Abstract
Tumor progression and metastasis depend on the ability of cancer cells to initiate angiogenesis and ensure delivery of oxygen, nutrients, and growth factors to rapidly dividing transformed cells and provide access to the systemic circulation. In addition to well established growth factors and inflammatory mediators that promote capillary sprouting and endothelial cell growth and migration, an emerging body of evidence supports a previously unrecognized function for axon guidance molecules in regulation of blood vessel development. Here we show that semaphorin 4D (Sema4D), a protein originally shown to regulate axonal growth cone guidance in the developing central nervous system through its receptor, plexin-B1, is highly expressed in cell lines derived from head and neck squamous cell carcinomas (HNSCCs) at both the protein and message level. Immunohistochemical analysis of a large collection of HNSCC specimens revealed high levels of Sema4D in a cell surface pattern in invading islands of transformed epithelial cells, but not in normal and noninvasive dysplastic epithelium. A similar pattern was observed in malignant cells from prostate, colon, breast, and lung cancer tissues. When shed from HNSCC cells, Sema4D stimulates endothelial cell migration, which can be prevented by Sema4D-blocking antibodies and by Sema4D knockdown. Furthermore, knocking down Sema4D by lentiviral expression of Sema4D shRNA reduces dramatically the size and vascularity of HNSCC tumor xenografts. These findings indicate that expression of Sema4D is a frequently used strategy by which a wide variety of carcinomas may promote angiogenesis, and therefore is a possible therapeutic target for the treatment of these malignancies.
Keywords: head and neck cancer, c-Met protein, plexin, chemotaxis, endothelium
Tumor growth and metastasis require induction of angiogenesis, the growth and remodeling of new blood vessels from a preexisting vascular network, to ensure the delivery of oxygen, nutrients, and growth factors to rapidly dividing transformed cells and to provide the tumor cells access to systemic circulation (1–3). Without the ability to induce angiogenesis, most neoplasms would fail to grow >2 mm in diameter or metastasize (3). Vascular endothelial growth factor (VEGF), a pro-angiogenic factor whose expression increases upon exposure of cells to low oxygen tension, plays a crucial role, together with inflammatory mediators and additional tumor and stromal-released growth factors, in stimulating the switch form a hypoxic, avascular phenotype to a pro-angiogenic phenotype in growing tumors (4–6).
Emerging evidence suggests that certain molecules implicated in axonal pathfinding in the developing nervous system also play a role in blood vessel development. We now know that proteins involved in transmitting axonal guidance cues, including members of the netrin, slit, eph, and semaphorin families, are widely expressed in tissues outside of the nervous system and play a critical role in blood vessel guidance and endothelial precursor cell homing during physiological and pathological blood vessel development (7, 8). For example, secreted class III semaphorins, which regulate axonal growth during the development of the central nervous system (9, 10), are now known to act through their receptors, the neuropilins and the A-family plexins, to initiate signaling events in a variety of tissues that influence vascular morphogenesis and endothelial cell motility (7, 8, 11–13). We have recently observed that plexin-B1, the receptor for the class IV, membrane-bound, semaphorin, semaphorin 4D (Sema4D), is highly expressed in endothelial cells and promotes endothelial cell migration and tubulogenesis (14). These findings, later confirmed by others (15), suggest that the class IV semaphorins may regulate angiogenesis in vivo, and raise the intriguing possibility that Sema4D could play a role in tumor-induced angiogenesis. Indeed, Sema4D-mediated activation of plexin-B1 has been shown to recruit and activate the hepatocyte growth factor receptor c-Met, a potent promoter of cell survival, migration, and angiogenesis (16).
While performing a gene array analysis in a panel of cell lines derived from primary and metastatic head and neck squamous cell carcinomas (HNSCCs), we detected levels of Sema4D message as high as those found in brain tissues, and Sema4D protein was readily detectable in HNSCC-derived cell lines but almost undetectable in nontumorigenic epithelial cell lines. Immunohistochemical analysis of Sema4D in a large tumor sample collection revealed that Sema4D overexpression is a very frequent event in SCC of the oral cavity and head and neck, as well as in prostate, colon, breast, and lung cancers. Furthermore, we observed that Sema4D can be shed into the medium by HNSCC cancer cells, thereby inducing endothelial cell migration in vitro, which was prevented by Sema4D-blocking antibodies or by Sema4D knockdown. Knocking down Sema4D expression in HNSCC cells with Sema4D shRNA before grafting into nude mice caused a dramatic reduction in tumor vascularity and tumor growth. These results suggest that expression of Sema4D may represent a widely used strategy by tumors to induce angiogenesis, thereby enhancing their growth, survival, and metastatic potential, and thus may represent a new therapeutic target for cancer treatment.
Results
Sema4D Is Highly Expressed in HNSCC.
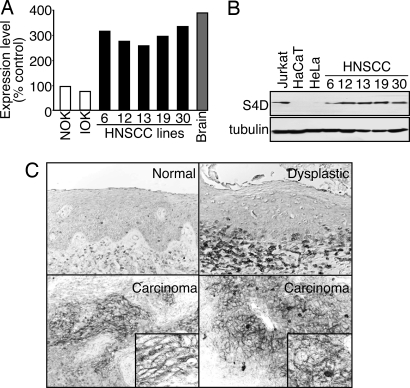
HNSCC represents the sixth-most common neoplasm in the developed world, with an overall 5-year survival rate among the lowest of the major cancers (17). We have examined gene expression profiles for this aggressive cancer type in a representative panel of HNSCC cells by using recently developed long oligonucleotide arrays. Our results showed that Sema4D was highly expressed in all HNSCC cells, comparable to that seen in human brain, which was used as the positive control, but was expressed only at very low levels in primary normal human oral keratinocytes (Fig. 1A). In contrast, immortalized, nontumorigenic oral keratinocytes did not exhibit enhanced levels of Sema4D message, suggesting that increased Sema4D expression is a characteristic of malignant HNSCC cells. To confirm these findings, we examined the protein levels of Sema4D, by using Jurkat cells, a T cell line that demonstrates high levels of Sema4D, as a positive control (18). As shown in Fig. 1B, all HNSCC cells studied demonstrated high levels of Sema4D protein, whereas Sema4D was barely detectable in the immortal nontumorigenic keratinocyte cell lines HaCaT (19) and in the human papillomavirus-16-immortalized cervical cancer cell line, HeLa (Fig. 1B). Tubulin was used as a loading control (Fig. 1B). These observations prompted us to explore whether Sema4D is expressed in HNSCC tumors. We failed to detect Sema4D expression in normal oral parakeratinized stratified squamous epithelium (Fig. 1C). Dysplastic oral squamous epithelium exhibited teardrop-shaped rete ridges, cellular crowding, disorganization at the basal and parabasal levels, and persistence of large nuclei into the upper spinous layer but failed to exhibit Sema4D expression in the epithelial cells. Sema4D was detected in the associated chronic inflammatory cells accumulating at the epithelial–connective tissue interface (Fig. 1C). In contrast, we detected robust immunohistochemical staining for Sema4D in the cytoplasm and cell surface in invading islands of transformed epithelial cells (Fig. 1C), which is more clearly seen at a higher magnification (Fig. 1C Insets). Taken together, these results demonstrate Sema4D expression in HNSCC tumors and their derived cells lines but not in normal and noninvasive oral epithelium.
Fig. 1.
Sema4D is highly expressed in HNSCC. (A) RNAs from controls and HNSCC cell lines were compared to reference RNA by microarray analysis. Sema4D mRNA levels are expressed relative to normal human oral keratinocytes (NOK). RNA from brain tissues was used as the positive control. IOK, immortal human oral keratinocytes. (B) Immunoblot analysis for Sema4D shows a band at ≈125 kDa for endogenously expressed protein in lysates from Jurkat cells (positive control) and HNSCC cell lines, but not in the nontumorigenic epithelial-derived cell lines HaCaT or HeLa. Tubulin was used as a loading control. (C) Immunohistochemical analysis of Sema4D in HNSCC. No Sema4D was detected in normal parakeratinized stratified squamous epithelium (Normal) and in dysplastic stratified squamous epithelium, although chronic inflammatory cells were positive (Dysplastic). Sema4D immunoreactivity is seen in invading islands of transformed epithelial cells (Carcinoma). A cell surface-staining pattern is seen at a higher magnification (Insets).
Prostate, Colon, Breast, and Lung Carcinomas Express High Levels of Sema4D, Whereas Endothelial Cells Express Plexin-B1.
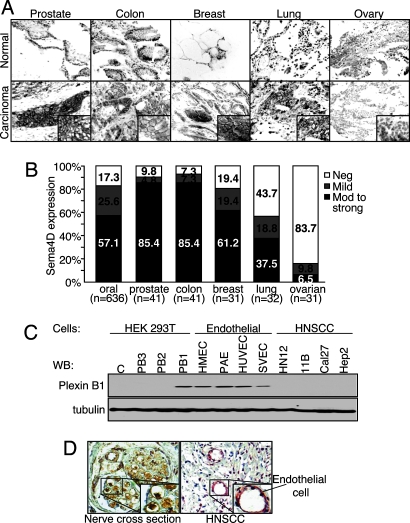
To determine whether Sema4D was also expressed in other tumor types, we performed immunohistochemical analysis of tumor tissue arrays containing representative cases of the most frequent human cancers and their matching normal control tissues. Normal tissue samples from these arrays exhibited no Sema4D expression, or small foci only in areas of chronic inflammatory cell accumulation (Fig. 2A). Tumor samples demonstrate abundant Sema4D staining in invading islands of transformed epithelial cells from the prostate, colon, breast, and lung, suggesting that Sema4D expression is a common feature of these highly frequent human carcinomas (Fig. 2A). At higher magnification, we observed generalized cytoplasmic immunoreactivity for Sema4D or a robust, cell-surface-specific staining pattern of expression in most adenocarcinomas of the prostate, colon, breast, and squamous cell carcinomas of the lung, similar to that observed in HNSCC (Fig. 2A). These results are expressed graphically in Fig. 2B. Ovarian tumors exhibited the poorest expression of Sema4D (Fig. 2B). These results suggest that Sema4D expression is a common feature in a wide variety of tumors of epithelial origin.
Fig. 2.
Sema4D is expressed in oral, prostate, colon, breast, and lung cancer, whereas plexin-B1 is expressed in tumor endothelial cells. (A) Tumor tissue arrays were analyzed for Sema4D expression. The photographs show representative areas of the indicated tumors and corresponding normal tissues. Normal tissues (Upper) exhibit no epithelial Sema4D expression or small foci in areas of areas of chronic inflammatory cell accumulation (normal colon). Tumor tissues demonstrate abundant Sema4D staining in invading islands of transformed epithelial cells in prostate, colon, breast, and lung carcinomas, although not in ovarian carcinomas (Lower). A robust cell-surface-specific staining pattern for Sema4D is seen at higher magnification (Insets). (B) The bar graph summarizes the data from Sema4D staining in tumor tissue arrays. Between 37.5% and 85.4% of oral, prostate, colon, breast, and lung carcinomas analyzed exhibit moderate to strong Sema4D expression. (C) Immunoblot analysis for plexin-B1 in the indicated endothelial and HNSCC cell lines. (Upper) Control transfected HEK293T cells or cells transfected with full-length plexin-B3, -B2, and –B1 demonstrate antibody specificity. (Lower) Tubulin was used as a loading control. (D) Immunohistochemical analysis of plexin-B1 in tumor tissues. (Right) Abundant plexin-B1 staining is seen in the endothelial cells lining the blood vessels. (Left) A cross section of peripheral nerve is shown as a positive control.
To determine whether Sema4D produced by these tumors could be acting through plexin-B1 on endothelial cells in the surrounding tumor stroma, we immunized rabbits with a peptide specific for intracellular plexin-B1. The antiserum generated was affinity purified and used for immunoblot analysis of endothelial cell lines in vitro and immunohistochemistry for plexin-B1 expression in the endothelium of the blood vessels present in tumor tissue samples. Plexin-B1 expression is seen in human embryonic kidney (HEK) 293T cells transfected with full-length plexin-B1, but not in control cells or cells transfected with other plexin-B family members, demonstrating the specificity of the antibody (Fig. 2C). Expression of plexin-B2 and plexin-B3 in the corresponding cell transfectants was confirmed with additional plexin-specific antiserum (not shown). The endothelial cell lines HMEC, PAE, HUVEC, and SVEC all express plexin-B1, whereas a representative panel of HNSCC cell lines fail to do so (Fig. 2C). Tubulin was used as a loading control (Fig. 2C). Immunohistochemical analysis of tumor tissue samples for plexin-B1 demonstrates expression in the blood vessels of these tumors (Fig. 2D). Close up views demonstrate cytoplasmic and cell surface expression of plexin-B1 in the endothelial cells lining these vessels (Fig. 2D). A peripheral nerve cross section was used as a positive control for plexin-B1 (10) (Fig. 2D).
Cells Expressing Sema4D Induce Chemotaxis and Stress Fiber Formation in Endothelial Cells.
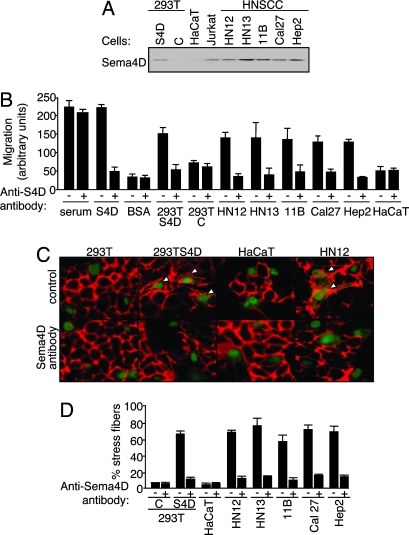
While the best-studied class III semaphorins exert soluble guidance signals capable of long-range effects through diffusion to their targets, there is available evidence suggesting that the membrane-bound class IV semaphorins may exist in a secreted form, leading us to suspect that the HNSCC cell lines expressing high levels of Sema4D may spontaneously shed this protein into the surrounding medium (20, 21). Fig. 3A demonstrates that a panel of HNSCC lines do, in fact, shed significant amounts of Sema4D into the tissue culture medium, equal to that seen for Jurkat cells, as previously reported (20, 21), and comparable to the level of Sema4D released by HEK293T cells transfected with SecTag2B Sema4D, a vector coding for a secreted form of Sema4D. In contrast, HaCaT and HeLa cells did not shed Sema4D (Fig. 3A). Therefore, we asked whether HNSCC cells can attract HUVEC in a Boyden chamber migration assay, preincubating the cell lines with and without Sema4D-blocking antibody as a specificity control. Endothelial cell migration was seen toward positive control wells containing medium with 10% FBS both with and without Sema4D blocking antibody, but not toward serum-free medium containing BSA, which served as a negative control. We observed significant chemotaxis toward wells containing purified soluble Sema4D and toward wells containing HEK293T expressing Sema4D, both of which were reduced to control levels upon incubation with the Sema4D-blocking antibody (Fig. 3B). Similarly, endothelial cell migration occurred toward wells containing HNSCC cell lines, which shed high levels of Sema4D into the surrounding medium, but not toward HaCaT cells (Fig. 3B). Moreover, HNSCC cell line-induced endothelial cell chemotaxis could be blocked with the addition of anti-Sema4D antibody, indicating that the observed migration depended on the Sema4D produced by these cells (Fig. 3B). These results suggest that Sema4D produced by HNSCC cells acts as a potent chemoattractant for endothelial cells.
Fig. 3.
Sema4D shed by HNSCC cells promotes stress fiber formation and chemotaxis in endothelial cells. (A) Immunoblot analysis for Sema4D in medium conditioned by the indicated cell lines and HEK293T cells transfected with Sema4D (S4D) or vector controls (C). Sema4D is seen in medium conditioned by the HNSCC cell lines but not in medium conditioned by HaCaT. Jurkat cells were used as a positive control. (B) Serum-free medium containing the indicated cell type or protein, with or without 10 μg/ml Sema4D-blocking antibody, were used as the chemoattractants for endothelial cells in a migration assay. Ten percent FBS and purified Sema4D were used as positive controls and 0.1% BSA was the negative control. HEK293T cells transfected with Sema4D (S4D) or vector (C) were used as additional controls. (C) HUVEC stably expressing EGFP were cocultured with HEK293T cells transfected with Sema4D (S4D) or vector (C), HaCaT, or HN12 cells with or without Sema4D-blocking antibody (C and Sema4D antibody, respectively), stained with Texas red-phalloidin and viewed in an immunofluorescence microscope. Endothelial cells exhibiting stress fibers are indicated with an arrowhead. (D) Quantification of stress fiber formation in EGFP-positive HUVEC cultured with the indicated cell lines and vector (C) or Sema4D (S4D) transfected HEK293T, with (+) and without (−) Sema4D-blocking antibody. The number of endothelial cells exhibiting stress fiber polymerization in the cocultures was counted and expressed as a percentage of the total endothelial cells in the cocultures.
Because angiogenic responses to a recombinant secreted form of Sema4D involves plexin-B1-mediated Rho activation and stress fiber formation in endothelial cells (14), we investigated whether cells expressing Sema4D naturally could affect endothelial cell behavior by coculturing endothelial cells with HNSCC cells. HUVEC stably expressing EGFP were grown on glass coverslips with HEK293T cells expressing Sema4D, or its vector control, as well as HNSCC tumor cell lines and the nontumorigenic epithelial cell line HaCaT, with and without anti-Sema4D as a specificity control. Although the overall morphology of the endothelial cells was affected by coculturing them with epithelial cells, there was a dramatic induction of stress fibers when cultured with HEK293T cells transfected with Sema4D but not with vector-transfected HEK293T cells, and this response was blocked by coincubation with Sema4D-blocking antibody (Fig. 3C). When cocultured with HaCaT cells, which exhibit little endogenous Sema4D, endothelial cells showed little to no stress fiber formation (Fig. 3C). However, when cocultured with HN12 cells, which produce high levels of endogenous Sema4D, endothelial cells showed an increase in stress fiber formation that was prevented by addition of anti-Sema4D antibody (Fig. 3C). Similar results were obtained by coculturing HUVEC with HN13, UMSCC 11B, Cal27, and Hep2 HNSCC cell lines and are presented graphically in Fig. 3D. Taken together, these findings suggest that cancer cells may promote a Sema4D-specific plexin-B1 response in endothelial cells (14) through the release of Sema4D.
Reduction of Sema4D Expression in HNSCCs Through RNA Interference Reduces Sema4D Shedding, Endothelial Cell Chemotaxis, and in Vivo Tumor Growth and Vascularity.
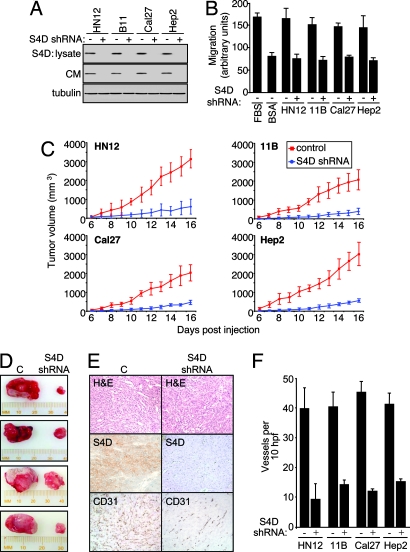
To further investigate whether Sema4D production by HNSCC cells was responsible for endothelial cell chemotaxis, we took advantage of the ability to interfere with the expression of endogenous Sema4D in cancer cells by the use of short hairpin RNAs (shRNAs) for Sema4D. Sema4D shRNAs were designed (22, 23), cloned into expression vectors, and transfected into HN12 cells. As shown in Fig. 5, which is published as supporting information on the PNAS web site, cells transfected with Sema4D shRNA exhibited reduced Sema4D levels in cell lysates and shed undetectable levels of Sema4D into the surrounding tissue culture medium, compared with controls. This procedure was then repeated in UMSCC 11B, Cal27, and Hep2 cells. As shown in Fig. 4A, cells transfected with Sema4D shRNA exhibited undetectable amounts of Sema4D in both lysates and conditioned medium compared to controls. We then used these same cell types as chemoattractants for endothelial cells in a migration assay. A robust chemotactic response was observed toward wells containing control transfected HN12, UMSCC 11B, Cal27, and Hep2 cells, but endothelial cell chemotaxis was prevented by the transfection of Sema4D shRNA (Fig. 4B). These results demonstrate the potent chemotactic activity toward endothelial cells of HNSCC-released Sema4D.
Fig. 4.
Sema4D knockdown reduces HNSCC-mediated endothelial cell chemotaxis and HNSCC tumor growth and vascularity. (A) Immunoblot analysis for Sema4D on cell lysates (S4D: lysate) and serum-free medium (CM) conditioned by the indicated cell types transfected with vector control (−) or Sema4D shRNA (+) (Top and Middle). Tubulin was used as a lysate loading control (Bottom). (B) Serum-free media conditioned by control (−) or Sema4D shRNA (+) transfected HNSCC cells were used as the chemoattractants for HUVEC in migration assays. Ten percent FBS and 0.1% BSA served as positive and negative controls. (C) The HNSCC cells HN12, UMSCC 11B, Cal27, and Hep2 were infected with control lentiviruses or lentiviruses coding for Sema4D shRNA and injected s.c. into nude mice. The results of tumor volume measurement from each day of the experiment are shown (n = 10). (D) Representative tumors derived from HN12 cells infected with EGFP or Sema4D lentiviruses are shown at the time of killing. (E) Tumors derived from HN12 cells infected with a control lentivirus (C, Left) and tumors from Sema4D shRNA-infected cells (S4D shRNA, Right) are shown in hematoxylin- and eosin-stained sections (H&E). Enhanced blood vessel infiltration is seen in tumors derived from control HN12 cells compared with tumors derived from Sema4D shRNA-infected cells (CD31). Tumors of HN12 cells infected with control lentiviruses also exhibit much higher levels of Sema4D staining compared with Sema4D shRNA-infected cells (S4D). (F) The number of vessels per 10 high-power fields in CD31-stained xenografts derived from Sema4D shRNA- and control-infected HNSCC cells was counted and averaged.
To determine whether Sema4D in HNSCC cells influences tumor growth in vivo, we engineered Sema4D shRNA lentiviral expression vectors for stable gene transfer into HN12, UMSCC 11B, Cal27, and Hep2 cells, which grow as tumor xenografts when injected into immunocompromised mice (24). The high infection efficiency of this system (14, 25) enabled us to use mass cultures of infected cells, thus avoiding the need to isolate cell clones. Growth of tumors composed of cells infected with control lentivirus greatly exceeded that seen for tumors from Sema4D shRNA-infected cells, particularly at later times (Fig. 4C), and were substantially larger for all animals at the time of killing (Fig. 4D and Fig. 6, which is published as supporting information on the PNAS web site). Immunohistochemical analysis of tumors for Sema4D demonstrated reduced staining in cells infected with Sema4D shRNA lentivirus, as expected (Fig. 4E), but it also showed greatly reduced vascularity, as demonstrated by anti-CD31 staining, when compared with infected controls (Fig. 4E). These results are quantified in the bar graph in Fig. 4F, which demonstrates significantly reduced vascularity in tumors derived from HN12, 11B, Cal27, and Hep2 HNSCC cell lines infected with Sema4D shRNA-expressing lentivirus, as determined by the number of CD31-positive vessels per 10 high-power fields. Taken together, these results indicate that expression of Sema4D in head and neck malignancies is important for tumor vascularity and growth.
Discussion
While genetic and biochemical studies have implicated semaphorins and their receptors, the plexins, in numerous aspects of neural development, mounting evidence suggests that the plexin–semaphorin signaling system can also control blood vessel growth and development (8). The class III semaphorins, which are secreted (10), reduce cell motility or act as chemorepulsants for endothelial cells (26), resulting in inhibition of angiogenesis (27, 28). Combined with the observation that Sema3F is often deleted in adenocarcinomas of the lung (29, 30), these findings support the hypothesis that the class III semaphorins act as tumor suppressors (31). In contrast, we have recently shown that endothelial cells express plexin-B1, the receptor for Sema4D, and that Sema4D promotes endothelial cell migration and tubulogenesis in vitro and the formation of blood vessels in vivo (14). The results of the present study support the biological significance of these findings and suggest that HNSCC in particular, and also other highly prevalent epithelial-derived tumors, might indeed exploit the pro-angiogenic effect of Sema4D to promote tumor growth and survival and enhance their metastatic potential.
In HNSCC, we detected robust Sema4D expression in the cytoplasm and the cell surface of invading islands of carcinoma cells but not in normal or dysplastic but noninvasive epithelial cells. These initial observations were extended by the analysis of a large number of HNSCC tissues, which revealed that Sema4D overexpression is a common feature in HNSCC, which was aligned with the high levels of Sema4D message and protein expression in a panel of representative HNSCC-derived cell lines. Significantly, the expression of Sema4D was not restricted to HNSCC, as a survey of carcinomas originating from prostate, colon, breast, and lung tissue also demonstrated Sema4D expression in a cell-surface and occasionally cytoplasmic pattern in invading islands of transformed epithelial cells compared with normal tissue controls (ovarian tumors exhibited the poorest expression of Sema4D). Thus, Sema4D expression is a common feature in some of the most prevalent tumors of epithelial origin.
To help determine the precise role of Sema4D in tumor angiogenesis, we took advantage of the high level of Sema4D expression in HNSCC cells and recently developed RNA interference techniques to knock down the levels of Sema4D in tumorigenic HNSCC lines. Expression of Sema4D shRNAs in HNSCC lines resulted in attenuation of the ability of HNSCC to induce endothelial cell migration and significantly reduced the growth of tumor xenografts. Furthermore, tumors derived from Sema4D shRNA-infected HNSCC cells also exhibited greatly reduced vascularity, thus collectively providing in vivo observations supporting a role for Sema4D in the control of tumor growth and angiogenesis.
Of interest, although it is not classified as a secreted semaphorin, we have observed spontaneous processing of membrane-bound Sema4D into a slightly smaller form that is shed by HSNCC, resembling the release of soluble Sema4D in T lymphocytes upon the cleavage of its membrane-bound form at a cysteine residue located immediately adjacent to the transmembrane domain (20). This observation raises the possibility that Sema4D may act both at a distance and locally, similar to the role of the different VEGF-A splice forms, which code for secreted proteins that exert their pro-angiogenic effects at a distance or locally, depending on their retention in the extracellular matrix based on their distinct affinities for heparin (32). Nonetheless, how the interplay between Sema4D, VEGF, and other pro-angiogenic factors promote the migration and proliferation of endothelial cells to areas of tumor growth is still poorly understood. In particular, VEGF and chemokines are sufficient to mobilize endothelial progenitor cells from the bone marrow and recruit circulating endothelial precursor cells to areas of hypoxia and inflammation (33–35). However, within the tumor, the elevated local concentrations of pro-angiogenic cytokines released by tumor, stromal, and inflammatory cells may prevent endothelial cells from sensing a chemoattractant gradient. In this scenario, we can hypothesize that Sema4D provides directional cues to guide endothelial cells specifically toward the cancer cells within the tumor microenvironment. On the other hand, results obtained from the use of Sema4D-blocking antibodies and Sema4D-knockdown techniques suggest that Sema4D is a critical endothelial chemoattractant under normoxic conditions. Thus, Sema4D may also promote the continued chemoattraction of endothelial cells to actively proliferating cancerous areas, even once the normal oxygen tension has been restored.
There is a great need to enhance our understanding of the molecular mechanisms involved in cancer cell survival, invasion, and metastasis to develop novel potential biological targets for therapy. Plexins and semaphorins were traditionally restricted to the central nervous system, but they are expressed in many other tissue types, where their function and mechanism of action require more study. Our present findings support the emerging concept that Sema4D may play a critical role in normal and tumor-induced angiogenesis in vivo, and thus could represent a new front of attack in the anti-angiogenic therapy of cancer.
Materials and Methods
Cell Culture.
Human umbilical vein endothelial cells (HUVEC) and microvascular endothelial cells (HMEC) were cultured in Endothelial Cell Basal Media (EBM) (Clonetics, Walkersville, MD); porcine aorta endothelial (PAE) cells were cultured in Ham’s F-12 medium (Sigma); the human T cell line Jurkat was grown in RPMI medium 1640 (Sigma); human embryonic kidney (HEK) 293T cells, the human keratinocytic cell lines HaCaT and HeLa, simian virus 40-transformed murine vascular endothelial cells (SVEC), and HNSCC cell lines (24) were grown in DMEM (Sigma). All media were supplemented with 10% FBS and 100 units/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B. HUVEC stably expressing EGFP were generated by transfection with the vector pCEFL-EGFP and selection in 1 mg/ml G418 (CellGro, Herndon, VA).
RNA Array Analysis.
Human microarrays consisting of 22,464 spots for the Operon Human Version 2.0 and composed by 70-mer oligonucleotides each were used. cDNA probes were synthesized as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. The fluorescence ratio of each gene was quantified relative to common reference RNA.
Immunoblot Analysis.
Cells were lysed and 100 μg of protein was subjected to SDS/polyacrylamide gel electrophoresis as described in ref. 36. Antibodies anti-Sema4D, and anti-tubulin were from BD Pharmingen. For anti-plexin-B1, a peptide specific to the intracellular domain of plexin-B1 (CLTKHVLRENQDYVPGERT) was conjugated to keyhole limpet hemocyanin and used to immunize rabbits (Spring Valley Laboratories, Sykesville, MD). The antiserum was purified against the peptide by using the SulfoLink Gel Kit (Pierce), and specificity was determined by Western blotting using HEK293 cells transfected with plexin-B1, -B2, -B3, and -D1 as controls.
Detection of Stress Fibers.
Endothelial cells stably expressing EGFP were cocultured with the indicated cell types on sterile glass coverslips in 35-mm six-well plates for 12 h, with or without Sema4D-blocking antibody (BD Transduction Laboratories). Stress fiber formation was determined as described previously (14). Briefly, the number of endothelial cells exhibiting stress fiber polymerization in the cocultures was counted, and the results are expressed as a percentage of the total number of endothelial cells counted.
Migration Assays.
Serum-free EBM containing the indicated cell type or chemoattractant was placed in the bottom well of a Boyden chamber, with or without 10 μg/ml Sema4D-blocking antibody (BD Transduction Laboratories), whereas serum-free EBM containing HUVEC was added to the top chamber. The migration assay was performed as described in ref. 14.
Sema4D shRNA.
The shRNA sequences for human Sema4D were obtained from Cold Spring Harbor Laboratory’s RNAi library (RNAi Codex; http://katahdin.cshl.org:9331/homepage/portal/scripts/main2.pl) (22, 23). Oligonucleotides (Invitrogen) based on the following sequence worked best to knock down Sema4D protein levels: 5′-GGCCTGAGGACCTTGCAGAAGA-3′. Oligonucleotides were digested with XhoI/EcoRI and cloned into pSHAG MAGIC2 (37, 38). Where indicated, pSHAG MAGIC2 or pSHAG MAGIC2 Sema4D shRNA was transfected into HNSCC cells by using Lipofectamine Plus (Invitrogen) supplemented with CombiMag transfection agent (Oz Biosciences, Marseille, France) to achieve high transfection efficiency.
Lentivirus Infections.
Because the pSHAG MAGIC2 vector is Gateway (Invitrogen) compatible, an LR reaction was performed according to the supplier’s instructions to transfer the Sema4D shRNA insert into pWPI GW, a Gateway-compatible CSCG-based lentiviral destination vector. Viral stocks were prepared and infections were performed as reported in refs. 14 and 25.
In Vivo Tumorigenesis Assay.
Two million HN12, UMSCC 11B, Cal27, or Hep2 cells infected with control lentivirus or virus coding for Sema4D shRNA were resuspended in 250 μl of serum-free DMEM with an equal volume of Cultrex basement membrane extract (Trevigen, Rockville, MD) and injected s.c. into nude mice. After tumor growth had been recorded, animals were killed and tumors were removed, photographed, and processed for microscopy as described in ref. 24.
Immunohistochemistry.
Slides were hydrated through graded alcohols, incubated in 3% hydrogen peroxide for 30 min and 1% horse serum for 1 h at room temperature, and treated with anti-CD31 primary antibody (anti-PECAM; BD Pharmingen; 1:100 dilution), Sema4D antibody (BD Transduction Laboratories; 1:50 dilution), or plexin-B1 antibody (Spring Valley Laboratories; 1:100 dilution), overnight at 4°C. The slides were washed in PBS, incubated with biotinylated secondary antibody (1:400 dilution; Vector Laboratories) for 1 h, and treated with ABC complex (Vector Stain Elite; Vector Laboratories) for 30 min at room temperature. The slides were developed in 3,3-diaminobenzidine (FASTDAB tablet; Sigma) and counterstained with hematoxylin. Images were taken by using a SPOT digital camera attached to an Axiophot microscope (Zeiss).
Human Tissues.
Multitumor tissue arrays, containing carcinoma samples as well as normal controls, were obtained from the Tissue Array Research Program (TARP) Laboratory, National Cancer Institute (TARP1; www.cancer.gov/tarp). Individual paraffin blocks of formalin-fixed tissues from normal/hyperplastic epithelium, dysplasias, and invasive squamous cell carcinomas of the oral cavity and HNSCC tissue arrays containing ≈700 samples of oral HNSCC and normal control tissues were obtained from the National Institute of Dental and Craniofacial Research, National Institutes of Health Oral Cancer Tissue Array Initiative (A. A. Molinolo and J.S.G., unpublished work). All samples from both arrays were obtained from paraffin-embedded samples and were processed by following TARP methodology (www.cancer.gov/tarp).
Supplementary Material
Acknowledgments
We thank Daniel Martin for design and production of lentiviral shRNAs; Panomwat Amornphimoltham, Lynn Cross, Vyomesh Patel, Ana Barac, Silvia Montaner, Akrit Sodhi, Nathalia Velloso, and Ivette Hernandez Negrete for support and critical comments; Alfredo Molinolo for providing the HNSCC tissue arrays; and Myung Hee Park for the normal oral keratinocyte and immortalized oral keratinocyte RNA.
Abbreviations
- HEK
human embryonic kidney cells
- HNSCC
head and neck squamous cell carcinoma
- HUVEC
human umbilical vein endothelial cells
- Sema4D
semaphorin 4D
- shRNA
short hairpin RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bergers G., Benjamin L. E. Nat. Rev. Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P. Nat. Med. 2000;6:389–395. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- 3.Folkman J. Nat. Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 4.Auguste P., Gursel D. B., Lemiere S., Reimers D., Cuevas P., Carceller F., Di Santo J. P., Bikfalvi A. Cancer Res. 2001;61:1717–1726. [PubMed] [Google Scholar]
- 5.Dor Y., Porat R., Keshet E. Am. J. Physiol. 2001;280:C1367–C1374. doi: 10.1152/ajpcell.2001.280.6.C1367. [DOI] [PubMed] [Google Scholar]
- 6.Neufeld G., Cohen T., Gengrinovitch S., Poltorak Z. FASEB J. 1999;13:9–22. [PubMed] [Google Scholar]
- 7.Autiero M., De Smet F., Claes F., Carmeliet P. Cardiovasc. Res. 2005;65:629–638. doi: 10.1016/j.cardiores.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Carmeliet P., Tessier-Lavigne M. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 9.Tessier-Lavigne M., Goodman C. S. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 10.Raper J. A. Curr. Opin. Neurobiol. 2000;10:88–94. doi: 10.1016/s0959-4388(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 11.Takashima S., Kitakaze M., Asakura M., Asanuma H., Sanada S., Tashiro F., Niwa H., Miyazaki J.-i., Hirota S., Kitamura Y., et al. Proc. Natl. Acad. Sci. USA. 2002;99:3657–3662. doi: 10.1073/pnas.022017899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao H. Q., Soker S., Feiner L., Alonso J. L., Raper J. A., Klagsbrun M. J. Cell Biol. 1999;146:233–242. doi: 10.1083/jcb.146.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gluzman-Poltorak Z., Cohen T., Herzog Y., Neufeld G. J. Biol. Chem. 2000;275:18040–18045. doi: 10.1074/jbc.M909259199. [DOI] [PubMed] [Google Scholar]
- 14.Basile J. R., Barac A., Zhu T., Guan K. L., Gutkind J. S. Cancer Res. 2004;64:5212–5224. doi: 10.1158/0008-5472.CAN-04-0126. [DOI] [PubMed] [Google Scholar]
- 15.Conrotto P., Valdembri D., Corso S., Serini G., Tamagnone L., Comoglio P. M., Bussolino F., Giordan S. Blood. 2005;105:4321–4329. doi: 10.1182/blood-2004-07-2885. [DOI] [PubMed] [Google Scholar]
- 16.Giordano S., Corso S., Conrotto P., Artigiani S., Gilestro G., Barberis D., Tamagnone L., Comogli P. M. Nat. Cell Biol. 2002;4:720–724. doi: 10.1038/ncb843. [DOI] [PubMed] [Google Scholar]
- 17.Mao L., Hong W. K., Papadimitrakopoulou V. A. Cancer Cell. 2004;5:311–316. doi: 10.1016/s1535-6108(04)00090-x. [DOI] [PubMed] [Google Scholar]
- 18.Kumanogoh A., Kikutani H. Trends Immunol. 2001;22:670–676. doi: 10.1016/s1471-4906(01)02087-7. [DOI] [PubMed] [Google Scholar]
- 19.Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elhabazi A., Delaire S., Bensussan A., Boumsell L., Bismuth G. J. Immunol. 2001;166:4341–4347. doi: 10.4049/jimmunol.166.7.4341. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Kumanogoh A., Watanabe C., Shi W., Yoshida K., Kikutani H. Blood. 2001;97:3498–3504. doi: 10.1182/blood.v97.11.3498. [DOI] [PubMed] [Google Scholar]
- 22.Siolas D., Lerner C., Burchard J., Ge W., Linsley P. S., Paddison P. J., Hannon G. J., Cleary M. A. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 23.Hannon G. J., Conklin D. S. Methods Mol. Biol. 2004;257:255–266. doi: 10.1385/1-59259-750-5:255. [DOI] [PubMed] [Google Scholar]
- 24.Amornphimoltham P., Sriuranpong V., Patel V., Benavides F., Conti C. J., Sauk J., Sausville E. A., Molinolo A. A., Gutkind J. S. Clin. Cancer Res. 2004;10:4029–4037. doi: 10.1158/1078-0432.CCR-03-0249. [DOI] [PubMed] [Google Scholar]
- 25.Naldini L., Blomer U., Gage F. H., Trono D., Verma I. M. Proc. Natl. Acad. Sci. USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bielenberg D. R., Hida Y., Shimizu A., Kaipainen A., Kreuter M., Kim C. C., Klagsbrun M. J. Clin. Invest. 2004;114:1260–1271. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler O., Shraga-Heled N., Lange T., Gutmann-Raviv N., Sabo E., Baruch L., Machluf M., Neufeld G. Cancer Res. 2004;64:1008–1015. doi: 10.1158/0008-5472.can-03-3090. [DOI] [PubMed] [Google Scholar]
- 28.Xiang R., Davalos A. R., Hensel C. H., Zhou X. J., Tse C., Naylor S. L. Cancer Res. 2002;62:2637–2643. [PubMed] [Google Scholar]
- 29.Xiang R. H., Hensel C. H., Garcia D. K., Carlson H. C., Kok K., Daly M. C., Kerbacher K., van den Berg A., Veldhuis P., Buys C. H., Naylor S. L. Genomics. 1996;32:39–48. doi: 10.1006/geno.1996.0074. [DOI] [PubMed] [Google Scholar]
- 30.Roche J., Boldog F., Robinson M., Robinson L., Varella-Garcia M., Swanton M., Waggoner B., Fishel R., Franklin W., Gemmill R., Drabkin H. Oncogene. 1996;12:1289–1297. [PubMed] [Google Scholar]
- 31.Neufeld G., Cohen T., Shraga N., Lange T., Kessler O., Herzog Y. Trends Cardiovasc. Med. 2002;12:13–19. doi: 10.1016/s1050-1738(01)00140-2. [DOI] [PubMed] [Google Scholar]
- 32.Cross M. J., Claesson-Welsh L. Trends Pharmacol. Sci. 2001;22:201–207. doi: 10.1016/s0165-6147(00)01676-x. [DOI] [PubMed] [Google Scholar]
- 33.Iruela-Arispe M. L., Dvorak H. F. Thromb. Haemostasis. 1997;78:672–677. [PubMed] [Google Scholar]
- 34.Buschmann I., Heil M., Jost M., Schaper W. Microcirculation. 2003;10:371–379. doi: 10.1038/sj.mn.7800199. [DOI] [PubMed] [Google Scholar]
- 35.Carmeliet P., Collen D. Ann. N.Y. Acad. Sci. 2000;902:249–262. doi: 10.1111/j.1749-6632.2000.tb06320.x. discussion 262–264. [DOI] [PubMed] [Google Scholar]
- 36.Basile J. R., Afkhami T., Gutkind J. S. Mol. Cell. Biol. 2005;25:6889–6898. doi: 10.1128/MCB.25.16.6889-6898.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paddison P. J., Cleary M., Silva J. M., Chang K., Sheth N., Sachidanandam R., Hannon G. J. Nat. Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 38.Paddison P. J., Caudy A. A., Sachidanandam R., Hannon G. J. Methods Mol. Biol. 2004;265:85–100. doi: 10.1385/1-59259-775-0:085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.