Abstract
The Rev1 protein lies at the root of mutagenesis in eukaryotes. Together with DNA polymerase ζ (Rev3/7), Rev1 function is required for the active introduction of the majority of mutations into the genomes of eukaryotes from yeast to humans. Rev1 and polymerase ζ are error-prone translesion DNA polymerases, but Rev1's DNA polymerase catalytic activity is not essential for mutagenesis. Rather, Rev1 is thought to contribute to mutagenesis principally by engaging in crucial protein–protein interactions that regulate the access of translesion DNA polymerases to the primer terminus. This inference is based on the requirement of the N-terminal BRCT (BRCA1 C-terminal) domain of Saccharomyces cerevisiae Rev1 for mutagenesis and the interaction of the C-terminal region of mammalian Rev1 with several other translesion DNA polymerases. Here, we report that S. cerevisiae Rev1 is subject to pronounced cell cycle control in which the levels of Rev1 protein are ≈50-fold higher in G2 and throughout mitosis than during G1 and much of S phase. Differential survival of a rev1Δ strain after UV irradiation at various points in the cell cycle indicates that this unanticipated regulation is physiologically relevant. This unexpected finding has important implications for the regulation of mutagenesis and challenges current models of error-prone lesion bypass as a process involving polymerase switching that operates mainly during S phase to rescue stalled replication forks.
Keywords: cell cycle, mutagenesis, translesion synthesis, DNA damage
The REV1 and REV3 genes of Saccharomyces cerevisiae were among the first genes known to be required for mutagenesis. Identified in 1971 in a screen for reversionless yeast strains (1), these genes play a central role in promoting mutagenesis from yeast to humans (2, 3). REV1 and REV3, together with REV7 (4), function in the “error-prone” branch of the RAD6 postreplication repair pathway (5). In contrast, RAD30, which shares homology with REV1, appears to function in parallel with REV1/3/7 in a separate “error-free” branch of the RAD6 epistasis group (5). After decades of genetic characterization, REV1, REV3/7, and RAD30 were shown to encode translesion DNA polymerases (6–9).
Rev1 possesses a unique enzymatic activity in vitro, displaying a marked preference for inserting only dCMP opposite a template G and several DNA lesions (6, 10, 11). The Rev3/7 heterodimer forms DNA polymerase ζ, which, although it is proposed to function mainly as an extender of mismatched primer termini (10), can also efficiently insert nucleotides across from lesions when stimulated by proliferating cell nuclear antigen (PCNA) (12). RAD30 encodes DNA polymerase η, which bypasses UV-induced lesions efficiently and accurately and, when mutated in humans, causes the cancer-prone syndrome xeroderma pigmentosum variant (13). Intriguingly, although Rev1's highly specialized catalytic activity has an effect on the spectrum of mutations generated (14, 15), its dCMP transferase activity is not required for its functions in induced mutagenesis or resistance to DNA damage (refs. 16 and 17 and unpublished data).
In contrast, Rev1's BRCT (BRCA1 C-terminal) domain is required for mutagenesis and resistance to DNA damaging agents in yeast (1), although it may be less important in higher eukaryotes (17, 18). BRCT domains mediate protein–protein interactions in many cell cycle and DNA repair proteins (19). Interestingly, the original loss-of-function rev1-1 mutant (1) carries a point mutation affecting the BRCT domain (2, 20). Because the purified Rev1-1 protein retains translesion synthesis (TLS) activity in vitro (21), whereas the rev1-1 mutant is nonmutable in vivo, the alteration of the BRCT domain is thought to disrupt key interactions.
In addition to the N-terminal BRCT domain and a central TLS polymerase domain, the Rev1 protein also contains a C-terminal region that, in mammalian cells, has been shown to interact with multiple other TLS polymerases (22–26). Rev1's C-terminal interaction region is required for resistance to DNA damaging agents in vertebrates (17) and in yeast (ref. 20 and L.S.W., S. D'Souza, and G.C.W., unpublished data). Additionally, the C terminus, as well as the BRCT and little finger domains, of yeast Rev1 were recently reported to interact with Rev7 (ref. 27 and S. D'Souza and G.C.W., unpublished data). Because Rev1's protein–protein interaction motifs are required for its function in vivo but its enzymatic activity is not, this enigmatic translesion polymerase is thought to play a predominantly structural role in assembling a TLS complex (16, 22, 25).
Polymerase switching during DNA replication has been proposed to be a fundamental mechanism by which cells control the action of TLS polymerases (28–30), all of which have low fidelity on undamaged DNA relative to replicative DNA polymerases (8, 31). Polymerase switching models suggest that when a replicative DNA polymerase stalls at a blocking lesion, a handoff allows one or more TLS polymerases access to the primer terminus, enabling lesion bypass and extension past the distortion. A further reciprocal switch would restore the highly processive and accurate replicative DNA polymerase to the primer terminus. Current models (13, 28) postulate that Rev1 plays a central role in the polymerase-switching mechanism during S phase to facilitate error-prone bypass of DNA lesions either itself by using its limited polymerase activity or by recruiting other TLS polymerases to bypass the lesion.
A recent report from Lopes et al. (32) shows that when yeast replication forks encounter a lesion, leading and lagging strand synthesis uncouple. Repriming events downstream of a DNA lesion then lead to persistent ssDNA gaps on both strands of the replication fork, which may remain throughout S phase into G2. Interestingly, deletion of all of the TLS polymerases did not further affect uncoupling or replication fork speed over damaged DNA; rather, it led to an increase in ssDNA gaps along replicated regions. These data strongly suggest that some component of TLS may occur behind replication forks and possibly postreplicatively outside of S phase.
We report here that Rev1 is expressed in a cell cycle-dependent manner and is highly up-regulated specifically during G2/M phase rather than during DNA replication in S phase. Rev1's G2/M expression pattern does not significantly change after DNA damage. Moreover, REV1 function is required for resistance to DNA damage differentially during the cell cycle. This finding suggests that Rev1-dependent TLS, and therefore much of mutagenesis, occurs to a significant extent, if not mostly, outside of S phase during G2/M.
Results
Rev1 Protein and mRNA Are Cell Cycle-Regulated and Reach Maximal Levels After Most Replication Is Completed.
To facilitate analysis of S. cerevisiae Rev1 regulation, a chromosomally located C-terminally tagged Rev1 construct was expressed from the native REV1 promoter. The tagged strain was indistinguishable from WT in its ability to survive DNA damage and to undergo mutagenesis (Fig. 5, which is published as supporting information on the PNAS web site).
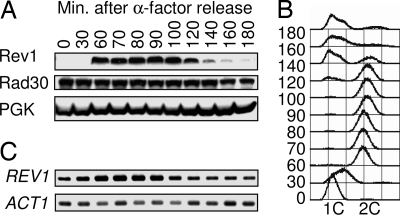
Our ability to visualize endogenous levels of Rev1 protein led to the unanticipated discovery that Rev1 is subject to pronounced cell cycle control (Fig. 1A). S. cerevisiae cells were arrested in G1 with α-factor, released, and allowed to proceed synchronously through the cell cycle. In α-factor arrested cells, Rev1 levels are almost undetectable. Surprisingly, Rev1 levels are very low in early S phase and rise only modestly as cells transit through S phase (also see Fig. 3). Substantial Rev1 accumulation occurs as most cells attain a G2 content of DNA (Fig. 1B), indicating that Rev1 levels do not peak as DNA is being synthesized; rather, the levels peak after most replication is completed. Using anti-tubulin immunofluorescence to monitor spindle length reveals that Rev1 is present at high levels as the chromosomes align during metaphase (Fig. 6A, which is published as supporting information on the PNAS web site) and is maintained at high levels even after most cells achieve fully extended spindles and completely separate their DNA masses (Fig. 6B). This observation implies that Rev1 is highly expressed throughout mitosis and that maximal protein levels are maintained until cells reenter G1. Levels of REV1 mRNA exhibit a pattern of cell cycle regulation similar to that of the protein, peaking slightly before the Rev1 protein levels in G2/M (Fig. 1C).
Fig. 1.
Rev1 is cell cycle-regulated and expressed maximally at G2/M phase. (A) Immunoblot against the protein A epitope shows Rev1 and Rad30 protein levels at indicated time points after release from G1 α-factor arrest. PGK (phosphoglycerate kinase) was used as a loading control. (B) FACS analysis of the DNA content of cells. (C) RT-PCR showing REV1 mRNA levels. ACT1 was used as a loading control.
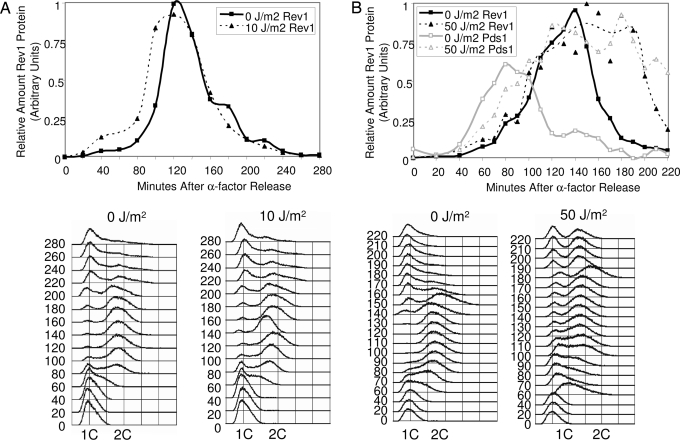
Fig. 3.
Cell cycle expression pattern of Rev1 is modestly altered after DNA damage. (A and B) Plot showing relative amount of Rev1 protein in arbitrary units as a function of cell cycle progression. G1 arrested cells were UV irradiated at 10 J/m2 (A) or 50 J/m2 (B) and released from α-factor block, and time points were taken as indicated. Immunoblots were quantitated and normalized to a standard dilution curve of Rev1 to allow comparison between blots. FACS data monitor cell cycle progression.
Peak levels of Rev1 protein in G2/M cells are ≈50-fold higher than the barely detectable Rev1 signal in G1 arrested cells (Fig. 6C), whereas we found only an ≈3-fold change between maximal and minimal levels of REV1 transcript (Fig. 6D). Thus, the cell cycle control of Rev1 levels is primarily posttranscriptional. The observed cell cycle regulation is not an α-factor-specific effect; cells synchronized by elutriation exhibit a similar pattern of Rev1 expression (Fig. 7, which is published as supporting information on the PNAS web site). An identically tagged Rad30 shows no change during the cell cycle (Fig. 1A), indicating that this type of cell cycle control is not a general property of all TLS polymerases.
Rev1 Protein Is Stably Present Throughout Mitosis.
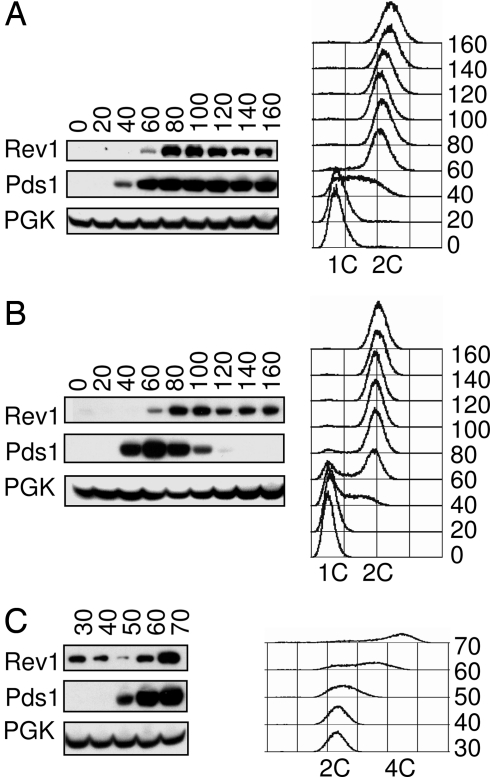
To analyze the timing of Rev1 accumulation more precisely, cdc23-1 and cdc15-2 temperature-sensitive strains were used to arrest cells at the metaphase-to-anaphase transition and at telophase, respectively (33, 34). Pds1 (securin) was used as a marker for cell cycle progression, because it is synthesized during S phase and is degraded at the metaphase-to-anaphase transition (35). Cells were synchronized in G1 with α-factor and then, upon removal of α-factor, shifted to the restrictive temperature to induce the second cell cycle arrest. We found that Rev1 levels do not rise until after Pds1 has accumulated during S phase, again demonstrating that Rev1 levels are low during much of DNA replication. Furthermore, Rev1 is present at high levels in cdc23-1 metaphase arrested cells (Fig. 2A), indicating that Rev1 accumulation begins during G2 before metaphase. Even more interestingly, Rev1 is also stable in cdc15-2 telophase arrested cells (Fig. 2B). The cdc15-2 allele produces a very late arrest in the cell cycle during exit from mitosis, just before reentry into G1 (34). After release from the cdc15-2 telophase block, Rev1 levels decrease as cells reenter G1 (Fig. 2C). Therefore, contrary to prevailing expectations for a TLS polymerase, we demonstrate that Rev1 is maximally present after the majority of DNA replication is finished, remains throughout all of mitosis, and is present even during exit from mitosis while cells reset for G1.
Fig. 2.
Rev1 protein is stable in both metaphase and telophase arrested cells. (A and B) Immunoblot showing Rev1-ProA and Pds1-HA in cdc23-1 (A) or cdc15-2 (B) arrested cells. Time points were taken every 20 min after release from α-factor and shift to the restrictive temperature. (C) Immunoblot showing Rev1 and Pds1 levels at the indicated times after release from the cdc15-2 block. Cells released from a cdc15-2 arrest fail to separate because of a cytokinesis defect and generate a 4C peak on FACS that is indicative of a second round of DNA replication.
DNA Damage Does Not Significantly Alter Rev1's Expression Pattern.
Taken together, these observations suggest that, in undamaged cells, the major physiological role of Rev1 in spontaneous mutagenesis occurs predominantly in G2/M. We wondered, however, whether exogenous DNA damage would significantly alter Rev1 expression so that it would accumulate mainly during S phase when the replication machinery would be actively encountering lesions. Because Rev1 is required for bypass of the 6-4 photoproduct induced by UV irradiation (21), we irradiated cells arrested in G1 and followed Rev1 levels through the cell cycle after DNA damage. Doses of UV irradiation of 10 J/m2 and 50 J/m2 resulted in ≈100% and 60% survival, respectively, of the tagged Rev1 strain and ≈75% and 1% survival, respectively, of an isogenic rev1Δ strain. We found that DNA damage did not result in a radical alteration of the overall pattern of Rev1 expression (Fig. 3). Despite the fact that replication forks would have encountered UV-induced lesions from the beginning of S phase, Rev1 levels were not dramatically increased early in S phase relative to an unirradiated strain. As observed with undamaged cells, Rev1 accumulated slowly through S phase, only reaching its peak when most of the cells were in G2.
Some changes in the timing of Rev1 accumulation, however, were discernable. After 10 J/m2 UV irradiation, low levels of Rev1 were still found in early S phase but began increasing slightly earlier to achieve higher levels during late S than in the absence of UV damage (Fig. 3A). After 50 J/m2 UV irradiation, this shift in Rev1 accumulation became more pronounced (Fig. 3B). The cells proceeded more slowly through the cell cycle after significant amounts of DNA damage, so direct comparisons of time courses by minutes after release do not reflect cell cycle stage. Despite this moderate shift in timing of Rev1 accumulation after substantial DNA damage, Rev1 protein is not present at high levels throughout S phase, as would be expected for a replication protein or an S phase repair protein. Instead, at high doses of UV irradiation, Rev1 accumulation appears to track slightly after the metaphase protein Pds1 (Fig. 3B). Additionally, as with undamaged cells, after UV irradiation, Rev1 still appears to persist into G2/M phase as the cells complete replication and enter mitosis.
REV1 Function Is Required Differentially During the Cell Cycle.
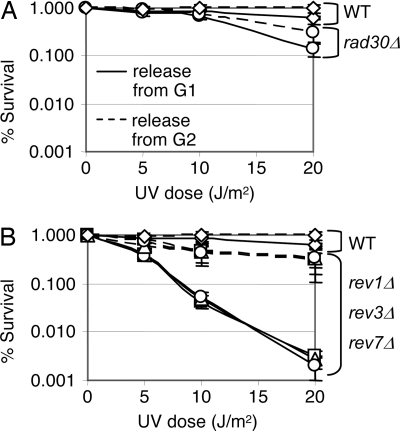
To analyze the possible biological significance of the observed Rev1 cell cycle regulation, we monitored survival after UV irradiation at different cell cycle stages. Cells were arrested in G1 with α-factor or in G2 with nocodazole, washed to remove the drugs, plated, and immediately UV irradiated. The WT strain was only slightly more sensitive to killing when it was UV irradiated just after release from G1 than when it was UV irradiated just after release from G2 (Fig. 4A), in agreement with previous reports (36). The mild sensitivity of a rad30Δ strain to killing by UV irradiation was likewise largely unaffected by the cell cycle stage during which the UV irradiation occurred (Fig. 4A), consistent with the observation that the Rad30 protein is constitutively expressed throughout the cell cycle (Fig. 1A).
Fig. 4.
REV1, REV3, and REV7 are required differentially throughout the cell cycle for survival after UV irradiation. (A) Percent survival of the WT (diamonds) and rad30Δ (circles) strains after release from G2 (dashed line) or G1 (solid line) arrests. (B) Percent survival of the WT (diamonds), rev1Δ (squares), rev3Δ (triangles), and rev7Δ (circles) strains after release from G2 (dashed line) or G1 (solid line) arrests. Note that rev1Δ, rev3Δ, and rev7Δ strains exhibit such similar survival that the strains can hardly be distinguished from each other.
In striking contrast, the rev1Δ strain was markedly more UV sensitive when irradiated after release from G1 than when irradiated after release from G2 (Fig. 4B), showing clearly that REV1 function is required differentially throughout the cell cycle. Because Rev1 is largely absent in G1 and present in G2, a plausible explanation is that irradiation after release from G1 results in replication forks encountering DNA lesions before they can be completely repaired by nucleotide excision repair (NER), causing leading and lagging strand uncoupling and repriming events downstream that lead to the generation of ssDNA gaps at lesions (32). Such ssDNA gaps would require TLS. In contrast, irradiation in G2 would allow a more prolonged period for NER before DNA replication, thereby reducing ssDNA gap formation and hence the need for TLS. Consistent with this explanation, microscopic examination of the plates revealed that, when irradiated with 20 J/m2 UV irradiation after release from G1, rev1Δ cells arrest predominantly as budded cells (data not shown). Arrest at this point indicates that the lethal event after UV irradiation in rev1Δ cells occurs after replication, rather than in G1 or at a random point several generations later.
Interestingly, although the protein levels of Rev3 and Rev7 do not vary during the cell cycle (S. D'Souza and G.C.W., unpublished data), the rev3Δ and rev7Δ strains display the same hypersensitivity to UV irradiation when irradiated after release from G1 as the rev1Δ strain (Fig. 4B). The striking similarity of the pattern of cell cycle-dependent UV sensitivity indicates that Rev1's cell cycle regulation is used to control the activity of DNA polymerase ζ (Rev3/7) during the cell cycle.
Discussion
We report here that in S. cerevisiae, Rev1 protein levels are dramatically cell cycle-regulated, being at least 50-fold higher in G2/M than in G1 and much of S phase. The remarkable dependence of the UV sensitivity of a rev1Δ mutant, but not a rad30Δ mutant, to the cell cycle stage in which UV irradiation occurs indicates that the cell cycle regulation of Rev1 is of major biological significance. Because Rev1 and polymerase ζ are required for ≈98% of the mutagenic events in a cell (10), the cell cycle regulation of Rev1 has profound implications for when mutagenesis takes place during the cell cycle.
We show that the amount of Rev1 protein is extremely low during G1 and rises slowly throughout early and mid-S phase. Rev1 levels only begin to increase rapidly in late S phase, reaching maximum levels in G2. The Rev1 protein is then maintained at a high intracellular concentration throughout mitosis until after telophase. DNA damage causes Rev1 to accumulate somewhat earlier in late S phase without significantly affecting the level reached in G2/M phase, but does not convert Rev1's expression pattern into that of a canonical replication protein, such as PCNA or a replicative DNA polymerase (37, 38). The observed pattern of cell cycle-dependent expression was initially surprising, given current models postulating that polymerase switching allows TLS to restart stalled replication forks during S phase (13, 28–30). In contrast, our unexpected finding that Rev1 is cell cycle-regulated with maximal expression during G2/M phase suggests that Rev1 acts predominantly in G2/M rather than during the active phase of DNA replication in S phase. This finding is consistent with the report from Lopes et al. (32), which challenges the assumption that the polymerase switch event occurs solely at blocked replication forks in S phase. We propose that Rev1 acts postreplicatively during G2 phase, and even during M phase, by carrying out its well established roles in mutagenic TLS. During this process, Rev1 could function as a DNA polymerase and also recruit other TLS polymerases to fill the ssDNA gaps that are left behind as a consequence of replication forks encountering lesions.
A rev1Δ strain is differentially sensitive to UV irradiation during the cell cycle, demonstrating that REV1 functions in a cell cycle-dependent manner. In yeast, DNA polymerase ζ (Rev3/7) (10, 27) does not display cell cycle-regulated protein levels (S. D'Souza and G.C.W., unpublished data), nor does the related Y family translesion DNA polymerase η (Rad30) (Fig. 1A). A rad30Δ strain showed no cell cycle dependence in its sensitivity to UV damage beyond that of the WT strain. However, the rev3Δ and rev7Δ strains were indistinguishable from the rev1Δ strain in their responses to UV damage after release from G1 or G2 arrests. Therefore, although the cell cycle regulation exhibited by the Rev1 protein appears to be unique among TLS polymerases, it is likely used to control the Rev1/3/7-dependent error-prone mode of TLS. Additionally, given that DNA polymerase ζ-dependent crosslink repair also shows cell cycle dependence (39), Rev1's cell cycle regulation may be used to coordinate the responses of other damage tolerance pathways as well.
This report provides direct evidence for cell cycle regulation of Rev1; other recent results are consistent with Rev1 and its partners Rev3/7 acting late in the cell cycle. For example, Rev1 functions in preventing chromosomal breaks in mouse ES cells and transformed chicken DT40 cells in late S/G2 phase (18, 40, 41). Similar to our observation with UV irradiation, rev3Δ cells progress through S phase normally but arrest permanently in G2 after cisplatin treatment (42). Analogous results have been observed in mouse and chicken cells with Rev1 BRCT−/− and Rev3−/− deficient lines (18, 41, 43). Our discovery that Rev1 levels are highest in G2, after a sister chromatid has been generated, is also consistent with the growing evidence for REV1/3/7 involvement in certain aspects of homologous recombination (HR) (40). Evidence consistent with Rev1 and Rev3/7 contributing to the processing of double-strand breaks during HR in meiosis includes the observations that, in yeast, each of the REV genes is up-regulated during sporulation (44–46) and that, in mammals, REV1 and all of the other TLS polymerases are at high levels in the testes (47). Further supporting an involvement in facets of HR repair, REV3 is required for the break repair-induced mutagenesis observed during double-strand break repair (48, 49). However, our demonstration that Rev1 persists until well after anaphase and sister chromatid separation suggests that, beyond any contribution to HR, Rev1 may play a role during mitosis after sister chromatids are physically separated and unable to synapse. These data, together with the observation that hREV7 (hMAD2B) inhibits the metaphase-to-anaphase transition through the spindle checkpoint in Xenopus extracts (50, 51), strongly indicate that Rev1/3/7 play a major role at the end of DNA replication and throughout mitosis.
Is it reasonable that the majority of Rev1-dependent mutagenic TLS could occur after most DNA replication is completed and extend throughout mitosis? The inhibition of many polymerases by DNA lesions in in vitro studies employing primed single-strand templates has contributed to a widespread impression that real replication forks can be similarly stalled by a single lesion. However, in vivo in both eukaryotes and prokaryotes, replication forks uncouple leading and lagging strand synthesis when they encounter lesions and leave gaps in their wake (32, 52–54), which may persist as cells enter G2 phase. The recent results of Lopes et al. (32) show that TLS defective S. cerevisiae cells do not further uncouple leading and lagging strands but have an increase in ssDNA gaps, consistent with the idea that TLS may occur behind the replication fork and even after bulk replication has been completed. Interestingly, after DNA damage, E. coli seems to delay mutagenic TLS by using the kinetics of the SOS-regulated UmuD → UmuD′ transition to impose a phase of largely accurate DNA repair and tolerance followed by a phase of error-prone lesion bypass (55). Restricting Rev1 to the latter part of the cell cycle may be a conceptually similar strategy to reserve REV1/3/7-dependent mutagenic TLS until after high-fidelity repair or damage-tolerance mechanisms have been attempted (Fig. 8A, which is published as supporting information on the PNAS web site).
In our model, the major site of Rev1 action is at inappropriate primer termini remaining in G2/M at gaps caused by lesions (Fig. 8B). A persistent gap in G2 may be recognized by Rev1 by its ability as a polymerase to bind primer termini or by using its BRCT domain, given that some BRCT domains can bind DNA, particularly single- or double-stranded breaks (56, 57). Therefore, the rev1-1 mutation might also inactivate Rev1's localization to aberrant DNA structures rather than exclusively disrupting a protein–protein complex. Additionally, because it is possible that modified forms of PCNA may persist at ssDNA gaps and serve as a marker for repair activities, Rev1 may recognize a ssDNA gap remaining in G2 by binding to a monoubiquitinylated PCNA through its UBM ubiquitin-interacting domains in a manner analogous to DNA polymerase ι in mammalian cells (58). Interaction with monoubiquitinylated PCNA also stimulates Rev1's catalytic activity (59). Once at the lesion, Rev1 may facilitate tolerance and gap-filling either by using its own dCMP transferase activity or by recruiting other TLS polymerases through its C-terminal region (22–26). Rev1 may also interact with other DNA repair or damage checkpoint signaling factors [for example, by using its BRCT domain to form a complex with other BRCT-containing proteins or indirectly through PCNA (17) or the alternative clamp 9-1-1 (60)]. Once a gap has been filled and the lesion has been bypassed, excision repair machinery could then be recruited by Rev1 to remove the lesion before the start of the next cell cycle.
We cannot exclude some contribution of Rev1 during S phase, as the low levels we observe during DNA replication may be sufficient for at least some TLS. However, the levels of Rev1 during S phase are likely significantly lower than those of replicative DNA polymerases, perhaps 10-fold or more. Asynchronous yeast cultures contain only ≈500 Rev1 molecules per cell, the majority of which are presumably due to the G2/M cells in the population, compared with ≈2,000 molecules per cell for Rad30 or the replicative polymerases (61). Furthermore, Rev1 and Rev3 are also thought to be present at a very low cellular concentration in higher eukaryotes (2, 3, 17, 62). This low level of Rev1, coupled with the cell cycle regulation we have observed, suggests that caution should be used in interpreting studies in which Rev1 is overexpressed (22, 63). The finding that overexpressed Rev1 localizes to replication forks may provide a rationale for why cells keep the amount of Rev1 low during S phase; if Rev1 were present at high levels, it might be recruited inappropriately to replication forks when not needed with mutagenic or lethal consequences. During S phase, relatively accurate TLS at stalled replication forks may be accomplished by TLS polymerases such as polymerase η [Rad30/XPV (xeroderma pigmentosum variant] recruited by monoubiquitinylated PCNA (13). In contrast, we suggest that Rev1 acts mostly outside of S phase, coordinating a more mutagenic usage of TLS polymerases later in the cell cycle.
Recently, a report appeared that showed that ectopically overexpressed hRev1 formed foci in S phase as well as in G1 (64). Although focus formation is frequently interpreted as indicating the site of a protein's major function, in this case, the most biologically significant action of Rev1 might not manifest itself as a focus. Whereas recruitment of many molecules of Rev1 to a replication factory or repair center would likely generate a focus, it is not clear that recruitment of Rev1 to multiple ssDNA gaps spaced out along replicated DNA would result in a high local concentration of Rev1.
Because most aspects of cell cycle control are shared between yeast and mammals, Rev1's cell cycle regulation may have general implications for TLS-dependent mutagenesis. Our results suggest that cells delay potentially mutagenic TLS until later in the cell cycle as a strategy for minimizing the mutagenic effects of DNA damage. In the environment, S. cerevisiae and other microorganisms likely spend most of their life in a quiescent, nonproliferating state. Most cells in higher eukaryotes are terminally differentiated and have withdrawn from the cell cycle. Thus, restricting Rev1 protein expression to G2/M may reflect a cellular mechanism for reducing mutagenesis in resting cells.
Materials and Methods
Strains.
Strains used were derivatives of W1588-4C, a W303 strain corrected for RAD5 (65). pYM10 was used to generate a C-terminal −TEV-ProA-7His tag (66). Strain information is available on request.
Immunoblots.
Whole-cell extracts were prepared by trichloroacetic acid precipitation (66). Antibodies used were rabbit PAP antibody (Sigma) against the protein A tag, anti-HA.11 (Covance, Richmond, CA), and anti-phosphoglycerate kinase (Molecular Probes). Quantitation was performed by using the Typhoon 9400 (General Electric) and imagequant software. Plots were generated by averaging two to four replicate immunoblots.
Flow Cytometry.
Cells were prepared essentially as described in ref. 67 and analyzed on a Becton Dickinson FACSCalibur flow cytometer.
Cell Synchronization.
Logarithmically growing bar1Δ yeast were arrested with 50 ng/ml α-factor for 4 h at 25°C and washed to remove α-factor. In Fig. 1, cells were resuspended in 25°C media, and α-factor was added back after 75 min. In Fig. 2, cells were resuspended in media prewarmed to 37°C. After 3 h at 37°C, cells were released from the cdc15-2 arrest by harvesting and resuspending cells in 25°C media. In Fig. 3, cells were resuspended in 20 ml of water, transferred to a 150 × 15-mm Petri plate, and irradiated. Aliquots were assayed for viability. Cells were diluted in 20 ml of 2× media to start the time course. In Fig. 3A, α-factor was added back after 90 min; in Fig. 3B, it was added after 80 min (0 J/m2 UV irradiation) or 100 min (50 J/m2 UV irradiation).
UV Survival Assay.
At least three independent cultures of each strain were arrested with 50 ng/ml α-factor or 15 μg/ml nocodazole for 3 h at 30°C and washed with water or 1% DMSO in yeast extract/peptone media to remove α-factor or nocodazole, respectively. Microscopic analysis of cells confirmed arrest. Cells were diluted appropriately in water, plated on synthetic complete media, immediately UV irradiated at 1 J/m2 per s by using a G15T8 UV lamp (General Electric) at 254 nm, and incubated for 3 days at 30°C in the dark. For further details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank A. Amon, S. Bell, H. Blitzblau, and A. Marston (all from Massachusetts Institute of Technology) for generous gifts of strains, protocols, and use of equipment; A. Amon, P. Beuning, S. Cohen, S. D'Souza, D. Jarosz, B. Minesinger, S. Simon, L. Simmons, F. Solomon, and R. Woodruff for critical reading of the manuscript; and A. Marston and L. Simmons for microscopy assistance. This work was supported by an American Cancer Society Research Professorship (to G.C.W.).
Abbreviations
- BRCT
BRCA1 C-terminal
- PCNA
proliferating cell nuclear antigen
- TLS
translesion synthesis.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lemontt J. F. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibbs P. E., Wang X. D., Li Z., McManus T. P., McGregor W. G., Lawrence C. W., Maher V. M. Proc. Natl. Acad. Sci. USA. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibbs P. E., McGregor W. G., Maher V. M., Nisson P., Lawrence C. W. Proc. Natl. Acad. Sci. USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrence C. W., Das G., Christensen R. B. Mol. Gen. Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 5.Kunz B. A., Straffon A. F., Vonarx E. J. Mutat. Res. 2000;451:169–185. doi: 10.1016/s0027-5107(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 6.Nelson J. R., Lawrence C. W., Hinkle D. C. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 7.Nelson J. R., Lawrence C. W., Hinkle D. C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 8.Goodman M. F. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 9.Johnson R. E., Prakash S., Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence C. W. Adv. Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 11.Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 12.Garg P., Stith C. M., Majka J., Burgers P. M. J. Biol. Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann A. R. FEBS Lett. 2005;579:873–876. doi: 10.1016/j.febslet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 14.Otsuka C., Kunitomi N., Iwai S., Loakes D., Negishi K. Mutat. Res. 2005;578:79–87. doi: 10.1016/j.mrfmmm.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Ross A. L., Sale J. E. Mol. Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Haracska L., Unk I., Johnson R. E., Johansson E., Burgers P. M., Prakash S., Prakash L. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross A. L., Simpson L. J., Sale J. E. Nucleic Acids Res. 2005;33:1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen J. G., Tsaalbi-Shtylik A., Langerak P., Calleja F., Meijers C. M., Jacobs H., de Wind N. Nucleic Acids Res. 2005;33:356–365. doi: 10.1093/nar/gki189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glover J. N., Williams R. S., Lee M. S. Trends Biochem. Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Larimer F. W., Perry J. R., Hardigree A. A. J. Bacteriol. 1989;171:230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson J. R., Gibbs P. E., Nowicka A. M., Hinkle D. C., Lawrence C. W. Mol. Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 22.Tissier A., Kannouche P., Reck M. P., Lehmann A. R., Fuchs R. P., Cordonnier A. DNA Repair (Amsterdam) 2004;3:1503–1514. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Ohashi E., Murakumo Y., Kanjo N., Akagi J., Masutani C., Hanaoka F., Ohmori H. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi R., Oshige M., Uchida M., Ishikawa G., Takata K., Shimanouchi K., Kanai Y., Ruike T., Morioka H., Sakaguchi K. Biochem. J. 2004;382:535–543. doi: 10.1042/BJ20031833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo C., Fischhaber P. L., Luk-Paszyc M. J., Masuda Y., Zhou J., Kamiya K., Kisker C., Friedberg E. C. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakumo Y., Ogura Y., Ishii H., Numata S., Ichihara M., Croce C. M., Fishel R., Takahashi M. J. Biol. Chem. 2001;276:35644–35651. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 27.Acharya N., Haracska L., Johnson R. E., Unk I., Prakash S., Prakash L. Mol. Cell. Biol. 2005;25:9734–9740. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedberg E. C., Lehmann A. R., Fuchs R. P. Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 29.Plosky B. S., Woodgate R. Curr. Opin. Genet. Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Prakash S., Prakash L. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 31.Kunkel T. A., Pavlov Y. I., Bebenek K. DNA Repair (Amsterdam) 2003;2:135–149. doi: 10.1016/s1568-7864(02)00224-0. [DOI] [PubMed] [Google Scholar]
- 32.Lopes M., Foiani M., Sogo J. M. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Irniger S., Piatti S., Michaelis C., Nasmyth K. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 34.Amon A. Methods Enzymol. 2002;351:457–467. doi: 10.1016/s0076-6879(02)51864-4. [DOI] [PubMed] [Google Scholar]
- 35.Zachariae W., Nasmyth K. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 36.Siede W., Friedberg E. C. Mutat. Res. 1990;245:287–292. doi: 10.1016/0165-7992(90)90158-g. [DOI] [PubMed] [Google Scholar]
- 37.Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., Eisen M. B., Brown P. O., Botstein D., Futcher B. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falconi M. M., Piseri A., Ferrari M., Lucchini G., Plevani P., Foiani M. Proc. Natl. Acad. Sci. USA. 1993;90:10519–10523. doi: 10.1073/pnas.90.22.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar S., Davies A. A., Ulrich H. D., McHugh P. J. EMBO J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okada T., Sonoda E., Yoshimura M., Kawano Y., Saya H., Kohzaki M., Takeda S. Mol. Cell. Biol. 2005;25:6103–6111. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson L. J., Sale J. E. EMBO J. 2003;22:1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grossmann K. F., Ward A. M., Moses R. E. Mutat. Res. 2000;461:1–13. doi: 10.1016/s0921-8777(00)00035-5. [DOI] [PubMed] [Google Scholar]
- 43.Zander L., Bemark M. DNA Repair (Amsterdam) 2004;3:743–752. doi: 10.1016/j.dnarep.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 44.Burns N., Grimwade B., Ross-Macdonald P. B., Choi E. Y., Finberg K., Roeder G. S., Snyder M. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 45.Singhal R. K., Hinkle D. C., Lawrence C. W. Mol. Gen. Genet. 1992;236:17–24. doi: 10.1007/BF00279638. [DOI] [PubMed] [Google Scholar]
- 46.Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P. O., Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 47.Laan R., Baarends W. M., Wassenaar E., Roest H. P., Hoeijmakers J. H., Grootegoed J. A. Int. J. Androl. 2005;28:1–15. doi: 10.1111/j.1365-2605.2004.00505.x. [DOI] [PubMed] [Google Scholar]
- 48.Holbeck S. L., Strathern J. N. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rattray A. J., Shafer B. K., McGill C. B., Strathern J. N. Genetics. 2002;162:1063–1077. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfleger C. M., Salic A., Lee E., Kirschner M. W. Genes Dev. 2001;15:1759–1764. doi: 10.1101/gad.897901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J., Fang G. Genes Dev. 2001;15:1765–1770. doi: 10.1101/gad.898701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pages V., Fuchs R. P. Science. 2003;300:1300–1303. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- 53.Cordeiro-Stone M., Schumacher R. I., Meneghini R. Biophys. J. 1979;27:287–300. doi: 10.1016/S0006-3495(79)85218-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rupp W. D., Howard-Flanders P. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 55.Opperman T., Murli S., Smith B. T., Walker G. C. Proc. Natl. Acad. Sci. USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamane K., Tsuruo T. Oncogene. 1999;18:5194–5203. doi: 10.1038/sj.onc.1202922. [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson A., Smith A., Bullard D., Lavesa-Curto M., Sayer H., Bonner A., Hemmings A., Bowater R. Biochim. Biophys. Acta. 2005;1749:113–122. doi: 10.1016/j.bbapap.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 58.Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., et al. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 59.Garg P., Burgers P. M. Proc. Natl. Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabbioneda S., Minesinger B. K., Giannattasio M., Plevani P., Muzi-Falconi M., Jinks-Robertson S. J. Biol. Chem. 2005;280:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- 61.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 62.Lin W., Xin H., Zhang Y., Wu X., Yuan F., Wang Z. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mukhopadhyay S., Clark D. R., Watson N. B., Zacharias W., McGregor W. G. Nucleic Acids Res. 2004;32:5820–5826. doi: 10.1093/nar/gkh903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murakumo Y., Mizutani S., Yamaguchi M., Ichihara M., Takahashi M. Genes Cells. 2006;11:193–205. doi: 10.1111/j.1365-2443.2006.00938.x. [DOI] [PubMed] [Google Scholar]
- 65.Zhao X., Muller E. G., Rothstein R. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 66.Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 67.Lau A., Blitzblau H., Bell S. P. Genes Dev. 2002;16:2935–2945. doi: 10.1101/gad.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.