Abstract
Cytoplasmic dynein, a minus-end-directed microtubule motor, has been implicated in many fundamental cellular processes; however, little is known regarding the underlying molecular machinery that regulates its stability. In Aspergillus nidulans, nuclear distribution gene C (nudC) has been implicated in the regulation of dynein-mediated nuclear migration. Here, we characterize a previously undescribed mammalian NudC-like protein (NudCL). The expression and phosphorylation of NudCL are increased during mitosis. Depletion of NudCL by RNA interference in HeLa cells inhibits cell growth and induces mitotic arrest with multiple mitotic defects, which subsequently result in cell death. Unexpectedly, the majority of NudCL depletion-induced mitotic defects may result from loss of dynein function; this interpretation is supported by the failure to recruit sufficient γ-tubulin to spindle poles and the mislocalization of the dynein complex from kinetochores, spindle microtubules, and spindle poles during mitosis. Depletion of NudCL also results in the aggregation of dynein intermediate chain throughout the cytoplasm during mitosis. NudCL was shown to bind to the dynein complex, and its depletion induces degradation of dynein intermediate chain, a process suppressed by MG132, a proteasome inhibitor. Taken together, these data suggest a previously undescribed mechanism whereby NudCL appears to influence the stabilization of dynein intermediate chain.
Keywords: chaperone, mitosis, proteasome, degradation, aggregation
Cytoplasmic dynein is a microtubule-associated minus-end-directed motor involved in many fundamental cellular processes, including nuclear migration, mitosis, maintenance of the Golgi apparatus, and intracellular trafficking (1, 2). Mammalian cytoplasmic dynein is a massive multisubunit complex composed of two heavy chains, several intermediate chains, light-intermediate chains, and various light chains (2, 3). This complexity allows cytoplasmic dynein to be regulated in subtle ways at multiple levels. At a molecular level, the regulation of cytoplasmic dynein has two major modes: regulation of its subunits and regulation through accessory proteins (2, 3). Among these associated regulators, the multisubunit dynactin complex plays a general regulatory role in cargo binding and processive movement of dynein (2).
Studies of nuclear migration in the fungus, Aspergillus nidulans, have uncovered at least seven different nuclear distribution (nud) genes (4). The characterization of nud mutants in A. nidulans that prevent nuclear migration into the mycelium revealed that the cytoplasmic dynein/dynactin complex is a major contributor to these processes (4). In mammalian cells, most nuclear distribution proteins are components of the cytoplasmic dynein/dynactin complex or appear to regulate dynein function (4, 5). The nudA, nudI, and nudG genes encode cytoplasmic dynein heavy, intermediate, and light chain, respectively, and the proteins encoded by nudK and nudM are actin-related protein 1 and p150 dynactin, elements of the dynactin complex (4, 6). The mammalian homolog of nudF is lissencephaly 1 (Lis1), a mutation that causes human lissencephaly (7). Lis1, together with mNudE/NdeI and NUDEL/Ndel1, mammalian homologues of NudE, seems to regulate cytoplasmic dynein (7, 8). A mutation in Aspergillus nudC induces the reduction of the protein level of NudF at restrictive temperature (9). Mammalian NudC associates with Lis1 and the dynein/dynactin complex (10, 11); however, the function of mammalian NudC remains largely unknown.
Recently, we demonstrated that mammalian NudC is required for mitosis and cytokinesis (11). Data from A. nidulans show that the nudC3 mutation greatly reduces the protein level of NudF (9). We sought to determine whether similar events occur in mammalian cells. Our data show that the protein level of Lis1 was not significantly affected by depletion of NudC in HeLa cells, suggesting another homolog of Aspergillus nudC in mammalian cells. Here we report a previously undescribed protein that shares significant homology with Aspergillus and human NudC. Our data show that this NudC-like protein (NudCL) plays an essential role in mitosis and contributes to stabilization of the dynein complex.
Results
Characterization of NudCL Protein.
To identify previously undescribed mammalian NudC homologs, we used the protein sequence of human NudC to search the database of National Center for Biotechnology Information with tblastn. We found an uncharacterized human sequence (GenBank; BC035014) homologous to human NudC and cloned this cDNA by RT-PCR with total RNA extracted from HeLa cells. It encoded a deduced 361-aa protein (GenBank; AAH35014) with a predicted molecular mass of 41 kDa; this protein shares significant homology with human NudC (identity, 26.0%; similarity, 59.0%) and Aspergillus NudC (identity, 32%; similarity, 50.4%), therefore, we named it NudC-like protein, NudCL. As is the case for Aspergillus and human NudC, NudCL has a predicted coiled-coil domain and a nuclear movement domain (pfam03593; National Center for Biotechnology Information) that contains a p23 domain (cd00237; National Center for Biotechnology Information; Fig. 1A).
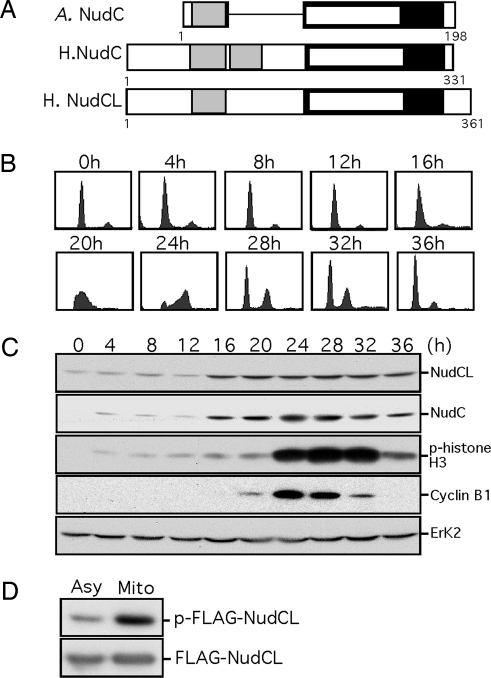
Fig. 1.
NudCL expression and phosphorylation are regulated during cell cycle progression. (A) Schematic comparison of human NudCL and Apergillus and human NudC. Light gray bars indicate coiled-coil domains, filled bars show nuclear movement domains, and open bars inside filled bars indicate p23 domains. A. NudC, Aspergillus NudC; H. NudC, human NudC; H. NudCL, human NudCL. (B) FACS profiles of T98G cells. Cells were serum-starved for 72 h and subsequently stimulated with 20% serum for the indicated times. DNA content is plotted versus cell number. (C) Immunoblot analyses of the indicated proteins during cell cycle progression. (D) NudCL is highly phosphorylated during mitosis. Asynchronous or mitotic HeLa cells transfected with pCMV-FLAG-NudCL were labeled with [32P]orthophosphate for 4 h. Cell lysates were immunoprecipitated with anti-FLAG antibody, resolved by SDS/PAGE and transferred onto membranes. Phosphorylated NudCL was visualized by autoradiography, and total immunoprecipitated NudCL was detected with anti-FLAG antibody. Asy, asynchronous; Mito, mitotic.
To characterize the function of NudCL, we raised a rabbit polyclonal antibody against GST-NudCL. After affinity purification, the anti-NudCL antibody specifically recognized NudCL protein (Fig. 7A, which is published as supporting information on the PNAS web site). Immunoblot analysis by using this antibody showed that NudCL protein is ubiquitously expressed (Fig. 7 B and C). We also found that, in interphase, NudCL colocalizes with both Golgi matrix protein GM130 and Golgi p58 protein in addition to some punctate distribution throughout the cell (Fig. 8 A and B, which is published as supporting information on the PNAS web site). NudCL diffused into the cytoplasm during mitosis and began to reassemble into vesicular structures, as did GM130 and Golgi p58 (Fig. 8C). Moreover, NudCL obviously was enriched in purified Golgi membranes, as was GM130 (Fig. 8D).
NudCL Is Regulated During Cell Cycle Progression.
Because mammalian NudC is required for mitosis (11), we sought to address whether NudCL also is regulated during cell cycle progression. T98G cells were rendered quiescent by serum deprivation for 72 h and then released into 20% FBS for cell cycle reentry. FACS analysis (Fig. 1B) showed that these cells synchronously progressed through G1 phase (4–12 h), G1/S transition (16 h), S phase (20 h), and G2/M transition (24 h). Synchronization also was confirmed by the phosphorylation of histone H3 and the expression of cyclin B1 (Fig. 1C). NudCL, which is constitutively expressed at a low level during G1, showed increased expression at the G1/S transition and maintained a high level throughout the cell cycle, displaying a pattern similar to that of NudC (Fig. 1C).
To assess whether NudCL is phosphorylated during mitosis, HeLa cells were transfected with pCMV-FLAG-NudCL, treated with nocodazole, and then labeled with [32P]orthophosphate in vivo. The data showed that FLAG-NudCL was more highly phosphorylated in mitotic cells than in randomly growing cells (Fig. 1D).
Depletion of NudCL Inhibits Cell Growth and Induces Cell Death.
To further characterize the function of NudCL, we depleted endogenous NudCL by vector-based RNA interference (RNAi; ref. 12). The vector, named pBS/U6-NudCL, targets human NudCL in the coding region. We cotransfected pBS/U6-NudCL or pBS/U6 with pBABE-puro, which contains a puromycin-resistance gene, at the ratio of 7:1 and added puromycin to the cell culture medium at 24 h after transfection to enable selection of transfection-positive cells. Forty-eight hours after transfection, most untransfected cells had been removed by selection with puromycin. The protein level of NudCL was greatly reduced from 48 h to 120 h after transfection, whereas the level of extracellular signal-regulated kinase 2 was unchanged (Fig. 2A). Using this strategy, we first determined whether NudCL is required for cell growth. Transfection with pBS/U6 and pBABE-puro did not influence cell proliferation, whereas silencing of NudCL significantly inhibited cell growth (Fig. 2B). Because deletion of nudC in A. nidulans causes morphological abnormalities and is lethal (13), we examined the viability of NudCL-depleted cells. Transfection with the control vector showed little effect on cell survival, whereas only 15% of NudCL-depleted cells remained attached to the culture dishes at 120 h after transfection (Fig. 2C), implying that NudCL is required for mammalian cell survival.
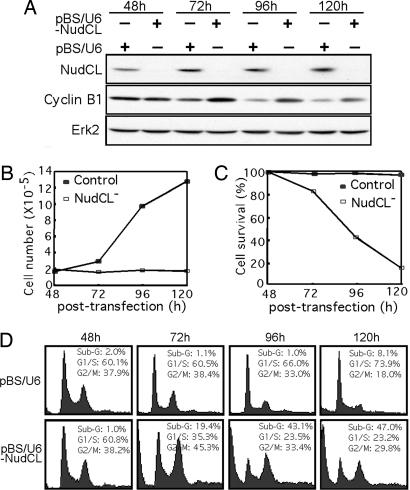
Fig. 2.
Depletion of NudCL inhibits cell growth and induces G2/M arrest and cell death. HeLa cells were transfected with either pBS/U6 or pBS/U6-NudCL and pBABE-puro at a ratio of 7:1. At 24 h after transfection, puromycin was added to select the transfection-positive cells. The cells were harvested at the indicated times and subjected to multiple analyses. (A) Western analysis shows the efficiency of depletion of NudCL by vector-based RNAi and the changes in the expression of cyclin B1. (B) NudCL depletion inhibits cell growth. The total number of cells was monitored at the times indicated. (C) NudCL depletion inhibits viability. The cells attached to the culture dishes were counted as viable cells, and the floating cells were counted as dead cells. (D) FACS analysis shows an increase in subgenomic and G2/M DNA content in cells depleted of NudCL.
Depletion of NudCL Induces Multiple Mitotic Defects.
Because the expression of NudCL protein begins to increase at G1/S and NudCL is phosphorylated during mitosis (Fig. 1), we assessed cell cycle progression in the NudCL-depleted cells. FACS profiles showed that at 72 h after transfection, the cell population with a G2/M DNA content was significantly increased relative to that with a G1 DNA content (Fig. 2D), indicating that down-regulation of endogenous NudCL may induce G2/M arrest. The increase of cyclin B1 in cells depleted of NudCL further suggests that there is a mitotic block in NudCL-depleted cells (Fig. 2A). Consistent with the data on cell survival, the percentage of cells with subgenomic DNA content started to increase dramatically at 72 h after transfection (Fig. 2D), suggesting that depletion of NudCL is lethal.
To explore the mechanism of mitotic arrest in NudCL-depleted cells, we performed immunofluorescence experiments. Close examination of mitotic cells revealed several defects: At metaphase, the majority of NudCL-depleted cells (63% of 695 cells) displayed misaligned chromosomes, whereas few of the control cells (6% of 645 cells) presented a similar phenotype (Fig. 3A); at metaphase and anaphase ≈3-fold more NudCL-depleted cells (9% of 783 cells) than control cells (3% of 710 cells) revealed multipolar spindles (Fig. 3B); and, additionally, a majority of NudCL-depleted cells (69% of 294 cells) with an elongated, disordered spindle showed a loss of focus of the spindle pole and appeared to have failed to completely separate chromosomes as compared with control cells (7% of 278 cells; Fig. 3C).
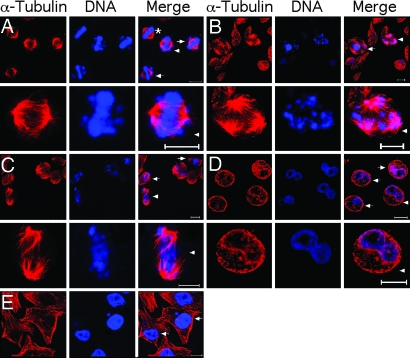
Fig. 3.
Multiple mitotic defects are induced by depletion of NudCL. HeLa cells were transfected with either pBS/U6 or pBS/U6-NudCL and pBABE-puro at a ratio of 7:1. At 24 h after transfection, puromycin was added for 3 days to select the transfection-positive cells. The cells were fixed with cold methanol and incubated with anti-α tubulin antibody. DNA was visualized by DAPI. (Scale bars: 10 μm.) (A) Chromosome misalignment. The cells are indicated with the arrowhead (a higher magnification of the cell is presented in Lower), and arrows show chromosome misalignment. The asterisk indicates a cell that exhibits normal morphology at metaphase. (B) Multipolar spindles. The cells indicated with an arrow and arrowhead (a higher magnification of the cell is presented in Lower) exhibit multipolar spindles. (C) Failure of chromosome segregation. The cells with arrows and arrowhead (a higher magnification of the cell is presented in Lower) display disordered and elongated mitotic spindles that appear to fail to separate chromosomes. (D) Formation of dumbbell-like DNA structures. The cells with dumbbell-like nuclei are indicated with an arrowhead (a higher magnification of the cell is presented in Lower) and arrows. (E) Accumulation of micronuclei. The cells with arrows display micronuclei.
To extend these results, we examined the nuclei of cells in interphase at 96 h after transfection of pBS/U6-NudCL. Approximately 30% of NudCL-depleted cells (941 cells) displayed a spherical shape with a dumbbell-like nuclear structure (Fig. 3D), implying that sister chromosomes had not separated completely during mitosis. Approximately 2% of the control cells (1,042 cells) showed a similar phenotype. In addition, micronuclei were more frequent in NudCL-depleted cells (14% of 941 cells) than in control cells (3% of 1,042 cells; Fig. 3E).
To rule out the off-target effects induced by the transfection of hairpin RNA vectors through a potential IFN response, we constructed a vector identical to pBS/U6-NudCL, except for a mutation of 2 nt within the targeting sequence, and performed rescue experiments. Our data show that almost all of the mitotic phenotypes in cells transfected with pBS/U6-NudCL are due specifically to the depletion of NudCL (Figs. 9 and 10, which are published as supporting information on the PNAS web site).
Depletion of NudCL Induces Dysfunction of the Dynein Complex.
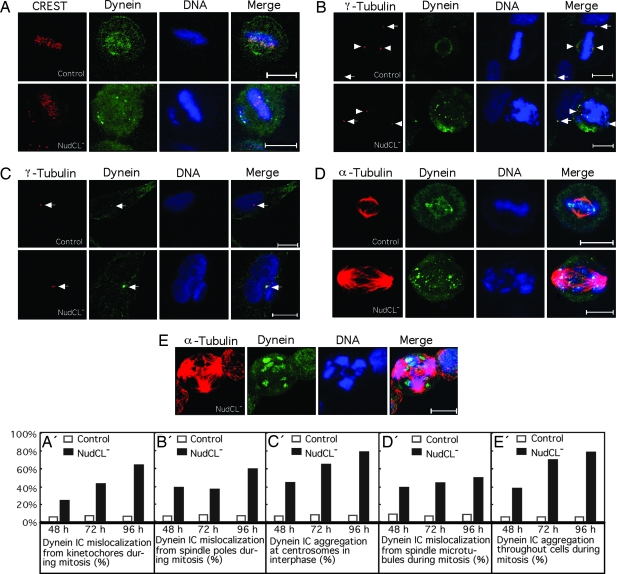
Because NudCL appears to be related to nuclear distribution proteins that contribute to the regulation of dynein complex (4, 5), we reasoned that NudCL might be involved in dynein function. First, we examined dynein localization at its mitotic-targeting sites. The dynein signal at most kinetochores was significantly decreased in cells transfected with pBS/U6-NudCL from 48 h after transfection compared with that in control cells (Fig. 4A and A′). Intriguingly, the dynein recruited to the spindle poles in cells depleted of NudCL was significantly decreased compared with that in control cells (Fig. 4 B and B′), whereas the dynein at the interphase centrosomes in NudCL-depleted cells was greatly increased (Fig. 4 C and C′). In general, the intensity of γ-tubulin at spindle poles during prometaphase and metaphase is at least 4-fold greater than that at centrosomes in interphase (14). In cells depleted of NudCL, the intensity of γ-tubulin signals at spindle poles was decreased considerably to a level at or below that at interphase centrosomes (Fig. 4 B and B′). Furthermore, the mitotic NudCL-depleted cells showed a significant increase in spindle length (from 8.6 ± 1.0 μm to 15.5 ± 3.8 μm; n = 18), a loss of spindle pole focus and dynein mislocalization from spindle microtubules (Fig. 4 D and D′). The mislocalized dynein appeared to aggregate during mitosis (Fig. 4 B, D, and E). Altogether, these data suggest that a dysfunctional dynein complex mislocalizes from the targeting sites during mitosis and appears to aggregate at centrosomes in interphase and throughout the cell during mitosis.
Fig. 4.
Depletion of NudCL induces the mislocalization and aggregation of the dynein complex. HeLa cells grown on the coverslips were transfected with either pBS/U6 or pBS/U6-NudCL and pBABE-puro at a ratio of 7:1. At 24 h after transfection, puromycin was added to select the transfection-positive cells. At different transfection times cells were fixed with cold methanol and incubated with anti-dynein IC antibody and anti-CREST anti-serum, anti-γ-tubulin, or α-tubulin antibodies. DNA was visualized by DAPI. (Scale bars: 10 μm.) (A and A′) Dynein mislocalization at kinetochores during mitosis. (B and B′) Dynein mislocalizes from mitotic spindle poles during mitosis. The spindle poles and centrosomes are indicated by arrowheads and arrows, respectively. (C and C′) Dynein appears to aggregate at the centrosomes in interphase. The centrosomes are indicated by arrows. (D and D′) Dynein mislocalizes from mitotic spindles. (E and E′) Dynein aggregates during mitosis in NudCL-depleted cells.
NudCL Associates with the Dynein Complex.
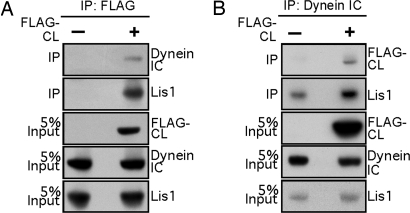
Because of the sequence homology between NudC and NudCL, we examined whether mammalian NudCL binds to the dynein complex as does NudC. Purified GST-NudCL protein bound to endogenous Lis1 and the dynein intermediate chain (IC) in cell extracts (Fig. 11, which is published as supporting information on the PNAS web site). To determine whether NudCL interacts with the dynein complex in vivo, we transfected pCMV-FLAG-NudCL vector into HeLa cells. The proteins were immunoprecipiated with anti-FLAG or anti-dynein IC antibody, and the associated proteins were detected by Western analysis. FLAG-NudCL coimmunoprecipitated with endogenous Lis1 and dynein IC and, moreover, was detected in the dynein complex immunoprecipitated by anti-dynein IC antibody (Fig. 5), indicating that NudCL associates with the dynein complex.
Fig. 5.
NudCL associates with Lis1 and the dynein complex. (A) FLAG-NudCL coimmunoprecipitates with Lis1 and dynein IC. At 40 h after transfection, HeLa cells transfected with control vector or pCMV-FLAG-NudCL were subjected to immunoprecipitation with anti-FLAG antibody. Immunoblotting analyses were performed with anti-FLAG, Lis1, and dynein IC antibodies. (B) FLAG-NudCL and Lis1 are present in the dynein complex. Lysates from HeLa cells transfected with control vector or pCMV-FLAG-NudCL were immunoprecipitated with anti-dynein IC antibody. Immunoprecipitated proteins were immunoblotted with anti-FLAG, Lis1, and dynein IC antibodies.
Depletion of NudCL Induces the Degradation of Dynein IC.
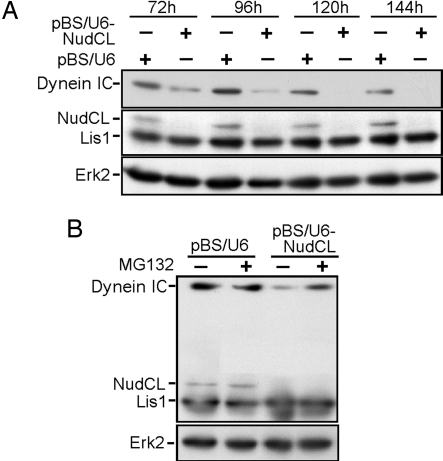
Because protein aggregation indicates the existence of misfolded proteins that are prone to degradation by the proteasome pathway (15), we examined the stability of the dynein complex. We found that dynein IC began to decrease significantly at 72 h after transfection of pBS/U6-NudCL and was detected at ≈15% of that in control cells at 120 h after transfection, whereas there was no significant change in the level of Lis1 and extracellular signal-regulated kinase 2 (Erk2) (Fig. 6A). To eliminate the possibility of NudCL depletion-induced dynein IC degradation by off-target effects of RNAi, we developed the lentivirus-based RNAi system with a targeting region of NudCL mRNA different from that of the vector-based RNAi. We found that dynein IC decreased considerably in lentivirus-based NudCL-depleted cells, whereas there were no significant changes of Lis1, p150 dynactin, and dynein heavy chain (Fig. 12, which is published as supporting information on the PNAS web site), suggesting that NudCL may be specifically required for the stability of dynein IC in mammalian cells.
Fig. 6.
Depletion of NudCL induces the degradation of dynein IC. HeLa cells were transfected with either pBS/U6 or pBS/U6-NudCL and pBABE-puro at a ratio of 7:1. At 24 h after transfection, puromycin was added to select the transfection-positive cells. (A) The degradation of dynein IC in cells depleted of NudCL. HeLa cells harvested at different times as indicated were subjected to immunoblotting analysis. (B) The inhibition of dynein IC degradation in cells depleted of NudCL by MG132, a proteasome inhibitor. At 96 h after transfection, HeLa cells were treated with MG132 for another 12 h and were subjected to immunoblotting analysis.
To assess whether the degradation of dynein IC in NudCL-depleted cells is related to the proteasome pathway, we added MG132, a proteasome inhibitor (16), into NudCL-depleted cells for 12 h and found that the level of dynein IC was significantly restored (Fig. 6B). Taken together, these results indicate that dynein IC appears to be destroyed via the proteasome pathway in cells depleted of NudCL.
Discussion
In this communication, we provide evidence that (i) depletion of NudCL inhibits cell growth, causes multiple mitotic defects, and induces cell death; (ii) NudCL associates with the dynein complex; and (iii) NudCL appears to be involved in the stabilization of dynein IC.
A Role of NudCL in Mitosis.
NudCL-depleted cells display a disordered mitotic spindle with chromosome misalignment and fail to separate sister chromatids normally, which is consistent with the accumulation of cells with dumbbell-like nuclei or micronuclei in interphase and elevated G2 DNA content. These data indicate a dysfunctional kinetochore and partial defects in the mitotic spindle checkpoint, generally consistent, to some extent, with effects induced by the depletion of dynein IC in various systems (17–19). During mitosis, dynein motors are recruited to kinetochores at prometaphase and drive spindle checkpoint proteins to spindle poles at metaphase to inactivate the spindle checkpoint, which is required for activation of anaphase promoting complex and correct chromosome segregation (18). Mislocalization of the dynein complex from the kinetochores during mitosis in NudCL-depleted cells may lead to mitotic arrest with activation of the spindle checkpoint and defects in chromosome segregation (1, 18, 19).
In general, the recruitment of the γ-tubulin ring complex, dynein/dynactin complex, and other spindle-promoting factors to the pericentriolar material by the dynein complex at late prophase is essential for mitotic spindle formation (20). In NudCL-depleted cells, both γ-tubulin and dynein complex at the spindle poles are obviously decreased during mitosis. We found a major proportion of the dynein complex detached from disordered and elongated spindles, with a loss of spindle pole focus in cells depleted of NudCL, implying an impaired mitotic spindle machinery. A number of studies using various model systems have shown that mitotic spindle length can be influenced by microtubule polymer dynamics, sister-chromatid cohesion, or mitotic motor activity at different levels (21, 22). In the Xenopus extract system, the dynein/dynactin complex regulates mitotic spindle length by targeting Kif2a to the spindle poles, which is a microtubule-depolymerizing regulator and plays a key role in bipolar spindle assembly and spindle microtubule length (21). The depletion of dynein IC by the addition of anti-dynein IC antibody into the Xenopus extract significantly increases the mitotic spindle length (21). In Aspergillus, a γ-tubulin mutant (mipAR63) in which NudI (dynein IC) does not localize to the spindle poles also shows a similar phenotype with much longer spindles (23), indicating that γ-tubulin is required for localization of the dynein to the spindle pole. Therefore, the reduction of dynein IC and γ-tubulin at mitotic spindle poles induced by NudCL depletion may contribute to the abnormal elongation of the mitotic spindle. Taken together, the majority of mitotic phenotypes in cells depleted of NudCL suggest that NudCL is involved in the dynein pathway in mammalian cells.
NudCL Is Required to Stabilize Dynein IC.
In mammalian cells, NudCL, as well as NudC, interact with Lis1 and the other components of the cytoplasmic dynein complex (4, 10, 11). In A. nidulans, extra copies of the nudF gene can rescue the nudC3 mutation phenotype by restoring the protein level of NudF (9), indicating that NudC may be upstream of NudF in Aspergillus. However, the protein level of NudF is not changed in A. nidulans in which NudC protein is down-regulated (13). Furthermore, mammalian cells individually depleted or codepleted of NudC or NudCL also showed no significant change in the level of Lis1/NudF protein (unpublished data). These data suggest that NudC and its mammalian homologs may not be upstream of NudF/Lis1. Recent data show that NudE may be upstream of NudF in A. nidulans, because NudF overproduction effectively rescues the abnormalities induced by deletion of the nudE gene (24), and similar results also have been obtained in yeasts and mammalian cells (25, 26).
Unexpectedly, dynein IC, a key scaffold protein for the assembly of dynein/dynactin complex (2), was greatly decreased in NudCL-depleted mammalian cells, implying that NudCL may play a role in stabilizing the dynein complex. This hypothesis is reinforced by the following data: First, in cells depleted of NudCL, dynein IC was mislocalized from its mitotic targeting sites and appeared to aggregate throughout the cytoplasm during mitosis. In interphase, dynein IC appeared as aggregates surrounding the microtubule-organizing center in cells depleted of NudCL, implying possible aggresome formation (15). Importantly, the degradation of dynein IC induced by NudCL depletion was significantly suppressed by MG132, a proteasome inhibitor. Mutations in the gene for cytoplasmic dynein IC, Dic19C, in Drosophila, result in larval lethality, demonstrating that the dynein IC serves an essential function (27), which was further supported by NudCL depletion-induced cell death reported here. Based on these observations, one possibility is that the dynein IC may be misfolded and is targeted to proteasomes for degradation in NudCL-depleted cells. This interpretation is consistent with the observation that dynein IC has a limited secondary and tertiary structure at near physiological solution conditions and requires other proteins to form a more ordered structure (28, 29).
We note that the nuclear movement domains in all NudC homologues contain a p23 domain that shows sequence similarity to a conserved cochaperone p23 that has a passive chaperone activity to specifically associate with partially folded proteins, preventing their aggregation and maintaining fold-competent conformations (30). Examination of the nuclear movement domain sequences of NudCL reveals that NudCL shares significant homology to p23 at its highly conserved positions (unpublished data), suggesting that NudCL may be involved in a chaperoning process. We speculate that NudCL may have a passive chaperone activity and bind to partially folded dynein ICs, preventing their aggregation until they are completely folded (Fig. 13, which is published as supporting information on the PNAS web site). Further investigation clearly is needed to determine whether NudCL has a passive chaperone activity to dynein IC.
Materials and Methods
Cloning of Human NudCL.
To identify human NudCL, the protein sequence of human NudC was used to search the National Center for Biotechnology Information database with tblastn. Based on the sequence of an expressed sequence tag (BC035014), one pair of primers, 5′-CGGGAGGCGACATGGAGCCA-3′ and 5′-GGCGAGGGTTTCCTTTCCTTC-3′, was designed to clone NudCL by RT-PCR from total RNA extracted from HeLa cells.
Plasmid Construction.
Full-length NudCL was subcloned into pGEX-5X-1 (Amersham Pharmacia) and pCMV-Tag2C (Stratagene). The targeting sequence in the construct pBS/U6-NudCL is 5′-GGGCATCAGGAAGTAGAGAAAG-3′, corresponding to the region 375–396 relative to the first nucleotide of the start codon.
Cell Culture, DNA Transfections, and FACS Analyses.
HeLa and T98G cells were maintained in DMEM containing 10% FBS. HeLa cells were transfected by GenePORTER transfection reagent (Gene Therapy Systems, San Diego). Propidium iodide staining and cell cycle analysis by using CellQuest were performed as described in ref. 12.
Anti-NudCL Antibody Production.
Rabbit polyclonal anti-NudCL antibody was generated by using bacterially expressed GST-NudCL as an antigen (Proteintech, Chicago) and was affinity purified.
Immunoprecipitation and Western Analyses.
Immunoprecipitation experiments were performed as described in ref. 11. The immunoprecipitates or total proteins isolated from mammalian cells were subjected to immunoblotting analyses with anti-NudCL, NudC (a gift from Li-yuan Yu-Lee, Baylor College of Medicine, Houston), cyclin B1, extracellular signal-regulated kinase 2 (Santa Cruz Biotechnology), dynein IC (Covance, Berkeley, CA), FLAG, Lis1 (Sigma) or phosphorylated histone H3 (Upstate Biotechnology) primary antibodies, and anti-mouse or anti-rabbit Ig horseradish peroxidase-linked secondary antibodies (Amersham Pharmacia). Immunoblotting analyses with extracellular signal-regulated kinase 2 were used as a loading control.
Immunofluorescence Staining.
The cells grown on coverslips were fixed with cold methanol (−20°C) and then stained with anti-dynein IC, α-tubulin, γ-tubulin (Sigma), NudCL antibodies, or anti-CREST (kinetochore) serum (Antibodies, Inc.) for 2 h at room temperature, followed by incubation with either Cy3- or FITC-conjugated anti-mouse or anti-rabbit Ig secondary antibody (Jackson ImmunoResearch) for 40 min. Finally, DNA was stained with DAPI (Sigma). The mounted coverslips were analyzed by confocal fluorescence microscopy (LSM510; Zeiss).
Supplementary Material
Acknowledgments
We thank Eleanor Erikson, Ming Lei, and Diana Kornet for critical comments. This work was supported by National Institutes of Health Grant GM59172. R.L.E. is the John F. Drum American Cancer Society Research Professor. T.Z. is supported by Natural Scientific Foundation of Zhejiang Province Grant R205291 from the People’s Republic of China.
Abbreviations
- IC
intermediate chain
- Lis1
lissencephaly 1
- NudCL
NudC-like protein
- RNAi
RNA interference.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Karki S., Holzbaur E. L. Curr. Opin. Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 2.Vale R. D. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 3.Mallik R., Gross S. P. Curr. Biol. 2004;14:R971–R982. doi: 10.1016/j.cub.2004.10.046. [DOI] [PubMed] [Google Scholar]
- 4.Morris N. R. J. Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wynshaw-Boris A., Gambello M. J. Genes Dev. 2001;15:639–651. doi: 10.1101/gad.886801. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J., Li S., Fischer R., Xiang X. Mol. Biol. Cell. 2003;14:1479–1488. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallee R. B., Tai C., Faulkner N. E. Trends Cell Biol. 2001;11:155–160. doi: 10.1016/s0962-8924(01)01956-0. [DOI] [PubMed] [Google Scholar]
- 8.Yan X., Li F., Liang Y., Shen Y., Zhao X., Huang Q., Zhu X. Mol. Cell. Biol. 2003;23:1239–1250. doi: 10.1128/MCB.23.4.1239-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang X., Osmani A. H., Osmani S. A., Xin M., Morris N. R. Mol. Biol. Cell. 1995;6:6297–6310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aumais J. P., Tunstead J. R., McNeil R. S., Schaar B. T., McConnell S. K., Lin S. H., Clark G. D., Yu-Lee L. Y. J. Neurosci. 2001;21:RC187. doi: 10.1523/JNEUROSCI.21-24-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou T., Aumais J., Liu X., Yu-Lee L. Y., Erikson R. L. Dev. Cell. 2003;5:127–138. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 12.Liu X., Yan S., Zhou T., Terada Y., Erikson R. L. Oncogene. 2004;23:763–776. doi: 10.1038/sj.onc.1207188. [DOI] [PubMed] [Google Scholar]
- 13.Chiu Y. H., Xiang X., Dawe A. L., Morris N. R. Mol. Biol. Cell. 1997;8:1735–1749. doi: 10.1091/mbc.8.9.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piehl M., Tulu U. S., Wadsworth P., Cassimeris L. Proc. Natl. Acad. Sci. USA. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Mata R., Gao Y. S., Sztul E. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee D. H., Goldberg A. L. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 17.Gaglio T., Dionne M. A., Compton D. A. J. Cell Biol. 1997;138:1055–1066. doi: 10.1083/jcb.138.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., Rieder C. L., Salmon E. D. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morales-Mulia S., Scholey J. M. Mol. Biol. Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blagden S. P., Glover D. M. Nat. Cell Biol. 2003;5:505–511. doi: 10.1038/ncb0603-505. [DOI] [PubMed] [Google Scholar]
- 21.Gaetz J., Kapoor T. M. J. Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goshima G., Wollman R., Stuurman N., Scholey J. M., Vale R. D. Curr. Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 23.Li S., Oakley C. E., Chen G., Han X., Oakley B. R., Xiang X. Mol. Biol. Cell. 2005;16:3591–3605. doi: 10.1091/mbc.E04-12-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efimov V. P. Mol. Biol. Cell. 2003;14:871–888. doi: 10.1091/mbc.E02-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Lee W. L., Cooper J. A. Nat. Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu T., Ayala R., Nguyen M. D., Xie Z., Gleeson J. G., Tsai L. H. Neuron. 2004;44:263–277. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 27.Boylan K. L. M., Hays T. S. Genetics. 2002;162:1211–1220. doi: 10.1093/genetics/162.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makokha M., Hare M., Li M., Hays T., Barbar E. Biochemistry. 2002;41:4302–4311. doi: 10.1021/bi011970h. [DOI] [PubMed] [Google Scholar]
- 29.Nyarko A., Hare M., Hays T. S., Barbar E. Biochemistry. 2004;43:15595–15603. doi: 10.1021/bi048451+. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Ranea J. A., Mirey G., Camonis J., Valencia A. FEBS Lett. 2002;529:162–167. doi: 10.1016/s0014-5793(02)03321-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.