction (16–18). On the basis of its specificity and precision of quantification, MS could, in principle, be an ideal method to quantify phosphorylation sites that correlate with pathway activation (19). However, currently used MS methods are not practical alternatives for the quantification of signaling pathways, because their sensitivities make these approaches unsuitable for many biological applications. Moreover, another important limitation of phosphoprotein analysis (either by immunochemistry or MS) as a surrogate measure for pathway activation is that this strategy also fails to take into account other covalent and allosteric modifications and protein–protein interactions that modulate enzymatic activity (20).
We present an alternative approach for the quantification of pathway activation that is based on mass spectrometric measurements of enzymatic activity, which is arguably the most important parameter of enzymes affecting cell biology (rather than their amounts or phosphorylation status). MS has been shown to be a useful tool for the determination of enzymatic activity (21), but, to the best of our knowledge, its utility for absolute quantification of protein kinase activity in tissue extracts has not been documented. In this study we used MS for the quantification of PI3K-dependent protein kinase activity toward a substrate that is highly selective for this pathway, thereby directly quantifying a biochemical property of protein kinases that has not been investigated by MS thus far to a significant extent. We have found that this approach allows for the specific and sensitive quantification of pathway activation in absolute units, an important requirement for clinical analysis and a limitation of other detection methods, such as those based on radioactive and fluorescence techniques (22, 23).
Results
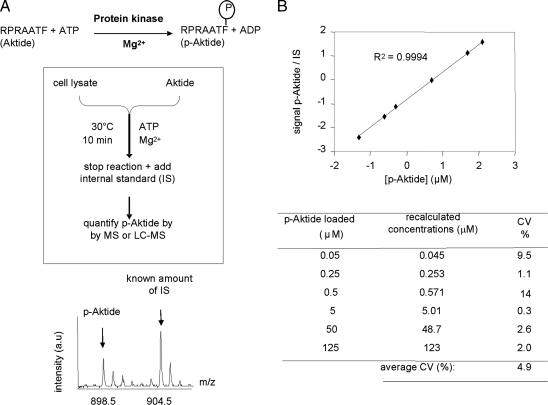
Our strategy (Fig. 1A) uses a nonradioactive isotope dilution approach and MS to quantify protein kinase activity toward the RPRAATF peptide (24) (hereafter named “Aktide”), a highly selective substrate of protein kinases, such as Akt and SGK, downstream of PI3K (22, 24–26), with Akt/PKB being particularly active toward this peptide. Indeed, protein kinases downstream of PI3K [chiefly Akt/PKB and SGK (22, 24–26)] phosphorylate Aktide with faster kinetics than do other protein kinases, thus making the contribution of nonspecific protein kinases to overall Aktide kinase activity in cell lysates negligible (see below and refs. 22 and 24). A known amount of phospho-Aktide (p-Aktide) labeled with stable isotopes (see Materials and Methods) is used as an internal standard (IS) to quantify the amounts of p-Aktide produced per unit of time. Standard curves constructed with synthetic peptides showed that this method can be used for absolute quantification of p-Aktide with great precision (Fig. 1B), with coefficients of variation being ≈5% on average.
Fig. 1.
Principle and precision of mass spectrometric quantification of protein kinase activity. (A) Principle of the method. (B) Precision of the method. Known amounts of synthetic p-Aktide were mixed with a fixed amount of IS peptide and analyzed by MALDI-TOF MS, LC-MS, or LC-MS/MS. Functions derived from standard curves (see Upper for an example) were used to recalculate the concentration of added p-Aktide and thus estimate coefficients of variation (CV) (Lower).
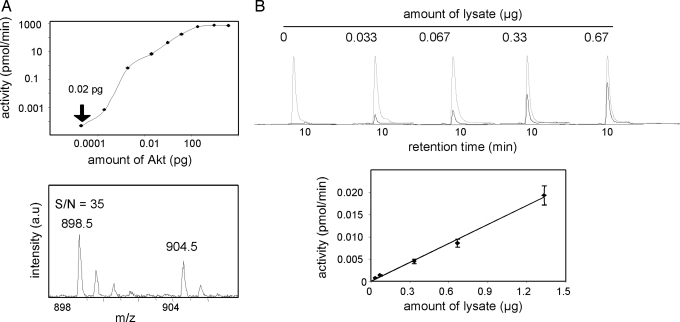
A combination of the amplification nature of enzymatic activity measurements and the sensitivity of MS contributed to sensitivities in the zeptomole range (1 zmol = 600 molecules) of recombinant Akt/PKB (Fig. 2A). Likewise, kinase activity toward Aktide could be measured in 30 ng of lysate of a B lymphoma cell line (Fig. 2B) with a signal-to-noise ratio, indicating that this approach allows for measuring signal transduction pathway activation with great sensitivity; the use of new generation commercial mass spectrometers, such as linear ion traps or Fourier transform ion cyclotron resonance instruments, should result in the achievement of even lower limits of detection. This sensitivity sharply contrasts with that afforded by currently used MS methods for the quantification of phosphoproteins in cells that require ≈107-108 cells per experimental point (16, 17, 27).
Fig. 2.
Sensitivity of Aktide kinase activity quantification by MS. (A) Quantification of Aktide kinase activity by using recombinant Akt/PKB. Decreasing amounts of recombinant Akt/PKB were used in activity assays with Aktide as a substrate. (Lower) Shown is a MALDI-TOF MS spectrum obtained by using 0.02 pg of Akt, which corresponds to ≈500 zmol (500 × 10−21 mol) of protein. S/N, signal-to-noise ratio. (B) Quantification of Aktide kinase activity in cell lysates. (Upper) Representative chromatograms from LC-MS runs obtained by using different amounts of WEHI-231 B cell lysate (gray and black lines correspond to IS and p-Aktide signals, respectively). (Lower) Shown is a plot of the quantitative data derived from these chromatograms.
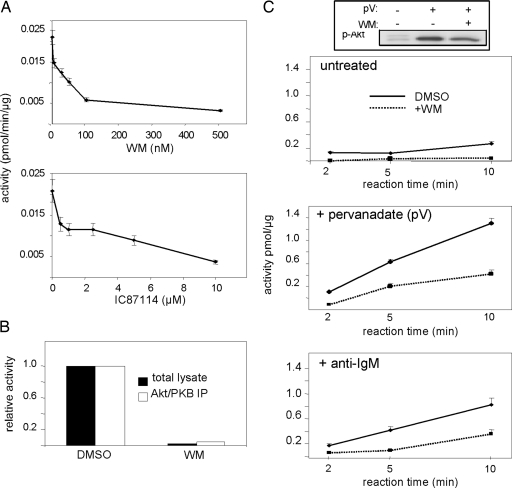
In agreement with previous studies (22), ≥80% of activity in cell lysates of an exponentially growing B lymphoma cell line was sensitive to pretreatment of the cells with PI3K inhibitors (Fig. 3A), with activity being equally inhibited in total cell lysates and Akt immunoprecipitates (Fig. 3B). Aktide kinase activity in B cell lysates increased upon treatment of the cells with broad-spectrum (pervanadate) or physiological (anti-IgM, to activate the B cell antigen receptor) activators of PI3K activity in a PI3K inhibitor-sensitive manner (Fig. 3C). Residual Aktide kinase activity in the presence of PI3K inhibitors correlated with residual Ser-473 phosphorylation on Akt/PKB detected by Western blotting (Fig. 3C Top). These results indicate that Aktide kinase activity is a specific readout for the PI3K/Akt pathway.
Fig. 3.
Specificity of Aktide kinase activity for the PI3K/Akt pathway. (A) Specificity in cell lysates. Cells were treated with different concentrations of wortmannin (WM, a pan PI3K inhibitor) or IC87114 (a PI3K inhibitor with selectivity for the p110δ isoform of PI3K) before lysis and MS activity measurements. (B) Aktide kinase activity in total cell lysates and Akt/PKB immunoprecipitates (IP) from untreated cells were equally sensitive to WM pretreatment. (C) Shown is kinase activity in WEHI-231 lysates at different in vitro kinase reaction times. Activities increased upon stimulation with PI3K/Akt pathway agonists [anti-IgM (Bottom) or pervanadate (Middle)] in a WM-sensitive manner. (Top) Shown is a Western blot of total cell lysates with anti-p-Akt/PKB (p-Ser-473) antibodies, demonstrating a correlation between Aktide kinase activity and p-Akt/PKB measurements.
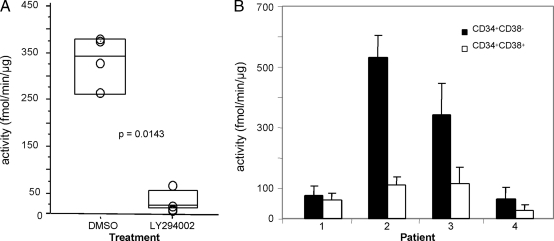
Potential applications of this method include the detection of malignancies with an overactive PI3K/Akt pathway, thus allowing for patient stratification and the quantification of the effects of PI3K/Akt pathway inhibitory agents in vivo. It has thus far been difficult to quantify PI3K/Akt pathway activation, especially in solid tumors. We found that mouse B16 melanoma tumor biopsies showed a robust Aktide kinase activity of 335 ± 53 fmol/min per μg (Fig. 4A), which decreased significantly (P = 0.0143) upon in vivo treatment of animals with the pan-PI3K inhibitor LY294002 (Fig. 4A). The sensitivity of our method also allows quantification of PI3K/Akt activity in the rare cancer stem cell population, which likely needs to be eradicated to achieve therapeutic success. Relatively small numbers of these cells can be isolated, and this has precluded the use of standard biochemical analysis. Cancer stem cells are best characterized phenotypically in acute myeloid leukemia and are present in the CD34+CD38− fraction (28). PI3K/Akt activity was readily quantified in the leukemic stem cell fraction from four individuals with acute myeloid leukemia with significant interindividual variation in absolute levels of activity (Fig. 4B).
Fig. 4.
Quantification of PI3K/Akt pathway activity in solid and liquid tumors. (A) B16/Bl6 solid tumors in mice were treated with DMSO (vehicle) or LY294002 (a pan-PI3K inhibitor) before excision and MS activity measurements. (B) Primary acute myeloid leukemia cells were sorted into CD34+CD38− stem cell and CD34+CD38+ bulk tumor fractions, and activity toward Aktide was measured by MS.
Discussion
MS is perhaps the most powerful analytical method thus far available for the identification and quantification of proteins and their sites of modification (29). Because of its specificity, MS is often preferred over immunochemical, radioactive, or optical methods for quantitative analyses, especially when the quantification is performed by liquid chromatography (LC)-MS/MS and reaction monitoring, which completely eliminates the possibility of artifacts due to nonspecific signal (30). The demands of proteomics have resulted in the development of mass spectrometers of increasing sensitivity, with typical limits of detection in the subfemtomole range. However, signaling proteins are most often present at considerably lower amounts in cells, and phosphorylation sites exist at very low stoichiometric levels. This fact means that even the most sensitive mass spectrometers struggle to detect sites of modification on regulatory proteins (quantification being even more challenging). Thus, MS-based proteomic studies aimed at investigating the mechanisms of signal transduction have used large numbers of cells (typically ≈108) per data point (16, 17, 27), overexpressed the protein under study (18), or performed analyses on recombinant proteins (31). Clearly, these strategies are not applicable to the analysis of all biological specimens, and it is extremely challenging for current mass spectrometric methods to detect regulatory proteins and their sites of modification in clinically relevant tissues such as biopsies.
An amplification method for proteins compatible with MS analyses should facilitate the implementation of MS methods for the analysis of these more challenging biological specimens. Exploiting the intrinsic activity of enzymes allows amplification of the signal of proteins with catalytic activity, thus facilitating the targeted mass spectral analysis of enzymes with great sensitivity and in a manner that, by including isotopically labeled ISs, also provides a quantitative measure of their activation status in absolute units. We have applied this analytical strategy to the PI3K/Akt pathway and show that it is possible to quantify signal transduction pathway activation (i) with great precision (Fig. 1), (ii) with high sensitivity (Fig. 2), and (iii) in a specific manner (Fig. 3).
Oncogenic somatic mutations in the p110α isoform of PI3K have been identified in many different types of cancer (6, 32, 33). Similarly, PTEN, a lipid phosphatase that degrades the phosphatidylinositol-3,4,5-trisphosphate lipid, is a tumor-suppressor gene that is inactivated or underexpressed in many cancer types (3, 4, 9–12). Recently, a systematic genetic analysis indicated that ≤40% of colorectal cancers may present mutations in PI3K/Akt pathway members (5). Many other studies have provided overwhelming cumulative data indicating that the PI3K/Akt pathway may be overactivated in 10–50% of most cancer types (for examples, see refs. 8 and 34–38), making this pathway one of the most frequent aberrantly activated signaling pathways in cancer. Thus, small molecule modulators of PI3K/Akt signaling offer enormous potential for the treatment of several types of malignancies, and some of them are already in the clinic or in preclinical stages of development (1). However, individual tumors with similar pathology may have a different spectrum of underlying genetic mutations in cancer-causing genes. Therefore, although modulators of PI3K/Akt will probably benefit a significant proportion of cancer patients, an even larger percentage of tumors may not respond to these inhibitors. Thus, development of methods that can identify the patient subpopulation with an aberrantly activated PI3K/Akt pathway has been recognized to be of paramount importance for successful therapeutic strategies that target this pathway (1, 39). The approach described here enables the robust measurement of signal transduction pathway activation in primary tumor tissues with the sensitivity, specificity, and precision needed for providing clinically useful information (Fig. 4).
Although we have initially focused on the PI3K/Akt pathway because of its importance as a therapeutic target in cancer, this analytical strategy could easily be applied to any enzyme whose product is amenable to mass spectrometric detection, provided that at least one substrate specific for the target enzyme is available. In addition, the specificity of MS offers the opportunity of measuring several reaction products simultaneously in a fast format that can be easily automated for clinical implementation. Such an approach could be applied to identify the subpopulation of patients that may benefit from targeting a specific pathway, avoiding unnecessary potential side effects, and also to the pharmacodynamic validation of new drugs by enabling an accurate assessment of target inhibition in primary tissues. This technique has the potential to significantly contribute to making the concept of personalized cancer therapy into a clinical reality.
Materials and Methods
Cell Culture.
The mouse WEHI-231 B lymphoma cell line was cultured as described in ref. 17. When indicated, cells were stimulated with anti-IgM (1 μM, 5 min) or pervanadate (500 μM, 30 min). Cells were treated with PI3K inhibitors for 30 min before lysis.
Preparation and Sorting of Primary Tumors.
Seven days after intradermal injection of 2 × 105 B16/Bl6 melanoma cells, tumors were injected with 50 μl of 10 μM LY294002 or vehicle 2 h before surgical excision. Tumor biopsies were snap frozen in liquid nitrogen and kept at −80°C until the day of analysis. Frozen primary acute myeloid leukemia samples were rapidly thawed, washed, and allowed to recover in RPMI medium 1640 with 10% FCS at 37°C in 5% CO2 for 3 h. Cells were then incubated with phycoerythrin-conjugated anti-CD34 (BD Pharmingen) and FITC-conjugated anti-CD38 (DAKO) monoclonal antibodies for 30 min on ice. Cells were sorted in PBS into CD34+CD38− (stem cell) and CD34+CD38+ fractions on an EPICS Elite flow cytometer (Beckman-Coulter). After centrifugation, cell pellets were resuspended in RPMI medium 1640 with 10% FCS and allowed to recover at 37°C in 5% CO2 for 1–2 h. Typical cell yields ranged from 5 × 103 to 7 × 104 stem cells per patient. Samples were obtained with informed consent and Ethics Committee approval.
Preparation of Biological Material for Analysis.
Cultured cells were lysed in lysis buffer (1% Triton X-100/150 mM NaCl/1 mM EDTA/Tris·HCl, pH 7.4/1 mM DTT) containing protease and phosphatase inhibitors. Frozen, solid tumors were homogenized in lysis buffer by using a pestle. After centrifugation at 20,000 × g, cell lysates and tissue homogenates were ready to use as enzyme source.
Enzymatic Assays.
Peptides used as substrate (Aktide) or IS (RP*RAApTF, where P* is l-proline-13C5, 15N, a 6-Da heavier version of l-proline) and to construct standard curves (RPRAApTF, p-Aktide) were synthesized by C. Hyde (Wolfson Institute for Biomedical Research, University College London). Reactions were performed by using either recombinant Akt/PKB (Upstate, Dundee, U.K.) or cell lysates as enzyme sources. A volume of 5.0 μl of substrate mix (150 mM ATP/150 mM Aktide/7.5 mM MgCl2/0.15 mM EGTA/7.5 mM β-glycerol phosphate/0.1 mM sodium orthovanadate/0.1 mM DTT) was mixed with 2.5 μl of enzyme source and incubated for 2–10 min at 30°C. When using recombinant enzyme, reactions were performed at different incubation times, from 2 min to 18 h. Reactions were stopped by adding 7.5 μl of 1% trifluoroacetic acid containing a known amount of IS (typically 1 pmol/μl).
MS Quantification of Enzymatic Reactions.
The product of the above reaction was quantified by MALDI-TOF MS (Ultraflex; Bruker, Billerica, MA) or LC-MS/MS [Ultimate HPLC (Dionex) connected to a Micromass (Manchester, U.K.) Q-Tof instrument]. The LC-MS/MS method consisted of monitoring the parent–daughter ion transition of m/z 449.7–400.3 for p-Aktide and m/z 452.7–403.3 for the IS. Reaction products were analyzed by LC-MS/MS without further treatment. Samples for MALDI-TOF MS analysis were prepared by solid-phase extraction with a modified ZipTip (Millipore) or by strong cation exchange over a polySULFOETHYL A resin (Poly LC, Columbia, MD). Standard curves showed exquisite linearity with correlation coefficients and slopes of virtually 1 (Fig. 1B). Therefore, the amounts of p-Aktide formed during the reaction were calculated by multiplying the amount of IS in the reaction vessel by the ratio of p-Aktide-to-IS signal responses (spectral peak areas in MALDI-TOF MS experiments or chromatographic peak areas of fragment ions for LC-MS/MS analyses).
Acknowledgments
We thank Khaled Ali, Barbara Geering, and Klaus Okkenhaug for helpful input and Arnold Pizzey for carrying out flow cytometric cell sorting. This work was supported by the International Association for Cancer Research (to P.R.C. and B.V.), the European Molecular Biology Organization (to M.G.), the European Union Fifth Framework MAIN Consortium (to W.P.), and the Ludwig Institute for Cancer Research.
Abbreviations
- IS
internal standard
- LC
liquid chromatography
- p-Aktide
phospho-Aktide
- PI3K
phosphoinositide 3-kinase
- PKB
protein kinase B
- WM
wortmannin.
Footnotes
Conflict of interest statement: M.W. is a director of PIramed Ltd.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hennessy B. T., Smith D. L., Ram P. T., Lu Y., Mills G. B. Nat. Rev. Drug Discovery. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B., Kinzler K. W. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3.Bader A. G., Kang S., Zhao L., Vogt P. K. Nat. Rev. Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 4.Cully M., You H., Levine A. J., Mak T. W. Nat. Rev. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 5.Parsons D. W., Wang T. L., Samuels Y., Bardelli A., Cummins J. M., DeLong L., Silliman N., Ptak J., Szabo S., Willson J. K., et al. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- 6.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S. M., Riggins G. J., et al. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 7.Sansal I., Sellers W. R. J. Clin. Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 8.Sun M., Wang G., Paciga J. E., Feldman R. I., Yuan Z. Q., Ma X. L., Shelley S. A., Jove R., Tsichlis P. N., Nicosia S. V., et al. Am. J. Pathol. 2001;159:431–437. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S. I., Puc J., Miliaresis C., Rodgers L., McCombie R., et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 10.Liaw D., Marsh D. J., Li J., Dahia P. L., Wang S. I., Zheng Z., Bose S., Call K. M., Tsou H. C., Peacocke M., et al. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 11.Furnari F. B., Lin H., Huang H. S., Cavenee W. K. Proc. Natl. Acad. Sci. USA. 1997;94:12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson G. P., Furnari F. B., Miele M. E., Glendening M. J., Welch D. R., Fountain J. W., Lugo T. G., Huang H. J., Cavenee W. K. Proc. Natl. Acad. Sci. USA. 1998;95:9418–9423. doi: 10.1073/pnas.95.16.9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 14.Ali K., Bilancio A., Thomas M., Pearce W., Gilfillan A. M., Tkaczyk C., Kuehn N., Gray A., Giddings J., Peskett E., et al. Nature. 2004;431:1007–1011. doi: 10.1038/nature02991. [DOI] [PubMed] [Google Scholar]
- 15.Foukas F., Claret M., Pearce W., Okkenhaug K., Meek S., Peskett E., Sancho S., Smith A., Withers D., Vanhaesebroeck Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 16.Blagoev B., Ong S. E., Kratchmarova I., Mann M. Nat. Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 17.Cutillas P. R., Geering B., Waterfield M. D., Vanhaesebroeck B. Mol. Cell Proteomics. 2005;4:1038–1051. doi: 10.1074/mcp.M500078-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Ballif B. A., Roux P. P., Gerber S. A., Mackeigan J. P., Blenis J., Gygi S. P. Proc. Natl. Acad. Sci. USA. 2005;102:667–672. doi: 10.1073/pnas.0409143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. Proc. Natl. Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du K., Tsichlis P. N. Oncogene. 2005;24:7401–7409. doi: 10.1038/sj.onc.1209099. [DOI] [PubMed] [Google Scholar]
- 21.Gao H., Leary J. A. J. Am. Soc. Mass Spectrom. 2003;14:173–181. doi: 10.1016/S1044-0305(02)00867-X. [DOI] [PubMed] [Google Scholar]
- 22.Bozinovski S., Cristiano B. E., Marmy-Conus N., Pearson R. B. Anal. Biochem. 2002;305:32–39. doi: 10.1006/abio.2002.5659. [DOI] [PubMed] [Google Scholar]
- 23.Shults M. D., Janes K. A., Lauffenburger D. A., Imperiali B. Nat. Methods. 2005;2:277–283. doi: 10.1038/nmeth747. [DOI] [PubMed] [Google Scholar]
- 24.Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., Cohen P. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi T., Cohen P. Biochem. J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 26.Park J., Leong M. L., Buse P., Maiyar A. C., Firestone G. L., Hemmings B. A. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 28.Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M. A., Dick J. E. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 29.Aebersold R., Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 30.John H., Walden M., Schafer S., Genz S., Forssmann W. G. Anal. Bioanal. Chem. 2004;378:883–897. doi: 10.1007/s00216-003-2298-y. [DOI] [PubMed] [Google Scholar]
- 31.Steen H., Jebanathirajah J. A., Springer M., Kirschner M. W. Proc. Natl. Acad. Sci. USA. 2005;102:3948–3953. doi: 10.1073/pnas.0409536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang S., Bader A. G., Vogt P. K. Proc. Natl. Acad. Sci. USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuels Y., Diaz L. A., Jr, Schmidt-Kittler O., Cummins J. M., DeLong L., Cheong I., Rago C., Huso D. L., Lengauer C., Kinzler K. W., et al. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Rostan G., Costa A. M., Pereira-Castro I., Salvatore G., Hernandez R., Hermsem M. J., Herrero A., Fusco A., Cameselle-Teijeiro J., Santoro M. Cancer Res. 2005;65:10199–10207. doi: 10.1158/0008-5472.CAN-04-4259. [DOI] [PubMed] [Google Scholar]
- 35.Levine D. A., Bogomolniy F., Yee C. J., Lash A., Barakat R. R., Borgen P. I., Boyd J. Clin. Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 36.Velho S., Oliveira C., Ferreira A., Ferreira A. C., Suriano G., Schwartz S, Jr, Duval A., Carneiro F., Machado J. C., Hamelin R., et al. Eur. J. Cancer. 2005;41:1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 37.Bachman K. E., Argani P., Samuels Y., Silliman N., Ptak J., Szabo S., Konishi H., Karakas B., Blair B. G., Lin C., et al. Cancer Biol. Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 38.Sun M., Paciga J. E., Feldman R. I., Yuan Z., Coppola D., Lu Y. Y., Shelley S. A., Nicosia S. V., Cheng J. Q. Cancer Res. 2001;61:5985–5991. [PubMed] [Google Scholar]
- 39.She Q. B., Solit D. B., Ye Q., O’Reilly K. E., Lobo J., Rosen N. Cancer Cell. 2005;8:287–297. doi: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]