Abstract
Centrosomes are the major microtubule-organizing center in animal cells. They are composed of a pair of [9(3) + 0] centrioles surrounded by a relatively ill-defined pericentriolar matrix, provide the ciliary centriole–kinetosome (basal body) progenitor, and organize the assembly of microtubules into the mitotic spindle during cell division. Despite >100 years of microscopic observation and their obvious significance, our understanding of centrosome composition, dynamic organization, and mechanism of action is limited when compared with that of other cellular organelles. Centrosomes duplicate only once per cell cycle to ensure development of a normal bipolar spindle. The initial event in centrosome duplication is centriole replication, which is generative, semiconservative, and independent of the nucleus. Such observations led to the proposal that centrosomes contain their own complement of nucleic acids, possibly representative of an organellar genome comparable with those described for mitochondria and chloroplasts. The consensus in the field is that centrosomes lack DNA but may contain RNA. We isolated centrosomes from oocytes of the surf clam, Spisula solidissima, and purified from them a unique set of RNAs. We show here by biochemical means and subcellular in situ hybridization that the first transcript we analyzed is intimately associated with centrosomes. Sequence analysis reveals that this centrosome-associated RNA encodes a conserved RNA-directed polymerase domain. The hypothesis that centrosomes contain an intrinsic complement of specific RNAs suggests new opportunities to address the century-old problem of centrosome function, heredity, and evolution.
Keywords: centriole, centrosomal RNA, microtubule-organizing center
The process of centrosome duplication is tightly regulated and under the control of both cytoplasmic and intrinsic factors (1, 2) to ensure that a normal bipolar spindle is formed during mitosis and that the genome is divided equally between daughter cells. The first observable event in centrosome duplication is centriole replication. Like mitochondria and chloroplasts, centriole replication in embryos is independent of the nucleus, but unlike the former two classes of organelles, centrioles replicate not by fission but in a semiconservative, generative manner. These and other observations have led some investigators to propose that the centrosome (or more specifically, the centriole) is of endosymbiotic evolutionary origin and, like mitochondria and chloroplasts, may contain its own genome (3).
The question of whether centrosomes or centrioles contain nucleic acids has remained unresolved since biochemical studies of the mitotic apparatus in the 1950s revealed “pentosenucleic acid” in the polar regions (4–8). The consensus, expressed in the most recent review on the subject, concludes that evidence for DNA in centrosomes is lacking and that significant evidence exists to the contrary (9). The existence of RNA in these organelles, however, remains an open question, although much of the historical evidence for the existence of centrosomal RNA is indirect (10–18).
The size and developmental dynamics of surf clam (Spisula solidissima) oocyte centrosomes permits isolation of relatively large quantities of material for direct analysis. We have extracted and cloned a unique set of RNAs from Spisula oocyte centrosomes, which we refer to as cnRNAs (centrosomal RNAs). We report on five of these molecules, focusing on the molecular characterization and subcellular localization of cnRNA11, an ≈4-kb transcript with a conserved reverse transcriptase domain.
Results
We report on five clones (cnRNAs 11, 102, 113, 170, and 184) that range in size from 638–869 bp. Exhaustive analysis revealed no database matches for these sequences. The average E value of the best matches obtained was 2.7e−1. By comparison, a smaller E value of 1.0e−1 was obtained in a blast analysis of a manually generated, random 600-nt oligomer. We examined whether negative blast reports might have resulted from the divergence of surf clam sequences from other well represented species in protein and nucleic acid databases by arbitrarily selecting 600-bp fragments of 12 known Spisula sequences (of 20 available) for analysis: 18s rRNA, cdc20, cyclin A, cyclin B, cytochrome B, eIF4a, G protein-coupled receptor, histone H3, p41, p53-like transcription factor, poly(A)-binding protein, and ribonucleotide reductase. Reports for these RNAs returned with significant matches to a variety of species. Probability values for the best non-Spisula matches averaged e−82, with a range of 4e−20 (cdc20) to e−138 (eIF4a), excluding 18s rRNA (0.0e00). We conclude that the lack of database matches for the five cnRNAs was a result of their uniqueness rather than species differences.
To determine whether these cloned cnRNAs were actually enriched in centrosomes, we isolated RNA from centrosomes and from whole oocyte lysates, reverse-transcribed 2 μg of each, and performed PCR using primers for the five putative cnRNAs as well as other, known Spisula cytoplasmic transcripts. If our cnRNAs are enriched in centrosomes, we would expect cnRNA PCR product to be generated from our centrosomal, but not cytoplasmic, template. Two notes regarding these cytoplasmic RNA preparations. First, they were made from whole oocyte lysate and therefore contain a normal complement of centrosomal components. Trace levels of cnRNA PCR product may therefore be expected using whole oocyte lysate for template. Second, oocyte lysates were prepared in the presence and absence of RNase inhibitors, which had no effect on the outcome of the experiment. This point is important because it ensures that RNAs from the cytoplasmic pool were not selectively protected by artifactual association with centrosomes while their cytosolic equivalents were exposed to degradation. Primers for S. solidissima poly(A)-binding protein and ribonucleotide reductase were used for controls as typical cytoplasmic mRNAs. Finally, we probed for Spisula 18s rRNA as a “loading” control, given the likelihood that it is a major contaminant in centrosome (or any other fractionated cell) preparations.
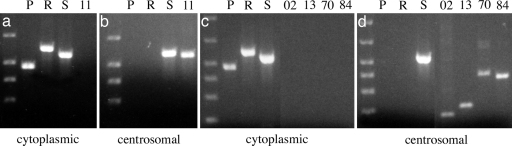
Ample quantities of cnRNA PCR product were generated from centrosomal template, whereas little or no cnRNA product was detected in whole oocyte cytoplasm (Fig. 1). Neither poly(A)-binding protein nor ribonucleotide reductase mRNAs were detected in centrosomal template, although both were well represented in cytoplasmic template. The loading control, 18s rRNA, appeared in similar amounts in both centrosomal and cytoplasmic templates. These results provide biochemical evidence that cnRNAs are concentrated in centrosomes. Regarding the distribution of 18s rRNA between the two cellular fractions, we reference two interesting articles suggesting a physiological association of ribosomes with centrosomes and the spindle pole (19, 20). It is possible that the presence of 18s rRNA in our centrosomal template sample represents something more than a simple case of contamination.
Fig. 1.
Differential expression of cnRNAs in centrosomes vs. whole oocyte cytoplasm. Equal quantities of RNA isolated from either whole oocyte cytoplasm or centrosomal fractions were reverse transcribed and used as template in PCR with primers directed against known cytoplasmic RNAs or putative cnRNAs. Conditions such as number of cycles, template input, and product sizes were chosen empirically to yield product in the linear range of amplification. The results of two separate experiments are shown. (a and b) The distribution of cnRNA11 between cytoplasmic and centrosomal fractions, respectively. (c and d) The distribution between cytoplasmic and centrosomal fractions of cnRNAs 102 (02), 113 (13), 170 (70), and 184 (84). The first lane in each panel is a 100-bp DNA reference ladder. P, poly(A)-binding protein RNA; R, ribonucleotide reductase; S, 18s rRNA subunit. Trace quantities of cnRNA PCR product can sometimes be seen in cytoplasmic template lanes but are not visible in these panels.
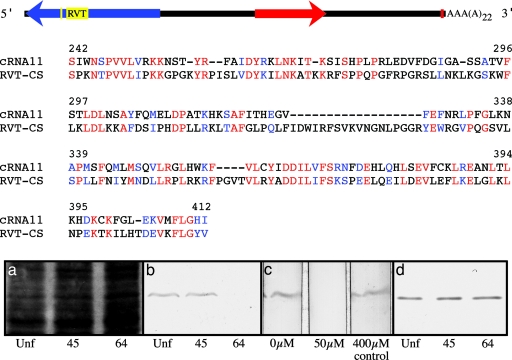
RACE was used to obtain the full-length sequence of cnRNA11 (diagrammed in Fig. 2 Top) which, at 3,863 nt, is relatively large for a cytoplasmic transcript. Our original cnRNA11 clone represents nucleotides 2,998–3,636 in the full-length sequence. The sense strand was defined by the presence of a poly(A) tail at the 3′ end and a canonical polyadenylation signal 29 nucleotides upstream. Despite extensive analysis, no bona fide ORF could be found in the sense strand, but a substantial ORF was readily mapped in the antisense strand. The predicted polypeptide encoded in this ORF includes a highly conserved 200-aa reverse transcriptase domain. This domain exhibits striking homology (P = e−75) to Zea mays putative polyprotein, a member of the RNA-dependent nucleotide polymerase family of proteins, Pfam00078. In addition, a ribonucleoprotein consensus RNA-binding site (RNP-1) was found downstream from the reverse transcriptase domain. We examined whether cnRNA11 protein is translated in cells using a polyclonal antibody generated against a predicted 14-aa oligopeptide from the reverse transcriptase domain (Fig. 2 Bottom). Although not useful for immunolocalization, this antibody recognized a single polypeptide of the predicted size in Western blots of whole cell lysates. The cnRNA11 protein was present in comparable quantities in both unactivated oocytes and zygotes at 45 min after fertilization. However, it was not detected after completion of first cleavage (65 min after activation), suggesting a cell cycle-dependent regulation of expression.
Fig. 2.
Structure of cnRNA11. (Top) A canonical polyadenylation signal 29 nucleotides upstream from the poly(A) tail is represented by the small red box near the 3′ end in the diagrammatic representation shown. The red arrow represents the 638-nt sequence originally cloned from centrosome preparations, 5′ to 3′ orientation (this region was used to generate probe for the in situ hybridizations shown in Fig. 3). An ORF predicting a 54-kDa polypeptide discovered in the antisense strand (arrowhead indicating direction from 3′ to 5′ relative to the sense strand) is shown in blue, with RNA-dependent polymerase (RVT) and RNP-1 domains shown in yellow. (Middle) Alignment of the cnRNA11 reverse transcriptase domain with the pfam00078 reverse transcriptase consensus sequence. Identities are shown in red, and conservative substitutions are shown in blue. Bit score = 94.7, P = 5e−21, GenBank accession no. DQ359732. (Bottom) Western blot analysis of cnRNA11 protein expression in early embryos. (a) A segment of Coomassie blue-stained polyacrylamide gel indicating relative protein loads (50 μg per lane) for lysed unfertilized (Unf) oocytes and 45 and 64 min zygotes. (b) A blot showing relative levels of cnRNA11 protein at these three developmental time points. Controls for specificity included preimmune serum as well as preabsorption of immune serum with the antigenic peptide. Results with preimmune serum were negative (not shown). Preabsorption controls are shown in c, which depicts antibody staining of three blot strips in the absence of peptide (0 μM), after incubation of serum with 50 μM antigenic peptide to preabsorb immunoreactivity, and after incubation with 400 μM nonrelevant peptide, which had no effect on immunoreactivity. (d) A Western blot using an antibody to an unknown Spisula protein and, in addition to a, serves as a loading control for the down-regulation of cnRNA11 protein depicted in b.
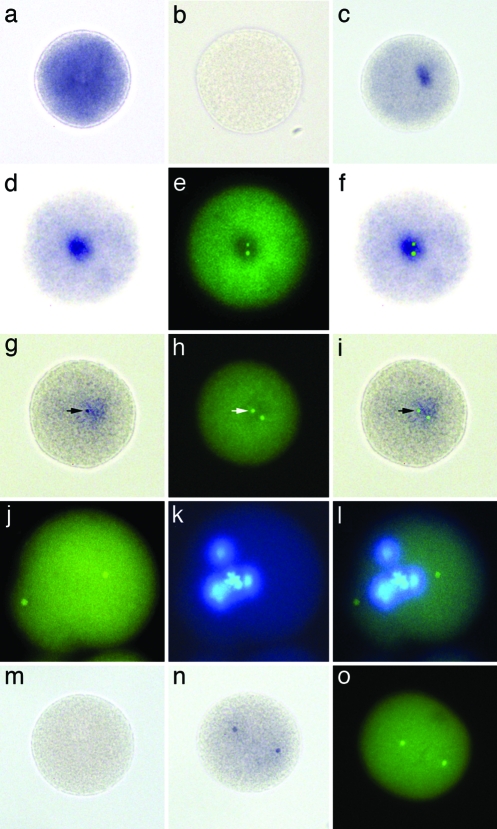
The development of two high sensitivity and high stringency in situ hybridization protocols was required to localize cnRNA11 in cells (see Materials and Methods). Both methods significantly impaired protein antigenicity, yet we successfully colabeled cnRNA11 with γ-tubulin protein using the first method described. Although the second (step-down) method ablated protein antigenicity, it had the advantage of partially preserving chromosome integrity. For controls, we used sense and antisense poly(A)-binding protein probes to reveal the distribution of a known, cytoplasmically distributed mRNA. As expected, poly(A)-binding protein was distributed throughout the zygote cytoplasm, independent of hybridization method (Fig. 3 a and b). Cells labeled with our cnRNA probe complementary to the ORF, by contrast, exhibited a discrete patch of staining in the cytoplasm (Fig. 3c). In cells double-labeled with this same cnRNA11 hybridization probe and antibodies to the centrosome marker protein, γ-tubulin, centrosomes were observed to be surrounded by the cnRNA11 patches (Fig. 3 d–f). This was true whether only a single γ-tubulin-stained focal point was evident (early zygotic time points) or when two foci were resolved in close proximity. It is likely that these patches observed in situ are slightly exaggerated in size due to diffusion of the colored alkaline phosphatase reaction product, especially in light of our PCR results (Fig. 1). More intensely stained zones, or distinct punctae, were sometimes observed within the cnRNA11 patches. By controlling the time of development of alkaline phosphatase reaction product in our in situ hybridizations, these densities were often more apparent and could be seen to coincide with the γ-tubulin-labeled centrosomes (Fig. 3 g–i). This finding confirms that cnRNA11 is intimately associated with centrosomes in situ.
Fig. 3.
In situ localization of cnRNA11. Spisula zygote labeled with antisense (a) and sense (b) probes for poly(A)-binding protein. Intense stain for poly(A)-binding protein mRNA is widely distributed in the cytoplasm. (c) Zygote hybridized with cnRNA11 probe complementary to the ORF shows single patch labeling pattern. (d–f) Another zygote labeled with this cnRNA11 hybridization probe (d) and colabeled with antibodies to the centrosome marker protein, γ-tubulin (e). An overlay of d and e is shown in f. The two γ-tubulin-labeled centrosomes are encased within the cnRNA11 hybridization patch. The cell depicted in g–i is from a sample developed for only 3 days as opposed to the typical 6-day development period for our in situ hybridizations. A densely stained cnRNA11 puncta is clearly visible (arrow in g), which coincides precisely with the γ-tubulin-labeled centrosome seen in h; an overlay of g and h is shown in i. The hybridization protocol used for labeling the cells (a–i) included 1% SDS in the prehybridization and hybridization steps. Chromatin structure and protein antigenicity are significantly compromised in these samples, although the monoclonal anti-γ-tubulin antibody used here was effective. (j–l) An embryo labeled (green) by fluorescent step-down in situ hybridization. Protein antigenicity is destroyed in the step-down method, but chromatin organization, although impaired, is better preserved than in SDS-treated embryos. A mitotic figure is discernable in the Hoechst-stained image of this cell (k). (l) Overlay of j and k. (m) Example of a zygote hybridized with cnRNA11 control probe; hybridization signal was not seen at any stage after activation. (n) Zygote fixed 16 min after activation and labeled with hybridization probe complementary to the cnRNA11 ORF by using the SDS protocol. Two distinct “kernel” structures are visible. These kernels did not stain with anti-γ-tubulin. Another 16-min zygote, prepared for routine immunocytochemistry and labeled with anti-γ-tubulin, is shown in o to compare the appearance and spatial organization of centrosomes with cnRNA11 kernels at the same time point. A nonspecific background signal is seen in all fluorescent images, attributable to the paraformaldehyde-fixed chorion.
In addition to the centrosome-associated cnRNA patches described above, one additional staining pattern was observed: that of two distinctly placed punctae, which we refer to as “kernels” (Fig. 3n). These kernels were of similar size and shape to centrosomes and exhibited similar spatial coordinates to centrosomes at later stages of separation. They did not stain with γ-tubulin antibodies. In the absence of tubulin staining, we do not have sufficient information at this time to be certain of the relationship of these paired structures to centrosomes, if any.
Discussion
We identified five RNAs derived from isolated surf clam centrosomes which we call cnRNAs. Exhaustive analysis revealed no matches to known nucleotide, translated nucleotide, or predicted protein sequences for the five cnRNAs, even though strong identity was found for 12 Spisula cytoplasmic RNAs that served as controls in our database analysis. We therefore conclude that the cnRNAs are a unique group. The lack of database matches is important because it has been proposed that, if centrosomes do contain RNA, it could represent a transient association with nuclear transcripts being shuttled to specific parts of the cell for localized translation (9). There are examples to suggest this occurs (21, 22). However, it is critical to note that in these specific examples, the transcripts identified were sufficiently abundant to have been detected in, and originally cloned, as part of the cytoplasmic mRNA pool. Their homologues are readily found in existing nucleotide and protein databases. The cnRNAs have not been identified as part of the nuclear gene pool, either in libraries based on RNA templates or through direct genome sequencing. This is consistent with the hypothesis that they are, at some level, intrinsic to the centrosome.
The identification of a reverse transcriptase domain in cnRNA11 might also prompt the question of whether cnRNAs are the product of a bacterial endosymbiont or if they are due to a viral infection of Spisula oocytes or contamination of isolated centrosomes. Wolbachia has been shown to interact with microtubules in Drosophila oocytes (23), and vaccinia virus can localize to centrosomes in cultured mammalian cells (24). However, previous EM analysis did not reveal bacteria or viral particles in isolated Spisula centrosomes (25, 26). Also, no significant cnRNA levels were revealed by PCR analysis in cytoplasmic extracts of Spisula oocytes. Similar levels of viral or bacterial sequences to those found in centrosomes would be expected. Finally, apart from the reverse transcriptase domain in cnRNA11, none of the cnRNA sequences resembled entries in current viral or bacterial databases.
The five cnRNAs copurify with centrosomes. Potential cytoplasmic mRNA contaminants, such as poly(A)-binding protein and ribonucleotide reductase, were not detected in centrosome fractions shown to be rich in cnRNAs. In situ hybridization experiments revealed the colocalization of cnRNA patches with γ-tubulin-containing centrosomes. Finally, cnRNA11 described here possesses a conserved reverse transcriptase domain, consistent with the 30-year-old speculation of Went that the primary genome of the centrosome is RNA that is replicated with the aid of a centrosome-associated reverse transcriptase (27). These observations suggest that the centrosome may carry at least a portion of its own genetic machinery.
Materials and Methods
cnRNA Library Development.
Centrosomes from activated S. solidissima oocytes were isolated as described (28–30). These centrosomes are functionally competent to form asters and duplicate and have been extensively characterized. RNA was extracted from the peak sucrose density gradient fraction by using Qiagen RNeasy reagents, reverse transcribed (RETROscript; Ambion, Austin, TX) and amplified by PCR by using the random primers and methods described by Froussard (31) and Von Eggeling and Spielvogel (32), and blunt-cloned into PCR-Script plasmid (Stratagene). To guard against the potential amplification and subsequent cloning of genomic DNA contaminants, equal quantities of non-reverse-transcribed cnRNA were used as a control template. Non-reverse-transcribed cnRNA yielded no detectable products from random PCR or, subsequently, cloned inserts.
RT-PCR Screening for Centrosome Enrichment.
Equal quantities of total RNA isolated from whole oocyte lysate and RNA isolated from peak centrosome fractions were reverse transcribed and used as PCR template to test for enrichment of putative cnRNAs. Internal controls, primer pairs for known Spisula cytoplasmic RNAs, included poly(A)-binding protein, ribonucleotide reductase, and 18s rRNA. PCR conditions determined in preliminary experiments to generate reaction product in the linear range of yield for putative cnRNAs were used: 20 ng of template; 300- to 500-bp amplification products; and 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72° for 1 min.
Molecular and Immunological Analysis of cnRNA11.
Full-length sequence data for cnRNA11 was obtained by RACE by using Invitrogen Gene Racer reagents. macvector software (Accelrys, Inc., San Diego) was used for ORF and structural analyses. Databases accessed for similarity searches included nonredundant eukaryotic, prokaryotic, and viral nucleotide (via blastn, blastx, and tblastxprograms), dbEST (via tblastx), human genome (blastn), human RefSeq protein (blastx), mouse genome (blastn), mouse RefSeq protein (blastx), Zebrafish mRNA and reference proteins (blastn and blastx), nematode mRNA and protein (blastn and blastx), plant (Triticum and Arabidopsis) DNA and protein, respectively (blastn and blastx), and insect (Drosophila), fungal (all), malarial (Plasmodium falciparum and Plasmodium yoelii), and microbial (all) DNA and protein databases (via blastn and blastx). Antipeptide polyclonal antibodies were generated against amino acids 272–285 (KSISHPLPRLEDVF) in the conserved reverse transcriptase domain of cnRNA11. The Western blot and immunofluorescence methods we used to stain cnRNA11 protein and γ- and α-tubulin are described in ref. 33.
In Situ Localization of cnRNA11.
Gravid S. solidissima were obtained from the Marine Resources Department at the Marine Biological Laboratory. Oocytes and zygotes were fixed for in situ hybridization for 2 hours at 4°C in 4% paraformaldehyde in 3-(N-morpholino) propanesulfonic acid (Mops) buffer (pH 7.4). They were subsequently washed by settling and resuspension twice in Mops buffer and once in 0.5 M NaCl, dehydrated in a graded series of ethanols to 70%, and stored at −20° until use. We developed two in situ hybridization methods to localize cnRNA11 at the subcellular level with high sensitivity and low background. One employs 1% SDS in the prehybridization and hybridization solutions [M. Martindale, personal communication; and Lee et al. (34)]. Chromatin structure was damaged by using this protocol, and endogenous protein antigenicity was greatly reduced. This problem is not uncommon during in situ hybridization, given the use of relatively high temperatures and proteinase K to expose target in most protocols, but was probably exacerbated by the inclusion of SDS at high temperatures for >3 days in this particular protocol. Our second method for in situ hybridization anticipated the possibility that target remained masked, either by components of the centrosome itself or by interaction between sense and antisense RNA strands. The protocol was therefore designed with exaggerated protease treatment to ameliorate persistent protein masking, and the temperature was raised to 94° for 10 min to melt duplex. Because the window available for hybridization was expected to close as the temperature was lowered into a range typical for in situ hybridization, the temperature was then reduced to final hybridization temperature in 5°C steps of 20 min each. Although protein antigenicity was destroyed by using this “step-down” method, chromatin organization was better preserved than in SDS-treated embryos. Samples were hybridized with digoxygenin-labeled RNA probes for 3 days at 60°C. Alkaline phosphatase reaction product was visualized after development for 6 days at 4°C with gentle rocking.
Acknowledgments
We thank Drs. Judith Venuti, David Adelson, and Conly Rieder for reading and commenting on the manuscript and Dr. Max Oeschger for enlightening and enthusiastic conversation on the subject matter. M.C.A. is supported by National Institutes of Health Grant GM075163, and R.E.P. is supported by National Institutes of Health Grant GM043264.
Abbreviation
- cnRNA
centrosomal RNA.
Footnotes
References
- 1.Sluder G., Hinchcliffe E. H. Biol. Cell. 1999;91:413–427. [PubMed] [Google Scholar]
- 2.Wong C., Stearns T. Nat. Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- 3.Chapman M. J., Dolan M. F., Margulis L. Q. Rev. Biol. 2000;75:409–429. doi: 10.1086/393621. [DOI] [PubMed] [Google Scholar]
- 4.Stich H. Chromosoma. 1954;6:199–236. [PubMed] [Google Scholar]
- 5.Mazia D. Symp. Soc. Exp. Biol. 1955;9:335–357. [Google Scholar]
- 6.Shimamura T., Ota T. Exp. Cell Res. 1956;11:346–361. doi: 10.1016/0014-4827(56)90111-2. [DOI] [PubMed] [Google Scholar]
- 7.Rustad R. C. Exp. Cell Res. 1959;16:575–583. doi: 10.1016/0014-4827(59)90125-9. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman A. Exp. Cell Res. 1960;20:529–547. doi: 10.1016/0014-4827(60)90122-1. [DOI] [PubMed] [Google Scholar]
- 9.Marshall W. F., Rosenbaum J. L. Curr. Top. Dev. Biol. 2000;49:187–205. doi: 10.1016/s0070-2153(99)49009-x. [DOI] [PubMed] [Google Scholar]
- 10.Dippel R. V. J. Cell Biol. 1976;69:622–637. doi: 10.1083/jcb.69.3.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rieder C. L. J. Cell Biol. 1979;80:1–9. doi: 10.1083/jcb.80.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiedemann S. R., Sander G., Kirschner M. W. Cell. 1977;10:337–350. doi: 10.1016/0092-8674(77)90021-6. [DOI] [PubMed] [Google Scholar]
- 13.Pepper D. A., Brinkley B. R. Cell Motil. Cytoskeleton. 1980;1:1–15. doi: 10.1002/cm.970010102. [DOI] [PubMed] [Google Scholar]
- 14.Snyder J. A. Cell Biol. Int. Rep. 1980;4:859–868. doi: 10.1016/0309-1651(80)90184-8. [DOI] [PubMed] [Google Scholar]
- 15.Zackroff R. V., Rosenfeld A. C., Weisenberg R. C. J. Supramol. Struct. 1976;5:577–589. doi: 10.1002/jss.400050412. [DOI] [PubMed] [Google Scholar]
- 16.Peterson S. P., Burns M. W. J. Cell Sci. 1978;34:289–301. doi: 10.1242/jcs.34.1.289. [DOI] [PubMed] [Google Scholar]
- 17.Klotz K., Dabauvalle M. C., Paintrand M., Weber T., Bornens M., Karsenti E. J. Cell Biol. 1990;110:405–415. doi: 10.1083/jcb.110.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartman H., Puma J. P., Gurney T. J. Cell Sci. 1974;16:241–259. doi: 10.1242/jcs.16.2.241. [DOI] [PubMed] [Google Scholar]
- 19.Hartman J. F., Zimmerman A. M. Exp. Cell Res. 1968;50:403–417. doi: 10.1016/0014-4827(68)90459-x. [DOI] [PubMed] [Google Scholar]
- 20.Kallenbach R. J. Biosci. Rep. 1983;3:1155–1162. doi: 10.1007/BF01120209. [DOI] [PubMed] [Google Scholar]
- 21.Groisman I., Huang Y. S., Mendez R., Cao Q., Theurkauf W., Richter J. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- 22.Lambert J. D., Nagy L. M. Nature. 2002;420:682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- 23.Ferree P. M., Frydman H. M., Li J. M., Cao J., Wieschaus E., Sullivan W. PLoS Pathog. 2005;1:11–24. doi: 10.1371/journal.ppat.0010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ploubidou A., Moreau V., Ashman K., Reckmann I., Gonzalez C., Way M. EMBO J. 2000;19:3932–3944. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel J. M., Stearns T., Rieder C. L., Palazzo R. E. J. Cell Biol. 1997;137:193–202. doi: 10.1083/jcb.137.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnackenberg B. J., Khodjakov A., Rieder C. L., Palazzo R. E. Proc. Natl. Acad. Sci. USA. 1998;95:9295–9300. doi: 10.1073/pnas.95.16.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Went H. A. J. Theor. Biol. 1977;68:95–100. doi: 10.1016/0022-5193(77)90230-2. [DOI] [PubMed] [Google Scholar]
- 28.Palazzo R. E., Vogel J. M. Methods Cell Biol. 1999;61:35–56. doi: 10.1016/s0091-679x(08)61974-3. [DOI] [PubMed] [Google Scholar]
- 29.Schnackenberg B. J., Palazzo R. E. Biol. Cell. 1999;91:429–438. [PubMed] [Google Scholar]
- 30.Schnackenberg B. J., Hull D. R., Balczon R. D., Palazzo R. E. J. Cell Sci. 2000;113:943–953. doi: 10.1242/jcs.113.6.943. [DOI] [PubMed] [Google Scholar]
- 31.Froussard P. Nucleic Acids Res. 1992;20:2900. doi: 10.1093/nar/20.11.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Von Eggeling F., Spielvogel H. Cell. Mol. Biol. 1995;41:653–670. [PubMed] [Google Scholar]
- 33.Alliegro M. C., Alliegro M. A. Dev. Dyn. 2005;232:216–221. doi: 10.1002/dvdy.20208. [DOI] [PubMed] [Google Scholar]
- 34.Lee P. N., Callaerts P., De Couet H. G., Martindale M. Q. Nature. 2003;424:1061–1065. doi: 10.1038/nature01872. [DOI] [PubMed] [Google Scholar]