Abstract
The drive toward more sensitive single-molecule manipulation techniques has led to the recent development of optical tweezers capable of resolving the motions of biological systems at the subnanometer level, approaching the fundamental limit set by Brownian fluctuations. One successful approach has been the dual-trap optical tweezers, in which the system of study is held at both ends by microspheres in two separate optical traps. We present here a theoretical description of the Brownian limit on the spatial resolution of such systems and verify these predictions by direct measurement in a Brownian noise-limited dual-trap optical tweezers. We find that by detecting the positions of both trapped microspheres, correlations in their motions can be exploited to maximize the resolving power of the instrument. Remarkably, we show that the spatial resolution of dual optical traps with dual-trap detection is always superior to that of more traditional, single-trap designs, despite the added Brownian noise of the second trapped microsphere.
Keywords: single molecule, subnanometer resolution, signal-to-noise ratio
Since the discovery that optical gradients could stably trap micrometer-sized dielectric particles (1), gradient optical traps, or optical tweezers, have been used in a large variety of applications ranging from microfabrication (2, 3) to the study of colloidal hydrodynamics (4–6) and nonequilibrium thermodynamics (7–10). In particular, the use of calibrated optical springs has provided unprecedented new insight on the mechanical properties of single biological molecules and the molecular motors that manipulate them in the cell (11–14). In a typical single-molecule experiment, a biopolymer is linked to a single, optically trapped dielectric microsphere at one end and to a fixed surface at the other. The length of this molecule is inferred from the force applied by the optical trap and the position of the microsphere relative to the fixed end of the polymer. In an ideal system with no measurement error, the ability to resolve changes in the length of the polymer, the spatial resolution of the optical tweezers, is limited only by the stochastic Brownian force induced on the microsphere by the surrounding solvent. This limit has been estimated previously (15, 16) and implies that it should be possible to resolve length changes on the angstrom scale with reasonable values of the experimental parameters, i.e., microsphere size, polymer stiffness, and averaging bandwidth. However, it has proven difficult to sufficiently isolate single-trap optical tweezers from environmental and instrumental sources of noise to reach this Brownian noise limit. Drift and low frequency fluctuations of the fixed surface relative to the optical trap typically obscure these small movements.
One method to increase the stability of optical tweezers is to introduce a second optical trap to hold the other end of the system, thereby decoupling it from movements of the fixed surface. Although such dual-trap optical tweezers were first envisioned more than 10 years ago (17, 18), it has not been until recently that the improvement in stability and isolation has been shown (19). As a demonstration of the possibility of such instruments, Abbondanzieri et al. (20) have observed the individual movements of RNA polymerase on dsDNA, ≈3.4 Å, using an ultrastable dual-trap optical tweezers in which the position of one microsphere in a weak trap is monitored and another microsphere in a second, very stiff trap replaces the fixed surface. Alternatively, the second trap in a dual-trap optical tweezers need not be a static attachment point. Meiners and Quake (21) monitor the motion of both microspheres in traps of equal stiffness and measure the femtoNewton dynamics of dsDNA by exploiting the correlations induced into the motion of the two microspheres. Such dynamic correlations allow measurement of some of the lowest forces to date despite the much larger force noise present in the individual motion of each microsphere.
In this paper, we demonstrate that these correlations can be exploited to improve the spatial resolution of optical tweezers. By solving the equations of motion of two microspheres of arbitrary size linked by a flexible polymer held in traps of arbitrary strength, we predict the ability to resolve equilibrium changes in the polymer length based on the motions of each microsphere or from linear combinations of these motions. We find that a coordinate that exploits the correlations between the microspheres provides an increase in spatial resolution over the movements of either microsphere separately. Furthermore, we find that with the proper choice of coordinate, the spatial resolution achievable with dual optical traps is always superior to that of a single trap. Finally, we verify these predictions with a Brownian noise-limited dual-trap optical tweezers.
Results
The Brownian Fluctuations of Two Coupled Microspheres.
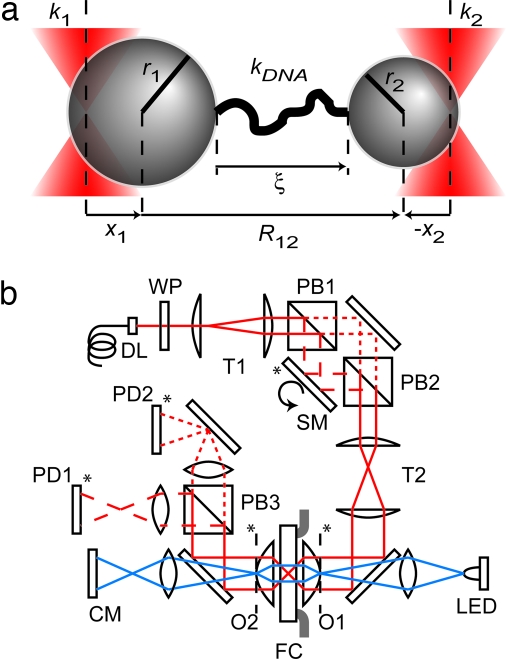
In this analysis, we describe the dynamics of two optically trapped microspheres attached via a single flexible polymer of contour length L, and held in a solvent bath at temperature T. In the interest of simplicity, we consider only displacements of the microspheres from equilibrium, x1 and x2, along a direction parallel to the tether and ignore orthogonal degrees of freedom, including Brownian rotation of each microsphere. Along this direction, we approximate the optical potential seen by each microsphere as harmonic, with spring constants k1 and k2. For our purposes, we assume that the polymer tether is dsDNA, although our analysis remains applicable for other choices of tether molecules. In the limit of small displacements, we treat the DNA as a linear spring with constant kDNA(ξeq) = dFDNA/dξ∣ξeq, where we expand its force-extension behavior (22–24) about the equilibrium extension ξeq. The drag coefficient of each microsphere of radius r1,2, in the absence of the other microsphere, is given by γ1,2 = 6πηr1,2, where η is the viscosity of the solvent (9 × 10−10 pNs/nm2 for water). The small frequency dependence of this drag coefficient (25, 26) and any modification due to nearby surfaces (26) are neglected (see instrument description below); however, the hydrodynamic modifications due to the proximity of the second microsphere can provide large corrections (4, 27) and will be included. The small effects of inertial terms, the finite relaxation time of the polymer tether, and its hydrodynamic interaction with the microspheres also are neglected. See Fig. 1a for a summary of this geometry.
Fig. 1.
Experimental configuration. (a) DNA–microsphere dumbbell geometry. Two microspheres of radii r1 and r2 tethered by a molecule of dsDNA of stiffness kDNA are held in separate optical traps of stiffnesses k1 and k2. At a given tension, the molecule is extended by ξ, and the microspheres are displaced from the center of each trap by x1 and x2 and separated by a distance R12. (b) Schematic layout of the dual-trap optical tweezers. The linearly polarized light of a 200-mW, 845-nm fiber-coupled laser (DL) (Lumics, Berlin) is magnified and collimated by the first telescope (T1). This beam is split by polarization at the first polarizing beam splitter (PB1). A computer-controlled, motorized half waveplate (WP) allows remote control of the relative power between the beams. The s-polarized beam is deflected by a two-axis piezo-steerable mirror (SM) (Mad City Labs, Madison, WI), whereas the p-polarized beam is deflected by a fixed mirror, and the two beams are then recombined at the second polarizing beam splitter (PB2). A second telescope (T2) provides the final magnification of the beams and images the plane (∗) of the steerable mirror onto the back focal plane of a 1.2 numerical aperture 60× water immersion objective (O1) (Nikon, Melville, NY). The objective focuses the beams to two diffraction-limited spots in the center of a 200-μm-thick fluidics chamber (FC). A second, identical objective (O2) collects the forward scattered light, which is split again by polarization at a third polarizing beam splitter (PB3) and imaged onto two separate position-sensitive photodetectors (PD1 and PD2) (Pacific Silicon Detectors, Westlake Village, CA). A light-emitting diode provides light for Kohler illumination, and the second objective and an additional tube lens image the specimen plane onto a charge-coupled device camera (CM).
In this model, the dynamics of the microspheres are governed by the balance between viscous, elastic, and stochastic Brownian forces and are described by a set of linear, coupled Langevin equations:
where
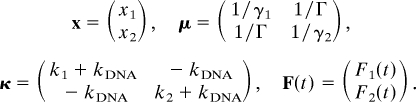 |
Off-diagonal terms in the stiffness matrix, κ, describe the coupling between the microspheres due to the DNA tether, whereas off-diagonal terms in the mobility matrix, μ, describe the hydrodynamic coupling of one microsphere with the fluid flow generated by movements of the other. This dynamic coupling is generally described by the Oseen tensor, which is approximately constant when the distance between the microspheres, R12, is much greater than their positional fluctuations (4) and the time it takes the fluid to propagate between them is sufficiently short (5). Furthermore, when R12 is much greater than the radii of the microspheres, Γ ≈ 4πηR12 (4). Finally, the fluctuating Brownian forces, F(t), have zero mean and are uncorrelated in time:
where ⊗ denotes the outer product of the force vectors and kB is the Boltzmann constant.
The fluctuations in x(t) are described by the correlation functions 〈x(t) ⊗ x(t′)〉 or their Fourier transform, Sx(f), the noise power spectra. As detailed in Supporting Text, which is published as supporting information on the PNAS web site, we solve for this quantity by finding a suitable normal coordinate that diagonalizes Eq. 1 and find that Sx(f) is a sum of Lorentzians with the characteristic frequencies λ+/2π and λ−/2π, associated with the two normal modes of Eq. 1. In a typical measurement, data are collected at a frequency higher than the highest characteristic frequency and then averaged to a frequency, fav, that is lower than the lowest. Thus, the power spectra are white over the measurement bandwidth, B = fav/2, and the mean squared noise in microsphere position over this bandwidth, 〈x ⊗ x〉B, is well approximated by the low-frequency power spectra Sx(0) multiplied by 2B. The explicit expressions for 〈x12〉B, 〈x22〉B, and 〈x1x2〉B are given in Eqs. 16 and 17 in Supporting Text.
Measured Signals.
On this fluctuating background, we consider a process that introduces a length change, ΔL, in the tether on a time scale sufficiently longer than the relaxation time of the microspheres in the optical traps, allowing the system to equilibrate before and after this length change. As a result, the first microsphere experiences an additional force kDNAΔLξ/L, and the second microsphere a force −kDNAΔLξ/L. The ratio ξ/L converts contour length to extension; for appreciable forces (>5 pN on dsDNA), this factor is close to unity and will be neglected. Time averaging the equations of motion, Eq. 1, in response to these forces yields the changes in the equilibrium position of each microsphere 〈Δx1〉 and 〈Δx2〉, listed in Eqs. 20 and 21 of Supporting Text. As expected for a system with finite compliance, 〈Δx1,2〉 ≤ ΔL.
The Spatial Resolution of Dual-Trap Optical Tweezers.
A DNA length change, ΔL, is revealed by the measured signal, i.e., the change in the equilibrium displacement of each microsphere from the center of its trap, and the noise in this measurement is given by the integrated fluctuations in each microsphere position over the measurement bandwidth. Thus, the spatial resolution of the instrument (its ability to resolve the change ΔL) is given by the ratio of the measured change in microsphere position to the root-mean-square of its fluctuations: the signal-to-noise ratio (SNR). For the microsphere in trap 1 this is
 |
SNR2 is found by exchanging indices, 1↔2.
To better understand Eq. 3, it is useful to compare it to the SNR for single-trap optical tweezers, which can be derived from SNR1 in the limit that k2 → ∞.
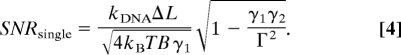 |
The expression derived previously in the literature (16) ignores hydrodynamic coupling and is recovered in the Γ−1 → 0 limit. The hydrodynamic coupling that appears in this equation reflects the fact that the mobility of the first microsphere is modified by the presence of the second, stationary microsphere. Similar modifications to this mobility can occur in the presence of a nearby surface (26).
Comparing Eqs. 3 and 4 for single and dual optical traps, one observes significant differences in behavior. SNRsingle is, remarkably, independent of trap stiffness and grows linearly with DNA stiffness. In contrast, for the dual optical trap, SNR1 depends on the stiffness of the second trap and becomes increasingly independent of DNA stiffness, asymptotically reaching the value
 |
for kDNA ≫ k2. This corresponds to the expected SNR for a dumbbell consisting of two microspheres connected by a rigid rod with a total drag coefficient of γ1 + γ2. Further inspection (see Supporting Text) shows that SNRsingle ≥ SNR1 for all experimental parameters, only approaching the equality in the trivial limit kDNA → 0.
At first glance this appears to be a fundamental limitation of the dual-trap optical tweezers relative to the single-trap optical tweezers. However, in only considering the displacements of the microspheres individually, we have ignored the correlations between them from the coupling by the DNA tether. Significantly, this correlation 〈x1x2〉B increases with DNA stiffness. If the deflections of both microspheres are monitored simultaneously, a new coordinate may be formed from a linear combination of x1 and x2 that exploits these correlations to produce a higher spatial resolution. The use of such a coordinate to measure the changes in the length of the DNA tether represents a significant advantage of a dual-trap optical tweezers in which the displacements of both microspheres are monitored.
To illustrate this point, consider the difference coordinate, x− = x1 − x2, for which we compute the small bandwidth noise, 〈x−2〉B = 〈x12〉B + 〈x22〉B − 2〈x1x2〉B and the signal 〈Δx−〉 = 〈Δx1〉 − 〈Δx2〉. The coupling due to DNA leads to a positive correlation in the positions of the microspheres 〈x1x2〉B, because one microsphere effectively pulls the other in the same direction via the DNA as it undergoes Brownian motion. The negative sign in the definition of x− thus implies that the small bandwidth noise 〈x−2〉B is decreased by this coupling. In contrast, the signal resulting from a change in DNA length corresponds to motions of the microspheres in opposite directions, and thus the negative sign in x− increases the signal 〈Δx−〉. The SNR in this coordinate is given by
 |
which, unlike SNR1, is linear in DNA stiffness and, thus, does not saturate with increasing kDNA. Unlike SNRsingle, however, SNRdiff depends on both trap stiffnesses and reaches a maximum under the condition
in which case the SNR is optimal, and given by the simple expression
 |
where γeff = γ1γ2/(γ1 + γ2). For a given DNA stiffness and bandwidth, the best possible resolution is achieved with identical microspheres of the smallest possible size in traps of identical stiffness; however, even for two microspheres of unequal size, the resolution in the difference coordinate can be maximized by tuning the trap stiffnesses according to Eq. 7.
The argument outlined above is valid for any arbitrary linear combination of x1 and x2, Ψ ≡ α1x1 − α2x2. However, in contrast to the SNR in the difference coordinate, the arbitrary amplitudes α1,2 can be chosen such that the SNR is maximized for all values of the trap stiffnesses. We compute the small bandwidth noise 〈Ψ2〉B = α12〈x12〉B + α22〈x22〉B − 2α1α2〈x1x2〉B and the signal 〈ΔΨ〉 = α1〈Δx1〉 − α2〈Δx2〉 and maximize the SNR in this coordinate, SNRΨ, with respect to the amplitudes α1,2. Because the SNR is independent of overall scaling, SNRΨ depends only on the ratio of the amplitudes α2/α1; thus, the condition that maximizes SNRΨ is
 |
Under this condition, SNRΨ is equal to SNRopt, Eq. 8. For any set of experimental parameters, Eq. 9 defines an optimal coordinate in which the ability to resolve changes in the tether length is maximized. However, a drawback compared with the difference coordinate is that this optimal coordinate requires knowledge of parameters that may change during the course of an experiment, most notably kDNA and Γ. An exception to this situation occurs when the experimental parameters obey the condition in Eq. 7, in which case the optimal coordinate given by Eq. 9 is the difference coordinate and is independent of kDNA. Furthermore, if identical microspheres are used, Eq. 7 is also independent of Γ.
By exploiting the correlations between the microspheres, we have derived an SNR, SNRopt, that does not saturate with DNA stiffness and that is remarkably similar in form to SNRsingle. However, because SNRopt depends on an effective drag coefficient, γeff, which is always smaller than either drag coefficient individually, it can be shown that SNRopt ≥ SNRsingle ≥ SNR1 for all values of the experimental parameters (see Supporting Text). The improvement of SNRopt over SNR1 reflects the fact that, despite the added Brownian noise due to the second trapped microsphere, only a fraction of the noise is relevant to the resolution of the signal. Because the microspheres move in a highly antisymmetric manner in response to change in the DNA length, only fluctuations that are similarly antisymmetric limit the resolution of this signal. By monitoring the motion of both microspheres, we can partition the symmetric and antisymmetric motions of the microspheres to improve the resolution. However, it may still seem surprising that the resolution of a system with two fluctuating degrees of freedom, SNRopt, is higher than that of a system with only one degree of freedom, SNRsingle. The reason is that resolution-limiting fluctuations relax faster in a system with two fluctuating microspheres, where both are involved in the dissipation of thermal energy, than in a system with only one fluctuating microsphere. The smaller effective drag coefficient γeff that results from the tether-mediated collective motion of the two microspheres reflects this faster relaxation mechanism.
Hydrodynamic coupling introduces correction terms as seen in Eqs. 3–9. Significantly, these corrections lower the spatial resolution in the difference and optimal coordinate. As demonstrated by Meiners and Quake (4), antisymmetric motions of two hydrodynamically coupled microspheres are longer-lived than symmetric motions. Thus, more noise power is contained in antisymmetric fluctuations and the net effect is to reduce the correlation 〈x1x2〉B and the SNR in the difference and optimal coordinates relative to that expected in the absence of coupling. Even for reasonable experimental parameters, the hydrodynamic coupling correction can be important and should not be ignored. Despite this correction, the optimal coordinate, in which the symmetric and antisymmetric fluctuations of the microspheres are best partitioned, continues to yield a higher spatial resolution than all other coordinates.
The fact that the trap stiffnesses do not appear in the optimal spatial resolution, Eq. 8, is significant and hints at the generality of this analysis. For example, it is common to run optical traps in force feedback, displacing the position of one trap relative to the other to maintain a constant force (28) or exploiting nonlinear trapping regimes in which the stiffness of the trap disappears (29). We expect, by the independence of SNRopt on the trap stiffnesses, that these systems should have spatial resolutions identical to those derived above. It is possible to verify this prediction directly by replacing the position of one of the microspheres with the position of the feedback trap, setting the stiffness of that trap to zero in Eq. 1, and deriving the appropriate coupled Langevin equations. An identical analysis yields the same SNR as derived above with the feedback trap stiffness set to zero, as expected. Thus, the best resolution, SNRopt, is still achieved by monitoring the movements of both microspheres, with no improvement to spatial resolution provided by force feedback.
Brownian Noise-Limited Dual-Trap Instrument.
To test the validity of this model, we have constructed a Brownian noise-limited dual-trap optical tweezers instrument with a dual-trap detection system (see Fig. 1b for details). The dual traps are formed from two orthogonally polarized beams generated by a single laser. One beam is steered relative to the other via a two-axis piezo mirror capable of rotating through 2 mrad with stability better than 20 nrad, corresponding to trap movements of ≈6 μm full range with ≈0.6-Å stability in the sample plane. Subangstrom determination of each microsphere position relative to its trap center is accomplished by back focal plane interferometry (30), in which the forward scattered light from each trapped microsphere is collected, separated by polarization, and imaged onto two separate position-sensitive photodetectors. With careful alignment, cross-talk between polarizations is maintained below 2%.
Despite the increased isolation due to the dual-trap geometry, significant efforts are also taken to isolate this instrument from different forms of environmental noise. The instrument is located in a basement room on a thick optical table levitated on air, and placed in a temperature controlled room with a typical stability of ±0.2°C, with all fanned cooled electronics placed outside the room. During experiments, the instrument is operated entirely remotely to eliminate thermal fluctuations due to human contact. All optical paths are isolated from background air currents via plastic enclosures. Abbondanzieri et al. (20) fill similar enclosures with helium gas, which has a lower index of refraction than air, to reduce Schlieren noise and improve spatial resolution. In contrast, in our instrument, differential movements of the two traps due to air density fluctuations have been reduced by minimizing the total differential path between the beams. We observe no improvement in spatial resolution by the introduction of helium, within the range of parameters probed here.
Spatial Resolution Measurements.
To test the above theory, a single flexible tether of dsDNA ≈5 kb in length, ≈1.7 μm, is formed between two polystyrene microspheres held in traps of variable stiffness (see Supporting Text). The observed noise spectra for these tethers are well described by the expected Lorentzian power spectra above ≈1 Hz for a large range of DNA tensions (see Fig. 5, which is published as supporting information on the PNAS web site). This result indicates that the predicted spatial resolution can be directly tested on bandwidths larger than ≈1 Hz. By modifying the distance between the traps, we produce equilibrium movements in the position of the microspheres that well approximate the expected signal for an identical change in the length of the DNA tether. By directly measuring these equilibrium changes and comparing them with measured noise values on a given bandwidth, we compute the SNR. To facilitate comparison to movements of different sizes and durations, we scale these SNR to that of a 1-nm step in a 1-Hz bandwidth.
In the first test of our predicted SNR, a DNA tether is formed between two 860-nm microspheres in traps of equal stiffness, 0.13 pN/nm. By stretching the tether to different tensions, thereby increasing its stiffness, we probe directly the dependence of the different SNRs on kDNA. In Fig. 2, we plot this dependence for the measured SNR for a single trap and the optimal coordinate (also the difference coordinate for a symmetric system) from six different DNA–microsphere dumbbells. Fig. 2 also contains the predicted values for SNR1 and SNRopt plotted as 1-σ confidence intervals to reflect the uncertainties in the experimental parameters. The measured values of SNR support the above predictions in both the single trap and the optimal coordinate for low values of DNA stiffness and demonstrate the improvement in SNR in the latter coordinate. As the tether is pulled to higher forces and, thus, higher stiffness, these measurements become increasingly sensitive to nonlinearity in the force and light deflection from each trap; thus, even the optimal coordinate effectively saturates to a maximum resolution. However, even at saturation, this coordinate still shows a 3-fold improvement in resolution over the single-trap coordinate.
Fig. 2.
Measured and predicted SNR for two tethered, 860-nm microspheres in traps of equal stiffness as a function of the DNA stiffness. The data and predictions are scaled by the square-root of the bandwidth divided by the step size and thus correspond to the SNR for a 1-nm step on a 1-Hz bandwidth. The SNR measured in the optimal coordinate (red squares) (also the difference coordinate) increases linearly with DNA stiffness, whereas the SNR measured in the single-trap coordinate (green diamonds) saturates. To reflect the uncertainty in the experimental parameters, the predicted SNR (Eqs. 3 and 8) are plotted as shaded, 1-σ confidence intervals. The observed spread in the data is dominated not by measurement error but by the inherent stochasticity in finite measurements of spatial resolution. Disagreement between measured and predicted SNR at high DNA stiffness results from systematic errors due to nonlinearities in the traps at high force. Data from six DNA–microsphere dumbbells are plotted, with one of three data points displayed.
To test the ability of the optimal coordinate to extract a higher spatial resolution when the stiffnesses of the traps are not equal, we repeat the above experiment with a range of trap stiffnesses. A DNA tether between two identical 860-nm microspheres is pulled to a specified tension, corresponding to the desired DNA stiffness, and the spatial resolution is measured as above. A half waveplate mounted on a rotary stage is then used to change the relative stiffness between the traps, and the measurement is repeated. Fig. 3a shows the measured values of SNR1, SNR2, SNRdiff, SNRopt for six DNA tethers at a DNA stiffness of 0.05 ± 0.005 pN/nm as a function of k1 and the corresponding k2. The range of stiffnesses that can be accurately probed is limited by the nonlinearities in stiffness and displacement conversions that become prominent at lower forces in the weaker traps; thus, the distinction between the optimal coordinate and the difference coordinate is only noticeable at the edge points where the difference in trap stiffness is large.
Fig. 3.
Measured and predicted SNR as a function of trap stiffness. (a) SNR for identical, tethered, 860-nm microspheres as measured in the trap 1 (green diamonds) and trap 2 (orange triangles) coordinates, the difference coordinate (blue circles), and the optimal coordinate (red squares) as the trap stiffnesses are changed. Measurements from six different DNA–microsphere dumbbells are plotted. (b) SNR for an 800-nm microsphere in trap 1 tethered to a 997-nm microsphere in trap 2 as the trap stiffnesses are changed. The coordinates are labeled as in a. Measurements from four different DNA tethers are plotted. The slight increase in SNRopt in a over that in b reflects the fact that γeff is smaller for two 860-nm microspheres than for one 800-nm and one 997-nm microsphere. The displayed SNR are scaled by the square-root of the measurement bandwidth divided by the step size as in Fig. 2, and the patterned areas are 1-σ confidence regions for the predicted SNR (Eqs. 3, 6, and 8).
To further test the predicted SNR, we form tethers between two microspheres of different diameters: 800 nm in trap 1 and 997 nm in trap 2. Again, we pull these tethers to a specified tension and measure the spatial resolution as a function of relative stiffness between the traps. Fig. 3b shows the results for four different DNA tethers. The agreement between the predicted and observed values is not quite as good as for equal microspheres, Fig. 3a. This effect is most likely due to slight imperfections in the sphericity of the 997-nm microspheres, which introduce additional noise. The power spectra for tethers made from two 997-nm microspheres have noticeable deviations from the predicted white noise on a larger frequency range (data not shown) for comparable tensions, confirming this possibility. If the origin of the additional noise were the imperfection of the 997-nm microsphere, the agreement between measured SNR and theory should improve for the optimal coordinate when the movements of the 800-nm microsphere are weighted more, i.e., for higher stiffness in trap 1 (see Eq. 9). As k1 increases, the measured SNR for the optimal coordinate agrees better with the predicted values.
Dual-trap detection should make it possible to detect smaller steps at larger bandwidths and lower DNA tensions than would be possible by only monitoring the motions of one microsphere. To demonstrate this point, we pull a 3.4-kb molecule of DNA tethered between two 860-nm microspheres in traps of equal stiffness to a tension of 7.3 ± 0.1 pN. The distance between the traps is then increased by ≈3.4 Å every 2 s, corresponding to a length change of dsDNA by 1 bp. Fig. 4a shows the displacements for a single trap and the optimal coordinate filtered to 100 Hz and 2 Hz. Whereas each of the seven steps can be seen clearly in the optimal coordinate, the presence of steps is not apparent in the displacements of a single microsphere. The pairwise distributions for the optimal coordinate and the single microsphere coordinate at 2 Hz (Fig. 4 b and c) confirm this result. Although a peak for all seven steps can be seen clearly for the optimal coordinate, there is no evidence of such peaks for the single microsphere. Because of finite trap stiffness, the peaks in Fig. 4b are separated by only 2.3 Å, the expected signal in the optimal coordinate for a 3.4-Å change in trap separation with the above experimental parameters. Comparable demonstrations of high spatial resolution have already been reported for dual-trap optical tweezers (20), although only one microsphere was monitored in those studies. The results presented here show, however, that monitoring the optimal coordinate makes it possible to resolve these movements with larger microspheres, longer DNA, and ≈2.5-fold less DNA tension than previously reported.
Fig. 4.
Improvement in spatial resolution from dual-trap detection. (a) Observed displacements for two 860-nm microspheres tethered by 3.4-kb dsDNA at a tension of 7.3 ± 0.1 pN as the distance between the traps is increased by 3.4 Å every 2 s. The displacement of the microsphere in trap 1 (lower trace) and the optimal coordinate (upper trace) are filtered with a sliding boxcar filter to 100 Hz (gray traces) and to 2 Hz (blue for the lower trace and red for the upper trace). (b) Pairwise distance histogram for the optimal coordinate in a averaged to 2 Hz. Peaks corresponding to the distance between each of the seven steps confirm the presence of these steps. (c) Similar peaks are not present in the pairwise distance histogram for the displacement of a single microsphere. The observed signals in the optimal coordinate are ≈2.3 Å, smaller than the 3.4 Å changes in the trap separation, as expected for finite trap stiffness (see Supporting Text).
Discussion
In this paper, we calculate the degree to which Brownian fluctuations will limit the ability to resolve equilibrium changes in the length of a flexible polymer held between two microspheres in two separate optical traps. By solving the equations of motion of each microsphere, we derive estimators for the spatial resolution of the system, i.e., the signal-to-noise ratios. Remarkably, we find that despite the addition of a second noisy degree of freedom, dual-trap optical tweezers actually have higher spatial resolution than single-trap systems, provided that the motions of both microspheres are monitored. Furthermore, we verify these predicted SNR by directly measuring the spatial resolution of a Brownian noise-limited dual-trap optical tweezers. The highest resolution is found with a perfectly symmetric system of the smallest possible microspheres, independent of trap stiffness.
Despite the good agreement between theory and experiment in the parameter range studied, it is useful to remember that several higher-order effects are ignored in the derivation of the above SNR. First, the DNA has a finite relaxation time, degrading the instantaneous correlation between microspheres that gives rise to the improvement in SNRopt. Using a simple Rouse model (31) with a drag coefficient extrapolated from experimental measurements (21), we estimate that this effect contributes no more than a 1% correction to the predicted SNR in Fig. 3. However, as the length of the DNA increases, this relaxation time increases and corrections due to DNA dynamics should become more important. Second, fluctuations of the microspheres in directions orthogonal to the DNA tether are coupled into the tether direction and become increasingly important as the DNA length decreases. The Brownian rotation of the microspheres also couple noise into this direction and become increasingly important as the microsphere diameter increases and DNA length decreases. We estimate that orthogonal movements and Brownian rotation degrade the SNR in Fig. 3 by ≈0.1% and ≈1%, respectively. Finally, we see no evidence in our measurements for partial shielding of the hydrodynamic coupling between the microspheres by the DNA as reported by Meiners and Quake (21). Our data agree well with the full 14% hydrodynamic correction in the predicted SNR, as given by Eq. 8. The absence of noticeable shielding is not surprising because the measurements of Meiners and Quake (21) are conducted with DNA that was ≈10 times longer than that used here, and we expect this effect to increase as the DNA length increases.
Practically, it has been shown that the greatest improvement provided by the addition of a second optical trap is the dramatic increase in isolation from the macroscopic environment (19, 20). Beyond this point, we have shown that there is a significant added benefit to monitoring the motions of both microspheres. In particular, the ability of the optimal coordinate to maximize the spatial resolution for a range of experimental parameters represents a significant increase in versatility of dual-trap systems. For example, the freedom to modulate the stiffnesses of the traps without adversely affecting the spatial resolution may be useful in certain applications that require smaller laser fluxes to minimize photoinduced damage or laser heating. Furthermore, many of the biological systems that manipulate nucleic acids or proteins on the subnanometer scale operate only in a limited range of forces. By exploiting correlations in the fluctuations of the microspheres, we have achieved subnanometer resolution at lower forces than previously reported, allowing the study of such systems under more permissive conditions.
Supplementary Material
Acknowledgments
We thank J. Choy for motivating conversations; J. Choy, S. W. Grill, and S. B. Smith for advice regarding instrumentation; O. Levy, E. A. Galburt, and A. Edelstein for help in DNA tether construction; O. Levy, W. Cheng, and E. A. Galburt (all from University of California) for plasmid DNA and functionalized microspheres; L. Bintu for help with calibration; and C. L. Hetherington and A. J. Spakowitz for a critical reading of the manuscript. J.R.M. was supported by the National Science Foundation’s Graduate Research Fellowship. Y.R.C. was supported by the Burroughs Welcome Fund’s Career Awards at the Scientific Interface.
Abbreviation
- SNR
signal-to-noise ratio.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ashkin A., Dziedzic J. M., Bjorkholm J. E., Chu S. Opt. Lett. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal R., Ladavac K., Roichman Y., Yu G. H., Lieber C. M., Grier D. G. Opt. Express. 2005;13:8906–8912. doi: 10.1364/opex.13.008906. [DOI] [PubMed] [Google Scholar]
- 3.Pauzauskie P. J., Radenovic A., Trepagnier E., Shroff H., Yang P., Liphardt J. Nat. Mater. 2006;5:97–101. doi: 10.1038/nmat1563. [DOI] [PubMed] [Google Scholar]
- 4.Meiners J. C., Quake S. R. Phys. Rev. Lett. 1999;82:2211–2214. [Google Scholar]
- 5.Henderson S., Mitchell S., Bartlett P. Phys. Rev. Lett. 2002;88:088302. doi: 10.1103/PhysRevLett.88.088302. [DOI] [PubMed] [Google Scholar]
- 6.Hough L. A., Ou-Yang H. D. Phys. Rev. E: Stat., Nonlin., Soft Matter Phys. 2002;65:021906. doi: 10.1103/PhysRevE.65.021906. [DOI] [PubMed] [Google Scholar]
- 7.Liphardt J., Dumont S., Smith S. B., Tinoco I., Jr, Bustamante C. Science. 2002;296:1832–1835. doi: 10.1126/science.1071152. [DOI] [PubMed] [Google Scholar]
- 8.Wang G. M., Sevick E. M., Mittag E., Searles D. J., Evans D. J. Phys. Rev. Lett. 2002;89:050601. doi: 10.1103/PhysRevLett.89.050601. [DOI] [PubMed] [Google Scholar]
- 9.Trepagnier E. H., Jarzynski C., Ritort F., Crooks G. E., Bustamante C. J., Liphardt J. Proc. Natl. Acad. Sci. USA. 2004;101:15038–15041. doi: 10.1073/pnas.0406405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collin D., Ritort F., Jarzynski C., Smith S. B., Tinoco I., Bustamante C. Nature. 2005;437:231–234. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta A. D., Rief M., Spudich J. A., Smith D. A., Simmons R. M. Science. 1999;283:1689–1695. doi: 10.1126/science.283.5408.1689. [DOI] [PubMed] [Google Scholar]
- 12.Bustamante C., Smith S. B., Liphardt J., Smith D. Curr. Opin. Struct. Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 13.Bustamante C., Bryant Z., Smith S. B. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 14.Bustamante C., Chemla Y. R., Forde N. R., Izhaky D. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 15.Svoboda K., Block S. M. Annu. Rev. Biophys. Biomol. Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 16.Gittes F., Schmidt C. F. Eur. Biophys. J. Biophys. Lett. 1998;27:75–81. [Google Scholar]
- 17.Finer J. T., Simmons R. M., Spudich J. A. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 18.Visscher K., Gross S. P., Block S. M. IEEE J. Quantum Electron. 1996;2:1066–1076. [Google Scholar]
- 19.Shaevitz J. W., Abbondanzieri E. A., Landick R., Block S. M. Nature. 2003;426:684. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbondanzieri E. A., Greenleaf W. J., Shaevitz J. W., Landick R., Block S. M. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meiners J. C., Quake S. R. Phys. Rev. Lett. 2000;84:5014–5017. doi: 10.1103/PhysRevLett.84.5014. [DOI] [PubMed] [Google Scholar]
- 22.Bustamante C., Marko J. F., Siggia E. D., Smith S. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 23.Odijk T. Macromolecules. 1995;28:7016–7018. [Google Scholar]
- 24.Wang M. D., Yin H., Landick R., Gelles J., Block S. M. Biophys. J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lifshitz E. M., Landau L. D. Fluid Mechanics. Oxford: Butterworth–Heinemann; 1987. [Google Scholar]
- 26.Berg-Sorensen K., Flyvbjerg H. Rev. Sci. Instrum. 2004;75:594–612. [Google Scholar]
- 27.Batchelor G. K. J. Fluid Mech. 1976;74:1–29. [Google Scholar]
- 28.Visscher K., Block S. M. Methods Enzymol. 1998;298:460–489. doi: 10.1016/s0076-6879(98)98040-5. [DOI] [PubMed] [Google Scholar]
- 29.Greenleaf W. J., Woodside M. T., Abbondanzieri E. A., Block S. M. Phys. Rev. Lett. 2005;95:208102. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gittes F., Schmidt C. F. Opt. Lett. 1998;23:7–9. doi: 10.1364/ol.23.000007. [DOI] [PubMed] [Google Scholar]
- 31.Rouse P. E. J. Chem. Phys. 1953;21:1272–1280. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.