Abstract
The repeatability of evolutionary change is difficult to quantify because only a single outcome can usually be observed for any precise set of circumstances. In this study, however, we have quantified the frequency of parallel and divergent genetic changes in 12 initially identical populations of Escherichia coli that evolved in identical environments for 20,000 cell generations. Unlike previous analyses in which candidate genes were identified based on parallel phenotypic changes, here we sequenced four loci (pykF, nadR, pbpA-rodA, and hokB/sokB) in which mutations of unknown effect had been discovered in one population, and then we compared the substitution pattern in these “blind” candidate genes with the pattern found in 36 randomly chosen genes. Two candidate genes, pykF and nadR, had substitutions in all 11 other populations, and the other 2 in several populations. There were very few cases, however, in which the exact same mutations were substituted, in contrast to the findings from conceptually related work performed with evolving virus populations. No random genes had any substitutions except in four populations that evolved defects in DNA repair. Tests of four different statistical aspects of the pattern of molecular evolution all indicate that adaptation by natural selection drove the parallel changes in these candidate genes.
Keywords: bacterial evolution, mutation, natural selection, parallel evolution
Parallel evolution and convergent evolution occur when two or more lineages independently evolve similar or identical features. Parallel evolution and convergent evolution are usually distinguished on the basis that parallelism involves changes in homologous features among closely related organisms, whereas convergence can involve changes in different antecedent features among more distantly related organisms (1–3). Both parallel evolution and convergent evolution provide strong evidence that the derived similarities resulted from adaptation by natural selection, provided the state-space of possible changes is so large that it is improbable that the observed similarities arose by a purely random process. There are many compelling examples of parallel evolution in nature, including recent studies of lizard morphology (4) and fish behavior (5), showing that certain phenotypes evolved repeatedly when separate populations independently colonized similar environments. Also, some pathogens exhibit striking parallel genomic changes, including multiple HIV lineages that substituted similar mutations conferring antiviral drug resistance (6) and several strains of Escherichia coli that independently acquired similar virulence factors by horizontal transfer (7).
Yet, despite these and other compelling examples of parallel evolution (8–10), it has proven difficult to quantify evolutionary repeatability. In principle, even the most basic quantification of parallel evolution would include the number of potential instances of parallel outcomes, which could be compared with the actual number seen. In practice, however, the number of potential instances is rarely given and difficult to ascertain. For example, undetected extinctions of other populations that had evolved different, but ultimately unsuccessful, adaptations might cause an upward bias to an estimate of the extent of parallelism. Also, comparative studies cannot generally exclude subtle differences in selective environments or in founding genotypes as causes of divergent evolutionary outcomes, which produce a downward bias in any estimate of evolutionary repeatability. Thus, it is difficult to know the denominator that corresponds to the number of potential cases of parallel outcomes to compare with the actual number observed. However, well designed evolution experiments overcome these problems because the number of independent populations is set by the experimenter, and systematic environmental differences are precluded by an appropriate design. Moreover, in experiments with microorganisms, replicate populations can be founded by single haploid individuals, such that there is no initial genetic variation and therefore any parallelism must depend on the independent origin, as well as fate, of variants (11, 12). Such experiments have now provided many examples of both parallel and divergent evolution affecting both phenotypic and genetic properties (13–31).
In a landmark study, Wichman et al. (24) examined parallel evolution at the genetic level in φX174, a DNA virus with 11 genes and a 5.4-kb genome. Two populations were propagated for 10 days on a novel host strain, and the viral genomes were sequenced before and after the experiment. That study found 29 mutations, of which 14 were identical in the two populations. Qualitatively similar results were obtained by Bull et al. (19) with several additional populations of φX174 by using a more complex experimental design. However, it is not known whether much larger genomes, encoding more complex organisms and having potentially many more targets of selection, would show similarly strong parallelism at the sequence level.
To address that issue, we sought to examine the extent of genetic parallelism in a long-term experiment with 12 populations of E. coli (11, 13, 25, 32), a bacterium with some 4,300 genes and a genome of ≈4,500 kb (33). We (27, 29, 31) have previously reported three cases of parallel substitutions affecting the rbs operon, the spoT gene, and two genes involved in DNA topology in these populations. Importantly, however, all three cases depended on first finding that there had been parallel phenotypic changes in ribose catabolism, global expression profiles, and DNA supercoiling, respectively. Therefore, although these cases provide clear examples of genetic parallelism, they represent a nonrandom sample that might not reflect the overall extent of parallel changes in the genome as a whole. In this study, by contrast, we pursue an approach that is completely independent of any known parallel phenotypic changes to address the extent of genetic parallelism in a statistically unbiased manner. Our specific approach is to compare the pattern of mutational substitutions, in several candidate genes in which we have found mutations of unknown effect in one population (34), with the pattern observed in many other genes that were chosen completely at random (35). Here, we use the idea of “candidate gene” to mean only that a mutational substitution was previously found in that gene in one population, not that the gene was investigated based on parallel phenotypic changes related to its function. Thus, they might also be called “blind” candidate genes.
The 12 replicate populations all were founded by the same ancestral strain and grown in identical environments for 20,000 generations. They evolved similar, but not identical, changes in various aspects of their performance, morphology, and physiology (11, 13, 15, 18, 25, 27, 29, 31, 32). Also, four of the populations became “mutators” by evolving defects in their DNA repair pathways, which caused large increases in their spontaneous mutation rates (20, 25); the distinction between mutator and nonmutator populations is important for some of our analyses. In previous work (34), we discovered and characterized four insertion mutations, which provide the basis for our present study, by comparing the genomic fingerprints of two evolved populations (neither a mutator) with their common ancestor. The position of these four mutations in the phylogenies of the populations in which they arose indicated that they were eventually substituted, which suggested that they either were beneficial themselves or, alternatively, had hitchhiked with unknown beneficial mutations (34, 36). Therefore, although we suspected these mutations might be beneficial, we chose each candidate gene based on a single mutation to avoid any bias toward parallel evolution. Population A+1 substituted IS150 insertion mutations in nadR, hokB/sokB, and upstream of pbpA-rodA; and population A−1 substituted an IS150 insertion in pykF. These four loci thus became the blind candidate genes for further investigation. In this study, they were sequenced in clones sampled from all 12 populations after 20,000 generations. The resulting sequence data were then used to test whether evolution was parallel at the levels of genes and the mutations within them and to quantify the extent of any parallelism.
Results and Discussion
Little Parallelism at the Level of Mutations in the Candidate Genes.
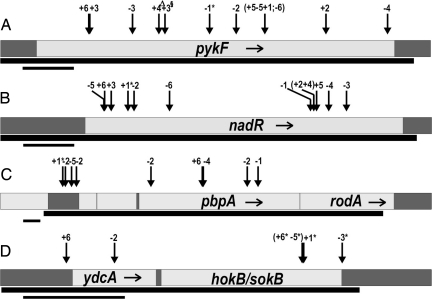
We sequenced the four candidate loci (pykF, nadR, hokB/sokB, and pbpA-rodA) in the ancestor and clones sampled at generation 20,000 from all 12 populations. Fig. 1 shows the physical extent of sequencing and marks the locations for all of the mutations found in the sequenced regions; some 7,150 bp were sequenced for each evolved clone. A total of 40 mutations were found including the 4 IS150 insertion mutations previously discovered and 36 additional mutations, all of which were confirmed by resequencing. It is impossible to prove that these mutations were absolutely fixed in these populations. In particular, frequency-dependent selection (37) and clonal interference (38) may sustain minority populations and give rise to situations in which beneficial mutations reach high frequency without being substituted. To examine this issue further, PCR and restriction fragment length polymorphism (RFLP) assays were developed to test numerous clones from multiple generations in two populations that together had seven mutations in candidate genes (R.W., unpublished data). All seven cases provide compelling evidence for substitution before 20,000 generations; for example, the pbpA mutation in population A−1 was present in all 48 clones tested from a sample taken at generation 5,000 (39). We conclude, therefore, that the vast majority of the mutations found in candidate genes were substituted. Table 4, which is published as supporting information on the PNAS web site, lists the precise locations and other molecular details for each mutation. Two of the 36 newly discovered mutations lie outside the 4 candidate genes and their known regulatory regions; both occur in or near ydcA, a gene of unknown function near the hokB/sokB locus. These two mutations were excluded from our main statistical tests because their relevance for quantifying parallel evolution was unclear, although this decision had no effect on these tests (as explained later).
Fig. 1.
Mutations substituted by 20,000 generations in four candidate genes in 12 experimental populations of E. coli. Lighter regions indicate protein-coding sequences for and near pykF (A), nadR (B), pbpA-rodA (C), and hokB/sokB (D). Long bars below indicate the range sequenced; short bars show scale (200 bp). Each arrow marks a mutation; the number shows the affected population. The mutations in and near ydcA are of unknown relevance. ∗, An IS150 insertion, and, for populations −1 and +1, these were the original mutations used to identify the candidate genes. ▵, A 1-bp deletion. §, A synonymous mutation. All others in the coding regions are nonsynonymous, except for a 1-bp insertion in ydcA in population −2.
Of the 38 mutations found in the candidate genes, there were 35 distinct mutations. Only two mutations were found in multiple populations: three populations had identical G→T substitutions at position 901 in pykF, and two other populations had the same A→G mutation at position 902 in nadR. The 66 possible pairs of the 12 E. coli populations shared, on average, only 2.1% of their mutations. By contrast, the two φX174 virus populations studied by Wichman et al. (24) shared almost half of their mutations. Parallelism at the level of mutations was evidently much less common in these evolving bacteria than in the previously studied viruses.
Extensive Parallelism at the Level of the Candidate Genes.
Turning from mutational identity to the level of the affected genes, the pattern is very different. Every population had exactly one nonsynonymous point mutation in both pykF (Fig. 1A) and nadR (Fig. 1B), with the exception of the focal populations that contained the previously discovered insertion mutations that led to identification of these candidate genes. One synonymous mutation was found in pykF, and none in nadR. The two other candidates also yielded mutations, although not in every population. Besides the focal population’s insertion upstream of pbpA-rodA, five others had mutations in the upstream region, the pbpA gene, or both (Fig. 1C). For hokB/sokB, three other populations had insertions similar to that in the focal population (Fig. 1D). The many independent substitutions in the candidate genes suggest parallelism, but it is necessary to demonstrate that the numbers are above those expected by chance. For example, perhaps most genes accumulated mutations after 20,000 generations, such that finding mutations in a candidate gene in several or even all of the populations is statistically unremarkable.
To that end, we compared the results of sequencing the candidate genes with the results of sequencing ≈500-bp regions in each of 36 randomly chosen genes for the same 12 populations (35). Only six substitutions in total were found in these random genes; three of these substitutions were synonymous point mutations, and the other three were nonsynonymous point mutations. Moreover, all of these substitutions in random genes were found in the four evolved mutator populations.
Statistical Tests Support Parallelism at the Level of Candidate Genes.
We performed four distinct tests of the hypothesis that natural selection drove parallel changes in the candidate genes. First, we compared the overall substitution rates between the candidate and randomly chosen genes, with an expectation of a higher rate in the candidates because they accumulated substitutions by selection as well as by drift. Consistent with that expectation, all 12 populations had higher substitution rates in the candidate genes (Table 1), which is highly unlikely by chance (sign test, P = 0.0002). This result is unaffected by excluding the insertions used to identify the candidates in populations A−1 and A+1; both of these focal populations had substitutions in one or more genes whose candidacy was identified in the other population, and neither of them had any substitutions in the random genes.
Table 1.
Number of mutations in random and candidate genes in 12 E. coli populations after 20,000 generations
| Population | Random genes (18,374 bp total) |
Candidate genes (7,150 bp total) |
||
|---|---|---|---|---|
| No. of mutations | Rate per 1000 bp | No. of mutations | Rate per 1000 bp | |
| A−1 | 0 | 0.000 | 3 | 0.420 |
| 2* | 0.280 | |||
| A−2 | 0 | 0.000 | 6 | 0.839 |
| A−3 | 0 | 0.000 | 3 | 0.420 |
| A−4 | 3 | 0.163 | 3 | 0.420 |
| A−5 | 0 | 0.000 | 4 | 0.559 |
| A−6 | 0 | 0.000 | 2 | 0.280 |
| A+1 | 0 | 0.000 | 4 | 0.559 |
| 1* | 0.140 | |||
| A+2 | 0 | 0.000 | 2 | 0.280 |
| A+3 | 1 | 0.054 | 3 | 0.420 |
| A+4 | 0 | 0.000 | 2 | 0.280 |
| A+5 | 0 | 0.000 | 2 | 0.280 |
| A+6 | 2 | 0.109 | 4 | 0.559 |
Numbers of mutations and substitution rates are pooled across 36 random genes and 4 candidate genes. Populations A−2, A−4, A+3, and A+6 became mutators; all others retained the ancestral mutation rate.
*Excluding the mutations (one in A−1, three in A+1) used to identify the candidate genes.
Second, if mutations in the candidate genes were beneficial, then we would expect to see an excess of nonsynonymous substitutions relative to synonymous substitutions. Three of 6 point mutations in the randomly chosen genes were synonymous, but only 1 of 27 was synonymous in the candidate genes (Table 2). This difference is significant (Fisher’s exact test, P = 0.0136) and also supports the hypothesis that the mutations substituted in the candidate genes were beneficial.
Table 2.
Candidate and random genes differ in relative abundance of synonymous and nonsynonymous point mutations
| Gene | Synonymous | Nonsynonymous |
|---|---|---|
| Candidate | 1 | 26 |
| Random | 3 | 3 |
Third, theory predicts that the substitution rate for neutral mutations should scale with the mutation rate (40), whereas the substitution rate for beneficial mutations is subject to diminishing returns in large asexual populations owing to clonal interference (41). Recall that four of the populations became mutators and had mutation rates ≈100-fold higher than the other eight populations. We expect to observe more nonbeneficial substitutions in the mutator populations and consequently a relatively higher proportion of beneficial mutations in the nonmutator populations. Indeed, all 6 substitutions in the random genes were found in the mutator populations, whereas half of the 30 point mutations in the candidate genes were substituted in the nonmutator populations (Table 3). This difference is significant in the direction expected if the candidate genes experienced selection favoring new alleles (Fisher’s exact test, P = 0.0279).
Table 3.
Candidate and random genes differ in relative abundance of point mutations in mutator and nonmutator populations
| Gene | Mutator | Nonmutator |
|---|---|---|
| Candidate | 15 | 15 |
| Random | 6 | 0 |
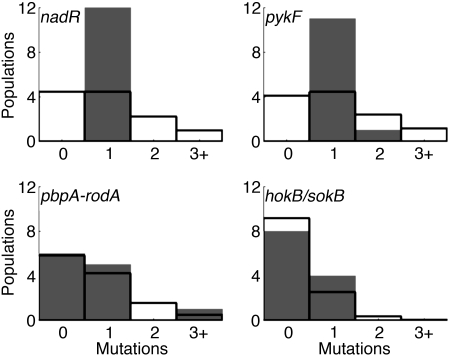
Fourth, if mutations in the candidate genes were neutral, then the numbers of substitutions in the populations should follow a Poisson distribution if all populations had the same mutation rate, or they would be substantially clumped in the mutator populations given the differences in mutation rates. By contrast, if the substituted mutations were beneficial, and if different mutations in the same gene conferred functionally similar benefits, then we would expect a more uniform distribution of mutations. Unlike the first three statistical tests, this test is independent of the evolutionary forces affecting the randomly chosen genes. The distributions are the most uniform possible given the numbers of mutations in two of the candidate genes (Fig. 2). For nadR, there were 12 substitutions, with each population having exactly 1; the likelihood of this distribution by chance is 12!/1212 < 0.0001. For pykF, the chance of 11 populations having 1 mutation and 1 having 2 mutations is 0.0004. Moreover, these calculations are very conservative because the four mutator populations should push strongly away from uniformity, making the observed distributions that much more unexpected. The other two candidate genes do not deviate significantly from the Poisson distribution, but that outcome may simply reflect fewer mutations in those genes and the very conservative nature of this test.
Fig. 2.
Distribution of numbers of substitutions in the 12 populations for the four candidate genes. Observed distributions are shaded. Poisson distributions with the same mean as the observed distribution are shown in outline.
Statistical Tests Are Robust with Respect to Criteria for Data Inclusion.
Regarding the excess of nonsynonymous substitutions in the candidate genes, Table 2 includes point mutations in protein-coding regions only, with 27 such mutations in the candidate genes and 6 in the random genes. Three additional nonpoint mutations occurred in the protein-coding regions of the candidate genes, including two IS-element insertions and one 1-bp deletion, and these mutations could be viewed as nonsynonymous because they change the resulting amino acid sequence. If these additional mutations are included in the analysis, the outcome remains significant (P = 0.0104).
With respect to the observed excess of substitutions in the nonmutator populations among the candidate genes, Table 3 includes 30 point mutations in the candidate genes and 6 point mutations in the random genes. There were eight additional mutations in candidate genes, including seven insertions and one 1-bp deletion. One of these insertions was in a mutator population, and all others were in nonmutator populations. If these additional mutations are included, the outcome remains significant (P = 0.0211).
Regarding the two mutations found in or near ydcA, we chose both candidate and random genes a priori, as explained. The ydcA mutations do not fit into either category, and therefore they were excluded from our main analyses. It is possible that ydcA is related to hokB/sokB, given its proximity and unknown functionality, but it is also possible these loci have nothing to do with each other. If we include ydcA with the random genes, it would not weaken any of our four tests and, in fact, strengthens one of them. The first test (Table 1) compares the density of mutations found in random and candidate genes; adding one random mutation to both A−2 and A+6 would not change the fact that all 12 populations have a higher substitution density in candidate genes. The second test (Table 2) is unaffected because neither ydcA mutation counts as synonymous or nonsynonymous; one is outside the coding region, and the other is not a point mutation. The third test (Table 3) compares the mutator and nonmutator populations. Both ydcA mutations are in mutator populations, and including it as a random gene would strengthen that already significant result. Finally, the fourth test (Fig. 2) is unaffected because noncandidate genes do not enter into the analysis. If, instead, we include ydcA with the hokB/sokB candidate locus, the third test would be slightly weakened but remain significant, whereas the other tests would not be affected.
Alternative Hypotheses Are Inconsistent with One or More Tests.
The four tests individually and collectively support the hypothesis that parallel evolution in the candidate genes was driven by natural selection favoring the mutant alleles, and their conclusions are robust when we use different criteria for including ambiguous data. The first two tests are also consistent with the alternative hypothesis that the candidate genes had relaxed selective constraints, such that they could accumulate mutations without adverse effects. However, the third and fourth tests clearly reject this alternative hypothesis because, if it were true, mutator populations should accumulate disproportionately more substitutions in candidate genes, and substitutions would not be uniform across populations. Another alternative is that the candidate genes might contain “hot spots” with mutation rates much higher than the genome-wide average. This alternative also runs counter to the test comparing mutator and nonmutator populations, unless one further supposes that these hypermutable sites are independent of the repair pathways that became defective in the mutators. However, substitutions in three of the four candidate loci (pykF, nadR, and pbpA) include transitions and transversions as well as the original insertions, whereas the substitutions in the random genes occurred only in the mutators and all of them had signatures reflecting specific defects in DNA repair (35). The extreme uniformity of substitution number in pykF and nadR, coupled with the multiplicity of mutational targets in these genes, also contradicts the hot-spot hypothesis. The several IS insertions in hokB/sokB, and the absence of other types of mutations, might indicate an increased rate of those mutations at that locus, but such a bias, if it exists, does not contradict the possibility that the substitutions are also beneficial (27).
Possible Functional Bases for Beneficial Effects of Mutations in the Candidate Genes.
The four tests collectively provide compelling evidence that the mutations that were substituted in the candidate genes are beneficial in the environment used in the evolution experiment. We do not know, however, the functional bases for their beneficial effects. At first glance, the fact that the candidates were first identified by IS-element insertions in the focal populations might suggest that the beneficial mutations are knockouts. However, a more nuanced view is appropriate for several reasons. First, most of the mutations found in the other populations are unlikely to act as knockouts, with the probable exception of the several IS insertions in hokB/sokB and one frameshift mutation in pykF (Table 4). Even if the originally discovered mutation in a gene were a knockout, other populations may have substituted mutations with more subtle effects. Second, the original IS insertion affecting pbpA-rodA is not in the reading frame but, instead, sits in the upstream regulatory region, where IS elements can exert subtle effects on gene expression (42). Third, in the case of nadR, the affected protein is bifunctional with both repressor and transport domains (34, 43). A knockout of one function could leave the other function intact; and, in the case of the repressor function, a knockout would elevate expression of the de-repressed genes.
The following hypotheses suggest how mutations in the candidate genes might enhance fitness in the environment of the long-term evolution experiment (34), although we emphasize that they require further testing and other explanations may also be plausible. The pykF gene encodes one of two pyruvate kinases that catalyze the conversion of phosphoenolpyruvate (PEP) and ADP into pyruvate and ATP. PEP is also used by the phosphotransferase system (PTS) to transport glucose into the cell. By slowing the conversion of PEP to pyruvate, mutations in pykF might make more PEP available to drive the PTS-mediated uptake of glucose, which is the limiting resource in the environment of the long-term evolution experiment. As noted, nadR encodes a bi-functional protein that is involved in several aspects of NAD metabolism, itself a key metabolite important in many different pathways. Several genes involved in NAD synthesis and recycling are repressed by the NadR protein, and mutations in nadR might increase their expression and the resulting intracellular concentration of NAD. The evolved bacteria have higher maximum growth rates as well as shorter lags after the daily transfers into fresh medium (15, 29), and increased levels of NAD might be beneficial in achieving one or both of these advantages (44). Alternatively, changes in NAD regulation might improve the control of oxidative stress in the experimental environment (45). The hokB/sokB locus is one of several loci in E. coli related to the hok/sok locus of plasmid R1; hok encodes a toxin and sok an antisense RNA that blocks translation of the toxin. Together, these activities kill any cells that lose the plasmid, a function that may benefit the plasmid but is obviously harmful to the bacteria. Inactivation of hokB/sokB would therefore benefit the bacteria (in the absence of plasmids), and indeed other copies of hok/sok loci in E. coli contain insertion elements that have presumably inactivated them (46, 47). Finally, the pbpA-rodA operon encodes two essential proteins that are involved with peptidoglycan synthesis and coupling cell-wall synthesis to the overall cell cycle (48). All 12 populations evolved much larger cell volumes (13), which may require altered rates of peptidoglycan synthesis, changes in the timing of its synthesis in relation to the cell cycle, or both.
Conclusions and Future Directions.
Our results demonstrate that evolution in these 12 E. coli populations was often parallel at the level of genes, but only rarely were the substitutions identical at the base pair level. This latter point stands in sharp contrast with results obtained in two replicate populations of virus φX174, where almost half of the substitutions were identical (24). We are tempted to suggest that this difference in parallelism reflects differences in genome size and complexity, but that explanation is by no means proven. Thus, it would be interesting to have comparably precise experiments for many other viruses and bacteria as well as archaea, single-celled eukaryotes, and multicellular animals and plants to evaluate comparatively whether increasing genomic and functional complexity leads to less repeatable outcomes of molecular and phenotypic evolution.
In this study, we also observed variation in the extent of parallelism among the candidate genes, with nadR and pykF exhibiting more evolutionary repeatability than pbpA and hokB/sokB. These differences might indicate important functional interactions among loci or, alternatively, that those genes under stronger selection may converge more quickly on beneficial substitutions than those experiencing weaker selection. Future work, including additional generations and genetic manipulations, may reveal whether the gene-level differences between populations will erode or be sustained. If the fitness effects of the mutations at these loci are additive, or at least do not change sign, then we would expect all of the populations to eventually get mutations at each of these loci. However, if there are epistatic interactions such that mutations at some of the genes are no longer beneficial on certain evolved backgrounds, then this gene-level divergence could be sustained indefinitely. Genetic manipulations that produce isogenic constructs differing by single known mutations will also be useful for examining the physiological mechanisms responsible for the beneficial effects of the mutations in the candidate genes (29, 31). Finally, recent technological advances have led to substantial reductions in the cost of whole-genome sequencing and resequencing (49, 50), so that it will become feasible to sequence entire genomes from several or all of the populations in this long-term evolution experiment.
Methods
Background on the Long-Term Evolution Experiment.
Twelve populations were started from the same ancestral strain of E. coli B, except that 6 of the populations were founded from an Ara− variant and 6 from an Ara+ variant. These populations are designated A−1 to A−6 and A+1 to A+6 according to this marker, which is neutral in the experimental environment (11). The populations were serially propagated in identical glucose-limited environments for 20,000 cell generations (3,000 days), with population sizes fluctuating daily between ≈5 × 106 and 5 × 108 cells. The populations achieved similar, although not identical, fitness gains (11, 13, 25). Also, all 12 populations evolved large increases in average cell size (13), certain catabolic abilities were lost in parallel (25, 27), and global gene-expression profiles showed similar changes in the two populations that were examined in this regard (29). Four populations evolved defects in DNA repair pathways, which caused ≈100-fold increases in their spontaneous mutation rates (20, 25). An in-depth review of this long-term evolution experiment can be found elsewhere (32).
Sequencing the Four Candidate Genes.
An earlier study (34) of two focal populations used restriction fragment length polymorphism-IS genomic “fingerprinting” to find mutations that were caused by IS-element insertions and that had been substituted in one focal population or the other. Four IS150 insertions were then characterized with respect to their site of insertion in pykF, nadR, hokB/sokB, and upstream of pbpA-rodA. These four genes became the blind candidate genes for this study, and each was sequenced in the ancestral strain and in clones sampled at generation 20,000 from all 12 evolved populations. Evolved clones were chosen at random from single-colony isolates. The candidate genes vary in length, with the extent of sequencing shown in Fig. 1. The ancestral sequences for these genes were deposited in GenBank with accession numbers AY849930–AY849933.
Sequences of Randomly Chosen Genes.
We previously chose 36 genes at random from the E. coli genome, and we sequenced ≈500-bp regions in each gene from the ancestor and 2 clones randomly sampled from each of the 12 populations after 20,000 generations (35). The ancestral nucleotide sequences for these regions were deposited in GenBank with accession numbers AY625099–AY625134. The total length sequenced from each clone was 18,374 bp. A total of eight mutations was found in the samples; the precise details of each mutation are provided elsewhere (table 3 in ref. 35). In cases where one or both clones had a mutation in a particular gene, that region was sequenced for three more clones randomly sampled from the same population. Four mutations, including three in population A−4 and one in population A+3, were present in all five clones. They are counted as substitutions in Table 1 of this paper. The other four mutations were in population A+6, and all were polymorphic, with two present in four of the five sampled clones and two others in only one clone. For the analyses in this paper, A+6 is considered to have two substitutions in the random genes, which corresponds to the number of cases in which a mutation reached majority status, and which also equals the summed frequency across these four polymorphisms.
Supplementary Material
Acknowledgments
We thank T. Cooper, D. Hall, F. Moore, E. Ostrowski, D. Schemske, and T. Schmidt for discussions and comments on our article. This work was supported by grants from the National Science Foundation.
Footnotes
References
- 1.Simpson G. G. The Major Features of Evolution. New York: Columbia Univ. Press; 1953. [Google Scholar]
- 2.Harvey P. H., Pagel M. The Comparative Method in Evolutionary Biology. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
- 3.Futuyma D. J. Evolutionary Biology. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 4.Losos J. B., Jackman T. R., Larson A., de Queiroz K., Rodríguez-Schettino L. Science. 1998;279:2115–2118. doi: 10.1126/science.279.5359.2115. [DOI] [PubMed] [Google Scholar]
- 5.Rundle H. D., Nagel L., Boughman J. W., Schluter D. Science. 2000;287:306–308. doi: 10.1126/science.287.5451.306. [DOI] [PubMed] [Google Scholar]
- 6.Crandall K. A., Kelsey C. R., Imamichi H., Lane H. C., Salzman N. P. Mol. Biol. Evol. 1999;16:372–382. doi: 10.1093/oxfordjournals.molbev.a026118. [DOI] [PubMed] [Google Scholar]
- 7.Reid S. D., Herbelin C. J., Bumbaugh A. C., Selander R. K., Whittam T. S. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 8.Stewart C. B., Schilling J. W., Wilson A. C. Nature. 1987;330:401–404. doi: 10.1038/330401a0. [DOI] [PubMed] [Google Scholar]
- 9.Conway Morris S. Life’s Solution. Cambridge, U.K.: Cambridge Univ. Press; 2003. [Google Scholar]
- 10.Jones C. D., Begun D. J. Proc. Natl. Acad. Sci. USA. 2005;102:11373–11378. doi: 10.1073/pnas.0503528102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenski R. E., Rose M. R., Simpson S. C., Tadler S. C. Am. Nat. 1991;138:1315–1341. [Google Scholar]
- 12.Elena S. F., Lenski R. E. Nat. Rev. Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 13.Lenski R. E, Travisano M. Proc. Natl. Acad. Sci. USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenzweig R. F., Sharp R. R., Treves D. S., Adams J. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasi F., Travisano M., Lenski R. E. Am. Nat. 1994;144:432–456. [Google Scholar]
- 16.Travisano M., Mongold J. A., Bennett A. F., Lenski R. E. Science. 1995;267:87–90. doi: 10.1126/science.7809610. [DOI] [PubMed] [Google Scholar]
- 17.Mongold J. A., Bennett A. F., Lenski R. E. Evolution. 1996;50:35–43. doi: 10.1111/j.1558-5646.1996.tb04470.x. [DOI] [PubMed] [Google Scholar]
- 18.Travisano M., Lenski R. E. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bull J. J., Badgett M. R., Wichman H. A., Huelsenback J. P., Hillis D. M., Gulati A., Ho C., Molineux I. J. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sniegowski P. D., Gerrish P. J., Lenski R. E. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 21.Rainey P. B., Travisano M. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 22.Burch C. L., Chao L. Genetics. 1999;151:921–927. doi: 10.1093/genetics/151.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferea T. L., Botstein D., Brown P. O., Rosenzweig. R. F. Proc. Natl. Acad. Sci. USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wichman H. A., Badgett M. R., Scott L. A., Boulianne C. M., Bull J. J. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 25.Cooper V. S., Lenski R. E. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- 26.Notley-McRobb L., Ferenci T. Genetics. 2000;156:1493–1501. doi: 10.1093/genetics/156.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper V. S., Schneider D., Blot M., Lenski R. E. J. Bacteriol. 2001;183:2834–2841. doi: 10.1128/JB.183.9.2834-2841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riehle M. M., Bennett A. F., Long A. D. Proc. Natl. Acad. Sci. USA. 2001;98:525–530. doi: 10.1073/pnas.021448998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper T. F., Rozen D. E., Lenski R. E. Proc. Natl. Acad. Sci. USA. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong S., Khodursky A., Dykhuizen D. E., Dean A. M. Proc. Natl. Acad. Sci. USA. 2004;101:11719–11724. doi: 10.1073/pnas.0404397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crozat E., Philippe N., Lenski R. E., Geiselmann J., Schneider D. Genetics. 2005;169:523–532. doi: 10.1534/genetics.104.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenski R. E. Plant Breed. Rev. 2004;24:225–265. [Google Scholar]
- 33.Blattner F. R., Plunkett G., Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 34.Schneider D., Duperchy E., Coursange E., Lenski R. E., Blot M. Genetics. 2000;156:477–488. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenski R. E., Winkworth C. L., Riley M. A. J. Mol. Evol. 2003;56:498–508. doi: 10.1007/s00239-002-2423-0. [DOI] [PubMed] [Google Scholar]
- 36.Papadopoulos D., Schneider D., Meier-Eiss J., Arber W., Lenski R. E., Blot M. Proc. Natl. Acad. Sci. USA. 1999;96:3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozen D. E., Schneider D., Lenski R. E. J. Mol. Evol. 2005;61:171–180. doi: 10.1007/s00239-004-0322-2. [DOI] [PubMed] [Google Scholar]
- 38.Gerrish P. J., Lenski R. E. Genetica. 1998;102/103:127–144. [PubMed] [Google Scholar]
- 39.Woods R. J. Ph.D. dissertation. East Lansing: Michigan State University; 2005. [Google Scholar]
- 40.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 41.De Visser J. A. G. M., Zeyl C. W., Gerrish P. J., Blanchard J. L, Lenski R. E. Science. 1999;283:404–406. doi: 10.1126/science.283.5400.404. [DOI] [PubMed] [Google Scholar]
- 42.Mahillon J., Chandler M. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penfound T., Foster J. W. J. Bacteriol. 1999;181:648–655. doi: 10.1128/jb.181.2.648-655.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grose J. H., Bergthorsson U., Roth J. R. J. Bacteriol. 2005;187:2774–2782. doi: 10.1128/JB.187.8.2774-2782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grose J. H., Joss L., Velick S. F., Roth J. R. Proc. Natl. Acad. Sci. USA. 2006;103:7601–7606. doi: 10.1073/pnas.0602494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen K., Gerdes K. Mol. Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 47.Schneider D., Duperchy E., Depeyrot J., Coursange E., Lenski R. E., Blot M. BMC Microbiol. 2002;2:18. doi: 10.1186/1471-2180-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Begg K. J., Donachie W. D. J. Bacteriol. 1998;180:2564–2567. doi: 10.1128/jb.180.9.2564-2567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shendure J., Porreca G. J., Reppas N. B., Lin X., McCutcheon J. P., Rosenbaum A. M., Wang M. D., Zhang K., Mitra R. D., Church G. M. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 50.Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y.-J., Chen Z., et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.