Abstract
The Agave (Agavaceae) are keystone species of semiarid to arid regions where the geographic center of origin is Mexico but whose populations spread from the southwestern U.S. through Central America, the Caribbean, and into northern South America. Our analyses indicate that Agave is a young genus, between 7.8 and 10.1 million years old, and yet it harbors the most species of any genera in the family. Of the eight genera in the family, Agave is paraphyletic with respect to three of them, and these four genera are often grouped into a genus termed Agave sensu lato, which harbors 208 of the 293 recognized species in the Agavaceae. In this article, we examine the phylogenetic limits of Agave sensu lato and present analyses elucidating the origin and rate of speciation in the group. These analyses lead to some new insights into the phylogenetic limits of Agave, indicate an estimated age of the family between 20 and 26 million years and an age of the Agave sensu lato of ≤10 million years. Furthermore, we estimate a high mean per-lineage rate of diversification for the genus and find that rates of speciation were significantly elevated between 8 and 6 million years ago and then again between 3 and 2.5 million years ago. We discuss the potential for both monocarpy and the evolution of a generalist pollination system, largely dependent on nectarivorous bat species, as possible driving factors in the radiation of the group.
Keywords: adaptive radiation, molecular phylogeny, monocotyledons, penalized likelihood
The Agave produce one of the largest known inflorescences in the plant kingdom (1) and are predominantly monocarpic (reproducing only once after many years and then dying). Agave spp. are of considerable ecological (as keystone species) and economic importance providing the bases for several important industries in Mexico (e.g., tequila and mezcal). The genus Agave includes ≈166 species and is the largest genus in the family Agavaceae that consists of 9 genera and ≈293 species. The genus Agave is paraphyletic to the genera Manfreda, Polianthes, and Prochnyanthes, and the entire clade of 208 species (in four genera) has been termed Agave sensu lato (2).
It has long been recognized that rates of speciation differ between families, or genera, of flowering plants. Broad-scale analyses have identified families with particularly high or low rates of diversification (3), and lower level studies have used molecular phylogenetic-based approaches to identify genera that exhibit rapid rates of speciation in plants (3–12). The ultimate goal of most of these studies is to identify which evolutionary processes are responsible for accelerating or decelerating speciation rates and, in particular, to identify whether radiations are adaptive (13). Schluter (13) describes four features that mark an adaptive radiation: common ancestry, rapid speciation, phenotype-environment correlation, and trait utility. The first two of these are typically identified by using statistical methods in molecular phylogenetics, the third is often demonstrated by showing independent correlations between phenotype and environment across taxa [for example by using independent contrasts (14)], and the fourth is shown by performing detailed experimental and theoretical work to demonstrate that a specific trait is associated with a rapid increase in speciation rates (13).
In this article, we examine the possibility of a radiation in the genus Agave sensu lato. Of the nine genera in the Agavaceae, two are distinctly more diverse than the others. The genus Yucca has an estimated 49 species, whereas Agave sensu stricto harbors ≈166 species and Agave sensu lato 208 species; thus together, Yucca and Agave include 257 of the 293 species in the family. Although basic aspects of the overall morphology, such as the existence of rosettes and a central spike bearing a large inflorescence, are shared between Yucca and Agave, the two differ remarkably in their reproductive ecology in ways that may have influenced the tempo and mode of speciation. Yucca species are long-lived iteroparous perennials, with the exception of Hesperoyucca whipplei (previously Yucca whipplei), which is monocarpic. Their flowers produce little or no nectar and are exclusively pollinated by specialized moths in the genera Tegeticula and Parategeticula. Pellmyr and Leebens-Mack (15) examined the molecular evolution and phylogenetics of the Tegeticula moths and estimated that the Yucca–Yucca moth coevolution began ≈40 million years ago (Mya) based on molecular clock analyses of Tegeticula mitochondrial genes. Gaunt and Miles (16) recently estimated this date to be between 30 and 32 Mya based on the divergence of the genera Tegeticula and Parategeticula. In either case, the unusual dynamic of Yucca–Yucca moth coevolution has been taken as an example of how exploitative interactions between species (in this case, plants and their pollinators) can mediate adaptive radiations (13).
On the other hand, the genus Agave sensu stricto is predominantly monocarpic and harbors the most dry-adapted (and succulent) members of the family Agavaceae. Agave sensu stricto is divided into two subgenera, Littaea (53 species) and Agave (113 species) based on the inflorescence (1). The three genera that are additionally included in Agave sensu lato, Manfreda (28 species), Polianthes (13 species), and Prochynanthes (1 species), are predominantly polycarpic, herbaceous, and inhabit a more temperate environment and are also mainly found in Mega-Mexico 3 (Mexico and continuous parts of southwestern U.S. and Central America that share similar flora) (2, 17).
In addition to their contrasting life-history strategy, polycarpy vs. monocarpy, the Yucca and Agave differ dramatically in their pollination ecology. Unlike Yucca, most species in the genus Agave have a broad coterie of pollinators including bees, birds, hawkmoths, and bats. Their flowers appear to have been selected by a bat pollination syndrome (1, 18, 19).
In this article, we present analyses to examine the first two features of an adaptive radiation as outlined by Schluter (13): the monophyly and rate of speciation in the group. To this end, we present analyses on (i) the phylogenetic limits of the Agave sensu lato, (ii) the time of origin of the family Agavaceae and Agave sensu lato, and (iii) the rate and timing of speciation events in Agave sensu lato. Although the nine genera fall into consistent patterns across these analyses, there are questions about the phylogenetic limits of the family and the phylogeny of Agave sensu lato. Additionally, using our estimates of the time of origin and rate of speciation in the genus Agave, we discuss the possible evolutionary forces mediating high diversification rates in Agave.
Results
Phylogeny of the Agavaceae and Agave.
The molecular clock hypothesis was firmly rejected for the rbcL data set but is only rejected at the 0.05% level for the trnL+trnL−trnF data (Table 1).
Table 1.
Comparison of likelihood scores under different phylogenetic estimators for two chloroplast regions: trnL + trnL−trnF and rbcL using the appropriate model of sequence evolution (Tamura Nei or GT + I + Γ)
| Taxa included (df) | Region | Tree | −Log L0 (nonclock) | −Log L1 (clock) | Λ | P |
|---|---|---|---|---|---|---|
| Agavaceae + monocots (59) | trnL + trnL−trnF | MP | 1,328.56 | 1,364.7 | 72.36 | <0.05 |
| ME | 1,331.8 | 1,367.74 | 71.80 | <0.05 | ||
| Monocots − grasses (242) | rbcL | MP | 61,964.3 | 62,847.9 | 1,767.2 | <0.0001 |
| ME | 62,847.6 | 63,607.2 | 1,507.2 | <0.0001 |
The likelihood ratio test is based on the difference between the two models. P values are based on a χ2 distribution with n− 2 degrees of freedom. MP, maximum parsimony; ME, minimum evolution.
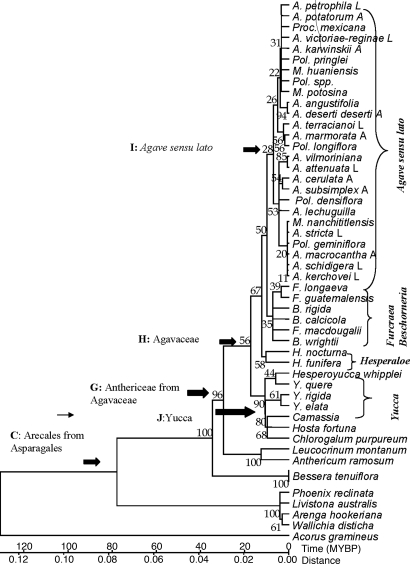
The topology of the minimum evolution tree based on the rbcL sequences is shown in Fig. 3, which is published as supporting information on the PNAS web site. A phylogenetic tree reconstructed based on the trnL+trnL−trnF sequence data using the minimum evolution method is depicted in Fig. 1. The tree depicted is one that has been linearized with respect to time. There is evidence that members of the Anthericaceae are a sister group to the Agavaceae and that Camassia, Hosta, and Chlorogalum are part of Agavaceae sensu lato. Both the methods of minimum evolution and maximum parsimony concur that there is low bootstrap, but are consistent with separation of Agave sensu lato, Furcraea, and Beschorneria, and give stronger support for the distinctiveness of Yucca (both analyses) and Hesperaloe (maximum parsimony analyses). Both analyses suggest that Hosta, Camassia, and Chlorogalum are paraphyletic within the Agavaceae as found by the Angiosperm Phylogeny Group (20).
Fig. 1.
Minimum evolution tree depicting the relationship among species in Agave sensu lato and other genera in the Agavaceae based on trnL+trnL−trnF. The tree was rooted using Acorus gramineus and calibrated using an estimated age of 132 million years for the crown age. Numbers at the nodes are bootstrap values generating from running 1,000 replicates of the tree. The tree was linearized with respect to time, and the time scale of evolution within this group of monocotyledons is indicated by a scale bar represented in Mya.
Estimating the Age of the Agavaceae and Agave sensu lato.
For the linearized trees method, a total of 134 sequences were eliminated for the rbcL analyses because they exhibited a significantly higher rate of sequence evolution; this method left 191 taxa for the analyses (including the 14 outgroup sequences), whereas for the trnL+trnL−trnF data a total of 11 sequences were removed (leaving 51 sequences). The trees were linearized with respect to time, and using a crown age of 132 million years, we established a substitution rate of r = 0.000373134 units/million years for rbcL and r = 0.0023148 units/million years for trnL+trnL−trnF and derived the SE for the time of divergence of the Agavaceae family using the method presented in Kumar et al. (21). For the nonparametric rate-smoothing (NPRS) method, a total of 242 rbcL sequences (including the 15 outgroup samples) and 54 trnL+trnL−trnF sequences were used to estimate divergence times.
The comparison of the age of nine internal nodes from our rbcL and trnL+trnL−trnF analyses with those of Wikström et al. (22) are presented in Table 2. Wikström et al. (22) used an internal absolute calibration date of 84 Mya and, consequently, estimate the age of the genus Acorus to ≈147 Mya. This age falls into the range of ages estimated by Sanderson and Doyle (23) using molecular estimates based on rbcL or 18S rDNA and is slightly older than the best fossil estimate of the origin of angiosperms based on a primitive dicotyledon. Because we calibrate our trees based on a fossil-date crown age of 132 million years, our dates could be expected to be younger than those obtained by Wikström et al. (22), but they are in fairly close agreement. Key points to note are that all of the molecular estimates of the age of Asparagales fall in the range of ≈60–69 million years, whereas the best fossil date for the age of the Asparagales is 37.5million years (3). Also, both this study and Wikström et al.'s (22) indicate a common estimate of the divergence of Agavaceae (including paraphyletically Camassia, Hosta, and Chlorogalum) from the Anthericaceae of ≈30–35 Mya. However, we obtain a younger estimate for the age of the Agavaceae family presumably because of the inclusion of more genera closely related to the group (see Fig. 3). Finally, it should be noted that we obtain similar ages for the origin of Agave sensu lato (see Table 2), between 7.9 and 9.8 Mya using both data sets and both methods of analyses but slightly more variable estimates of the age of the family Agavaceae spanning from 20.5 (trnL+trnL−trnF data using the linearized trees method) to 25.8 Mya (using the rbcL data set and the NPRS method) and dates for the origin of the Yucca between 13 and 18 Mya (Table 2). The difficulties inherent in estimating the origin of the family may be caused, in part, by the inclusion of different species in the two analyses: in particular with the trnL+trnL−trnF data, we include several species of Yucca including Y. whipplei, recently renamed H. whipplei, the monocarpic species of Yucca, with several other polycarpic Yucca's. H. whipplei is more closely related to the Agave than other species of Yucca are to the Agave, which would influence the estimated age of origin of the family. Additionally, this discrepancy may reflect differences in the rate or nature of molecular evolution of the two genes.
Table 2.
Estimated age of eight nodes on phylogenetic trees reconstructed with rbcL and trnL+trnL−trnF sequence data using either linearized trees method or NPRS and a comparison of these ages with those obtained by Wikström et al. (22) based on a combined data set of rbcL and atpB
| Node | Split | Linearized trees |
NPRS |
Wikström (ML) | ||
|---|---|---|---|---|---|---|
| rbcL | trn data | rbcL | trn data | |||
| A | Acorus | 132 | 132 | 134 | 132 | 147 |
| B | Alismatales | 126.1 | — | 112.2 | — | 118 |
| C | Arecales from Asparagales | 84.1 | 78.2 | 81.6 | 88.9 | — |
| D | Asparagales | 69 | — | 60 | — | 60 |
| E | Asparagaceae/Anthericaceae + Agavaceae | 47.3 | — | 39.3 | — | 49 |
| F | Behniaceae | 31.1 | — | 32.1 | — | 35 |
| G | Anthericaceae/Agavaceae clade+ | 32.6 | 29.1 | 31.7 | 34.2 | 35 |
| H | Agavaceae clade+ | 20.5 ± 2.1 | 23.4 ± 2.2 | 22.4 ± 1.7 | 25.8 ± 3.4 | — |
| I | Agave sensu lato | 8.8 ± 2.2 | 7.9 ± 1.7 | 8.3 ± 2.4 | 9.8 ± 3.3 | — |
| J | Yucca | 13.4 ± 2.7 | 18.3 ± 3.1 | 14.1 ± 3.0 | 17.2 ± 2.3 | — |
The nodes letters refer to those indicated in Fig. 3 (rbcL) and Fig. 1 (trnL+trnL−trnF). +, clade containing the Agavaceae also includes Hosta, Camassia, and Chlorogalum; ML, maximum likelihood.
Rates of Diversification in Agave sensu lato.
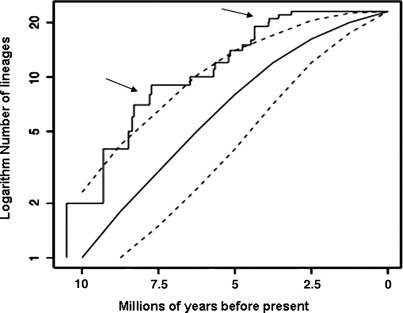
The rates of diversification Ŝ and D̂ in Agave sensu lato and D̂ in Yucca are presented in Table 3. This shows that the estimator of the per-lineage rate of diversification, Ŝ, is 0.32 in Agave sensu lato. Although it is a less accurate estimator, comparison of the values of D̂ in Agave sensu lato with those in Yucca show that rates of diversification have been ≈2 times greater in Agave sensu lato (0.21–0.27 in Yucca vs. 0.51 in Agave). The difference between the estimates of Ŝ and D̂ results from different information contained in the two estimates: Ŝ uses information in the branching pattern among the 26 species represented in our phylogeny, whereas D̂uses only the raw species data information. We calculate the γ statistic to be −4.401, which indicates that the rates of diversification have been significantly decelerating toward the present (a value of γ < −1.64 is significant at the 5% level). Using the program phylogen and constructing 1,000 phylogenies of 26 species from a total taxonomic pool of 208 extant species demonstrates that the expected estimate of γ is −1.56 with a 95% confidence limit of −3.46 to 0.23 This indicates that sampling only a subset of the true phylogeny biases the estimate of γ downwards, but not to the degree observed with our data set. By using a time of origin of Agave sensu lato of 10 Mya, the logarithm of the number of lineages through time (Fig. 2) indicates that the two periods during which speciation rates appear to have been significantly elevated against background levels are between ≈8 and 6 Mya and from 3–2.5 Mya.
Table 3.
Mean per lineage rate of diversification in Agave sensu lato or Yucca using two different estimators
| Group | No. of species | S, species/Mya | D, species/Mya |
|---|---|---|---|
| Agave sensu lato | 208 | 0.32 ± 0.08 | 0.51 ± 0.06 |
| Yucca | 50 | — | D14.1 = 0.27 ± 0.03 or D18.3 = 0.21 ± 0.02 |
Fig. 2.
A lineage through time plot of the logarithm number of species against millions of years before present generated from the phylogeny based on trnL+trnL−trnF for Agave sensu lato. The dashed lines represent the 95% confidence interval of the expected number of lineages present over time when 26 individuals are sampled from a total phylogeny of 208 individuals. Arrows point to the time intervals when the observed number of lineages is greater than expected under a pure birth–death model of diversification.
Discussion
Monophyly of Agave and the Estimated Age of Agavaceae and Agave sensu lato.
Our phylogenetic analyses concur with those obtained in other molecular studies, confirming the phylogenetic position of the Agavaceae within the Asparagales, paraphyletic with Camassia, Hosta, and Chlorogalum, and a sister group to the Anthericaceae (17, 24), although they raise questions about the phylogenetic limits of the family. We estimate the age of the family to be between 20 and 26 million years and the age of Yucca to be between 13 and 18 million years; both dates are considerably younger than the age of Yucca–Yucca-moth coevolution (at 41.5 Mya) as suggested by Pellmyr and Leebens-Mack (15), although the upper limit for our estimate of the family is closer to a recent estimate by Gaunt and Miles (16) of 30–32 million years for the divergence of Tegeticula and Parategeticula moths. Tidwell and Parker (25) identified a Yucca-like fossil in Mexico and suggest that it has an estimated age of 14 million years, whereas Yucca-like pollen has been reported in the range of 15–25 million years old (25–27). The oldest Asparagales fossil (the suborder including the family Agavaceae) is estimated to be 37.5 million years old (3); however, as this article indicates, molecular estimates of the age of the Asparagales are considerably older than this, ranging from 60–69 million years old. Although all of these fossil dates serve as minimum ages, phylogenetic analyses also suggest that an age of 41.5 million years is probably too old. For example, Wikström et al. (22) estimate the age of divergence of the Anthericaceae from the Agavaceae at 35 million years and Bogler et al. (24) suggest that the origin of the family Agavaceae is <40 million years old.
Our age estimates are determined by calibrating trees based on the crown age of the angiosperms of 132–143 million years old. Furthermore, rbcL does not conform to a constant molecular clock as found in other studies (23). Despite this phenomenon, our results suggest that an origin of Agave sensu lato of 8–10 Mya appears robust: We find good agreement between our analyses and those obtained by Wikström et al. (22). We obtain similar dates for all nodes using two independent methods of analyses, one that assumes a molecular clock and one that does not, and with two different genes that evolve at different rates (although they are linked).
Rate of Diversification in Agave sensu lato.
Using a conservative date of 10 Mya for the origin of Agave sensu lato, we obtain mean lineage diversification rates of Ŝ = 0.32 ± 0.08 species per million years, calculated from the origin of the Agave until the present. This value is within the range observed for other genera of plants that have been identified as undergoing rapid rates of relatively recent speciation. For example, lineage diversification rates from the start of the radiation to the present in genera of the Poaceae in the South African Cape Flora range from 0.12–0.39 species per Mya (11), although the rates range from 0.87–4.18 during the period of elevated radiation. Several other recent studies have identified remarkable rates of diversification rates including 0.56 in Silversword alliance (4), 0.717–0.832 in the oceanic island shrubs Gaertnera (9), and 0.85 in the South America radiation of Gentianella (12).
Because we do not have a detailed phylogeny of Yucca, we can only compare values of D̂ in Agave sensu lato and Yucca. Depending on whether we accept a date of 13 or 18 Mya for the age of the Yucca, we conclude that rates of diversification in the genus Yucca are 0.21 or 0.27 or approximately half of the rate of 0.53 in Agave sensu lato, suggesting that despite evidence that the extreme form of specialized pollination found in the Yuccas has, in part, driven high rates of speciation in Yucca, other forces have precipitated an even greater rate of diversification in the Agave.
Timing of Speciation in Agave sensu lato and Possible Mechanisms Driving Diversification.
Given the difficulties in obtaining adequate sampling to obtain reliable estimates of lineage diversification rates (28), one of the most useful applications of this approach is in detecting significant changes in the rate of diversification (10). Our analyses indicate that the distribution of the γ parameter is left skewed given our sampling regime, but we still obtain evidence that speciation rates have been significantly decelerating toward the present and that rates of speciation were elevated between 8 and 6 Mya and again between 3 and 2.5 Mya. Do these dates correspond to any specific changes in the geology or life-history of the group?
The peak in speciation rates in Agave sensu lato ≈8–10 Mya corresponds to an increase in dry conditions in central Mexico. Geological evidence suggests that volcanic activity between 30–15 Mya changed the climate within Mexico, leading to drier regions in the center of the country after this period and the Sonoran desert forming between 15 and 8 Mya (29). During this dry period, a basin running through central Mexico, the Mesa Central of Mexico, split: this event has been used to explain an early diversification of Goodeidae fishes in Central Mexico (30) at 11.5 Mya. Between 5.3 and 1.8 Mya, there was a general reversal toward more tropical wet climates in Mexico after which the climate became much colder and drier during glacial periods (29).
A recent article on the radiation of the tropical dry forest genus Bursera (Burseraceae) estimates that this old genus radiated in northwest Mexico 34–17 Mya and then into southwest Mexico 15–7.5 Mya (31), suggesting that the tropical dry forest in Mexico expanded north and then south during these time frames (31) or earlier, depending on whether a date of 95–100 Mya (the breakup of Gondwanaland) or 60 Mya (Paleocene) is the correct date to use for the divergence of African and New World genera of Burseraceae (32). These dates reveal that the first diversification of Bursera occurred reasonably close to our estimate for the origin of the family (20–26 Mya), and the timing of the second diversification is comparable to the date of diversification of Agave (<10 Mya). Indeed the most rapid phase of diversification for Bursera was between 13.5 and 7.5 Mya, calculated using the Gondwanaland calibration point. A recent analysis of the dry forest legume Leucaena also indicates that this genus underwent an endemic radiation beginning 10 Mya in Mexico through a similar region of the country to Bursera (33). Cumulatively, these analyses suggest that dry-adapted species have undergone parallel radiations within the last 10 million years in Mexico. The Agave exhibit the most adaptations to dry environments of all genera in the Agavaceae such as the formation of large succulent rosettes that funnel water and maintain extensive root systems (1).
If the early increase in speciation in the Agave was precipitated by colonization to arid environments, what factors may have influenced the rapid speciation from 3.0–2.5 Mya? Nectar feeding bats are probably the single most important pollinator in most species of the subgenera Agave (2, 34). In Table 4, we show a list of the genera in the Agavaceae and describe the predominant pollinators of each of the genera, as reviewed by Rocha et al. (17). Members of Agave sensu stricto and Manfreda (closely related to Agave) are the only genera in the family that are predominantly pollinated by bats, although bat pollination is also reported for one species of Hesperaloe (35). Moth/hawkmoth pollination appears to be the ancestral pollination syndrome in the Agavaceae, with bat pollination evolving more recently in the genus Agave. This finding raises the question of when and to what extent rates of diversification of Agave have been driven by diversification in their bat pollinators. The distribution of Agave species is largely coincident with the distribution of nectarivorous bats. Mexico is home to 12 species of nectarivorous bats (Phyllostomidae, subfamily Glossophaginae) (36). The two species of Leptonycteris, L. curasoae and L. nivalis, feed primarily on Agave spp. and columnar cacti (37, 38). Phyllostomid New World bats are estimated to have evolved ≤15 Mya (39), and bats of the genera Glossophaga and Leptonyceteris diverged from a common ancestor ≈2.4 Mya, whereas the two Leptonycteris species diverged ≈1.0 Mya (40). This finding suggests that the pollination ecology of Agave species has been influenced by nectar feeding bats only within the last 3 million years, although a generalist bat species could have influenced their evolution before that time.
Table 4.
Description of the number of species, life-history (monocarpic vs. polycarpic), and dominant pollinators in each genus in the Agavaceae, after Rocha et al. (17) and Eguiarte et al. (2)
| Genus | No. of species | Life-history | Dominant pollinators |
|---|---|---|---|
| Agave | 166 | Monocarpic | Bat, hawkmoth, bee, and bird |
| Manfreda | 28 | Polypcarpic | Hawkmoth, moth, and bat |
| Polianthes | 13 | Polycarpic | Hawkmoth and hummingbird |
| Prochnyanthes | 1 | Polycarpic | Hawkmoth or moth |
| Furcraea | 25 | Monocarpic | Moth and Hummingbird |
| Beschorneria | 7 | Polycarpic | Hummingbird and hawkmoth |
| Hesperaloe | 5 | Polycarpic | Hawkmoth, Hummingbird, and bat |
| H. whipplei | 1 | Monocarpic | Yucca moths |
| Yucca | 50 | polycarpic | Yucca moths |
Given the coincident distribution of Agave and columnar cacti with that of Leptonycteris species, authors have suggested that the Leptonycteris–columnar cacti–Agave interspecific relationships may be an example of coevolution (1, 41–44). This hypothesis is supported by phenological data for members of the subgenus Agave species and columnar cacti that demonstrate that bat migration follows their nectar corridor (1, 44–46). Columnar cacti belong to the tribe Pachycereeae, which harbor 58 species (47) of which ≈70% are suspected to have some bat pollination, many of them also are pollinated by Leptonycteris species (43). Recent phylogenetic data surprisingly indicate that the entire family Cactaceae is very young, probably on the order of 30 million years old (48) and that the group originated in South America, subsequently radiating northwards into Mexico. Taken together, the phylogenetic history of the columnar cacti and Leptonycteris bats suggests that Agave pollination ecology has been greatly influenced by the emergence of interspecific relations with these two groups only within the last 2–3 million years. Interestingly, both Pellmyr and Leebans-Mack (15) and Gaunt and Miles (16) estimate a radiation in yucca moths in the last 2–3 million years coincident with this most recent radiation in Agave.
In addition to the role that low interspecific competition and high ecological opportunity can contribute to radiations, Hodges and Arnold have stressed the role of key innovations (49). Table 4 describes the life-history characteristics of each of the genera in the Agavaceae and indicates that Agave and Furcraea are monocarpic genera with only one other species, H. whipplei, being identified as monocarpic in the family. The evolution of monocarpy in concert with changes in the physiology and morphology of the genus (long tap roots, funnel shaped rosette, etc.) were probably important factors in the ability of the Agave to colonize dry heterogeneous desert environments because the family is not pleisiomorphically dry-adapted to the extent of families like the Cactaceae. Monocarpy would also have been selected by bat pollination because the reproductive effort required to secure pollination by bats is high, as evidenced by the positive correlation between nectar production and inflorescence height with bat pollination in Agave (17, 50). By combining the rate analyses presented here with analyses aimed at mapping life-history and flowering characteristics onto phylogenetic trees, we aim to uncover the driving forces of rapid speciation in the Agave.
Materials and Methods
Study Organism and Samples.
Gentry (1) proposed that the subgenus Littaea was the ancestral form, but molecular data indicate that the Agave subgenus is paraphyletic (2, 51). Because the genus Agave is paraphyletic with respect to the genera Manfreda (28 species), Polianthes (13 species), and Prochnyanthes (1 species) at the molecular level (2, 51, 52), we use the term Agave sensu lato to refer to the clade including all four genera as recently proposed by Thiede (53) and retain the use of Agave sensu stricto for Agave in the traditional sense. We collected samples from 25 species of Agave sensu lato for phylogenetic analyses from the Jardín Botánico at the Universidad Autónoma de Mexico (Mexico City). In addition, we present previously uncharacterized sequence data for 12 additional species in the Agavaceae (see Table 5, which is published as supporting information on the PNAS web site).
Phylogeny of the Agave.
We combined new sequences from a 570-bp region of the intron in trnL that we sequenced using primers c and d from Taberlet (54) with a 378-bp region of sequence for the spacer between trnL and trnF using primers e and f (54) for all 37 species in the Agavaceae (we call this the trnL+trnL−trnF data set). These sequence data were combined with homologous sequences from 21 other species of monocotyledons obtained from GenBank. The final alignment length of the combined data sets was 1097 bp. We also obtained 325 DNA sequences representing most of the families in the monocotyledons for 1428 bp of the chloroplast rbcL gene from GenBank and from a previous paper (55) and two species of Acorus (A. calamus and A. gramineus), the recognized sister lineage to all other monocots, 18 primitive dicotyledons, and a conifer (Pinus radiata, in subsequent analyses) to root the tree.
The trnL+trnL−trnF and rbcL data sets were aligned with clustalx and then edited manually in bioedit (56). The best model of sequence evolution was determined by likelihood ratio tests as implemented in modeltest (57) and was determined to be GTR + I + Γ (a = 0.78) for trnL+trnL−trnF and Tamura–Nei + Γ (a = 76) for rbcL, where GTR is general time reversible. Phylogenies were reconstructed by using both minimum evolution and maximum parsimony optimality criterion using mega version 2.1 (58) and paup version 4.0 (59). Confidence in nodes was assessed using the bootstrap method with 1,000 replicates. The assumption of a molecular clock was tested for all analyses by comparing the log likelihood of a phylogeny using a clock-constrained model with that of one constructed with an unconstrained model using paup version 4.0 (59). The likelihood ratio G (−2[lnclock − lnno clock]) was assumed to be distributed as a χ2, with the degrees of freedom equal to the number of terminal sequences minus 2.
Origin of the Agavaceae and Agave.
We used both the rbcL data and the combined trnL+trnL−trnF data set to estimate the age of origin of the Agavaceae and Agave sensu lato using both the linearized trees (60) and NPRS (61) methods. For the former method, all of the slow and fast evolving branches were removed by using the two cluster test (60), and then a uniform rate of evolution was imposed on the phylogenetic tree reconstructed using the minimum evolution method as described above. The NPRS method allows independent molecular clock analyses across the tree but imposes a penalty for changing rates too quickly. For this analysis, phylogenetic trees were first constructed using maximum parsimony with paup version 4.1; the branch lengths on this tree were estimated using maximum likelihood and the GTR + I + Γ model of DNA sequence evolution. To estimate the appropriate penalty to impose for changing the rate of evolution along branches of the tree, we first estimated the smoothing parameter by a cross-validation procedure on a subset of the data and then performed the NPRS analyses using the TN (Truncated Newton) algorithm (for rbcL) or the POWELL algorithm (for trnL+trnL−trnF) on the full data set in the program r8s (61).
After the relative age (distance) of nodes was ascertained for both of the methods described above, a molecular clock was calibrated using a single reference point for the linearized trees method or, for NPRS, by constraining the age of multiple nodes to minimum and maximum ages. For the linearized trees method, we use the fossil estimate of 132 Mya as the crown age of the angiosperms (62) to calibrate the clock. To calibrate the clock for NPRS, we constrained the root of the tree, the crown age of the angiosperms, to between 132 (fossil estimate) and 143 Mya (molecular estimate) (23) and use fossil evidence to set the minimum age of the following groups after Magallón et al. (3): Asparagales, 37.5 Mya; Arecaeae, 84 Mya; Bromeliaceae, 37.5 Mya; and Liliales, 45.15 Mya. We compare the age of the nodes from our analyses with those presented in Wikström et al. (22).
Rate and Timing of Speciation Accelerated Speciation in the Agave.
To understand the rate and timing of speciation events in Agave sensu lato, we use the linearized minimum evolution tree constructed from the trnL+trnL−trnF data described above. We calculate the rate of speciation on this tree assuming a Yule process as Ŝ = (n − m)/B, where n and m are the number of the lineages at the end and beginning of the time (26 and 1, respectively) under consideration and B is the summed durations of all branches in the Agave sensu lato clade. This estimate assumes a constant rate of speciation but uses the phylogenetic information in the tree to estimate Ŝ. Two variances of the estimator Ŝ are commonly used, Kendall's as estimated by var(Ŝ) =S2/2(eST − 1) or Moran's as estimated by var(Ŝ) =b2/2(n − m); we used Kendall's, the more conservative estimate (4). One can also calculate a simpler statistic based on a pure-birth model for the rate of diversification, D. This statistic does not make use of the information in a phylogenetic tree and is simply calculated as D = [ln(Nt) − ln (N0)]/T, where T is the time of origin of the clade and N is the number of extant species (Nt = 208 and N0 = 1). We calculated this statistic for Agave sensu lato and compared it to the second most diverse group, Yucca, using the dates of origin of the genera as inferred from the linearized trees and NPRS analyses.
To determine whether speciation events are accelerating or decelerating toward the present, we calculate the statistic γ, developed by Pybus and Harvey (63) on an ultrametric tree generated using NPRS from the maximum parsimony tree with branch lengths estimated by maximum likelihood for the trnL+trnL−trnF sequence data. The statistic γ follows a standard normal distribution when there is constant speciation but takes on positive or negative values when there is an acceleration or deceleration of speciation rates toward the present. To visualize this progression, we plot the number of lineages through time using the NPRS estimates of node age at all internal nodes on the phylogeny. This method assumes that the complete phylogeny has been sampled: to understand the effect of sampling on the lineage through time plots, we used the program phylogen (version 1.1) (created by A. Rambaut, University of Oxford) to generate 1,000 phylogenies of 208 species that have undergone a constant rate of diversification and then sampled 26 species from the phylogenies and calculated γ and the lineage through time plot for these sampled phylogenies (63). The mean curve and 95% confidence interval of the lineage through time plots is shown on the same graph as our lineage through time plot.
Supplementary Material
Acknowledgments
We thank Abisaí García-Mendoza [Botanical Garden, Institute of Biology, Universidad Nacional Autónoma de México (UNAM)] for supplying most of the plants analyzed in the study and identifying the species; Arturo Silva-Montellano and Alejandro Martínez-Palacios for collecting some other plants; Aldo Valera for extracting most of the DNA and helping in the different stages of the project; Becky Gaut, Andy Peek, Peter Tiffin, and Maud Teneillon for help during the amplification and sequencing of the genes; Amanda Castillo for preliminary analyses of the sequences; Martha Rocha for illuminating discussions on the genus Agave; and Susanna Magallón for introducing us to the program r8s. This project was funded with support from Consejo Nacional de Ciencia y Technología (Mexico) (CONACYT) Projects 27983-N, 4647-Q, and 0028, Secretaría de medio ambiente y recursos naturales–CONACYT Project 2002-C01-0246, Natural Sciences and Engineering Research Council (Canada) funding (to S.V.G.-A.), and sabbatical scholarships from the Dirección General de Asuntos del Personal Académico, UNAM, and CONACYT (to V.S. and L.E.E.).
Abbreviations
- Mya
million years ago
- NPRS
nonparametric rate-smoothing.
Footnotes
References
- 1.Gentry H. S. Agaves of Continental North America. Arizona: University of Arizona Press; 1982. [Google Scholar]
- 2.Eguiarte L., Souza V., Silva-Montellano A. Boletin de la Sociedad Botanica de México. 2000;66:131–150. [Google Scholar]
- 3.Magallón S., Sanderson M. J. Evolution (Lawrence, Kans.) 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin B., Sanderson M. J. Proc. Natl. Acad. Sci. USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodges S. A. Int. J. Plant Sci. 1997;158:S81–S88. [Google Scholar]
- 6.Rauscher J. T. Am. J. Bot. 2002;89:1074–1084. doi: 10.3732/ajb.89.7.1074. [DOI] [PubMed] [Google Scholar]
- 7.Richardson J. E., Weitz F. M., Fay M. F., Cronk Q. C. B., Linder H. P., Reeves G., Chase M. W. Nature. 2001;412:181–183. doi: 10.1038/35084067. [DOI] [PubMed] [Google Scholar]
- 8.Linder H. P., Eldenäs P., Briggs B. Evolution (Lawrence, Kans.) 2003;57:2688–2702. doi: 10.1111/j.0014-3820.2003.tb01513.x. [DOI] [PubMed] [Google Scholar]
- 9.Malcomber S. T. Evolution (Lawrence, Kans.) 2002;56:42–57. doi: 10.1111/j.0014-3820.2002.tb00848.x. [DOI] [PubMed] [Google Scholar]
- 10.Shaw A. J., Cox C. J., Goffinet B., Buck W. R., Boles S. B. Evolution (Lawrence, Kans.) 2003;57:2226–2241. doi: 10.1111/j.0014-3820.2003.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 11.Verboom G. A., Linder H. P., Stock W. D. Evolution (Lawrence, Kans.) 2003;57:1008–1021. doi: 10.1111/j.0014-3820.2003.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 12.von Hagen K. B., Kadereit J. W. Organisms Divers. Evol. 2001;1:61–79. [Google Scholar]
- 13.Schluter D. The Ecology and Evolution of Adaptive Radiations. New York: Oxford Univ. Press; 2000. [Google Scholar]
- 14.Felsenstein J. Am. Nat. 1985;125:1–15. [Google Scholar]
- 15.Pellmyr O., Leebens-Mack J. Proc. Natl. Acad. Sci. USA. 1999;96:9178–9183. doi: 10.1073/pnas.96.16.9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaunt M. W., Miles M. A. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 17.Rocha M., Good-Avila S. V., Molina-Freaner F., Arita H., Castillo A., Garcia-Mendoza A., Silva-Montellano A., Gaut B., Souza V., Eguiarte L. In: Monocots: Comparative Biology and Evolution. Columbus J. T., Friar E. A., Porter J. M., Prince L. M., Simpson M. G., editors. Clairmont, CA: Rancho Santa Ana Botanical Gardens; 2006. in press. [Google Scholar]
- 18.Howell D. Ph.D. thesis. Tucson: University of Arizona; 1972. [Google Scholar]
- 19.Howell D. J. Am. Nat. 1979;114:23–49. [Google Scholar]
- 20.Angiosperm Phylogeny Group. Bot. J. Linnean Soc. 2003;141:399–436. [Google Scholar]
- 21.Kumar S., Balczarek K. A., Lai Z. C. Genetics. 1996;142:965–972. doi: 10.1093/genetics/142.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wikström N., Savolainen V., Chase M. W. Proc. R. Soc. London Ser. B.; 2001. pp. 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanderson M. J., Doyle J. A. Am. J. Bot. 2001;88:1499–1516. [PubMed] [Google Scholar]
- 24.Bogler D. J., Pires J. C., Francisco-Ortega J. In: Monocots: Comparative Biology and Evolution. Columbus J. T., Friar E. A., Porter J. M., Prince L. M., Simpson M. G., editors. Clairmont, CA: Rancho Santa Ana Botanical Gardens; 2006. in press. [Google Scholar]
- 25.Tidwell W. D., Parker L. R. Rev. Paleobot. Palinol. 1990;63:79–85. [Google Scholar]
- 26.Wells P. V. In: Transactions of the Symposium on the Biological Resources of the Chihuahuan Desrt Region: United States and Mexico. Riskind S. H., editor. Washington, DC: Depart. of the Interior, Natl. Park Service; 1974. pp. 67–83. [Google Scholar]
- 27.Palacios R., Rzedowski J. Acta Bot. México. 1993;24:1–96. [Google Scholar]
- 28.Sims H. J., McConway K. J. Evolution (Lawrence, Kans.) 2003;57:460–479. doi: 10.1111/j.0014-3820.2003.tb01538.x. [DOI] [PubMed] [Google Scholar]
- 29.van Devender T. R. In: A Natural History of the Sonoran Desert. Phillips S. J., Comus P. W., editors. Berkeley, CA: Univ. of California Press; 2000. pp. 61–69. [Google Scholar]
- 30.Doadrio I., Domínguez O. Mol. Phyl. Evol. 2004;31:416–430. doi: 10.1016/j.ympev.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Becerra J. X. Proc. Natl. Acad. Sci. USA. 2005;102:10919–10923. doi: 10.1073/pnas.0409127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dick C. W., Wrght S. J. Proc. Natl. Acad. Sci. USA. 2005;102:10757–10758. doi: 10.1073/pnas.0505013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavin M., Schrire B., Lewis G., Pennington R. T., Delgado-Salinas A., Thulin M., Hughes C., Wojciechowski M. F. Philos. Trans. R. Soc. London B. 2004;359:1509–1522. doi: 10.1098/rstb.2004.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slauson L. A. Am. J. Bot. 2000;87:825–836. [PubMed] [Google Scholar]
- 35.Engard R. G. Yucca. Saltillo Coahuila, Mexico: Centro de Investigaciones en Química Aplicada y Comisión Nacional de las Zonas Aridas; 1980. pp. 115–123. [Google Scholar]
- 36.Santos M., Arita H. T. In: Columnar Cacti and Their Mutualists. Flemming T. H., Valiente-Banuet , editors. Tucson: Univ. of Arizona Press; 2002. pp. 342–363. [Google Scholar]
- 37.Rojas-Martinez A., Valiente-Banuet A., Arizmendi M. C., Alcantara-Eguren A., Arita H. T. J. Biogeogr. 1999;26:1065–1077. [Google Scholar]
- 38.Arita H. T., Humphrey S. R. Acta Zool. Mex. 1988;29:1–60. [Google Scholar]
- 39.Proctor M., Yeo P., Lack A. The Natural History of Pollination. Portland, OR: Timber; 1996. [Google Scholar]
- 40.Wilkinson G. S., Fleming T. H. Mol. Ecol. 1996;5:329–339. [PubMed] [Google Scholar]
- 41.Valiente-Banuet A. Am. J. Bot. 1997;84:452–455. [Google Scholar]
- 42.Valiente-Banuet A. Rev. Chil. Hist. Nat. 2002;75:99–104. [Google Scholar]
- 43.Valiente-Banuet A., Arizmendi M. C., Rojas M. A., Domínguez-Canseco L. J. Trop. Ecol. 1996;12:103–119. [Google Scholar]
- 44.Arita H. T. J. Mammal. 1991;80:31–41. [Google Scholar]
- 45.Molina-Freaner F., Eguiarte L. E. Am. J. Bot. 2003;90:1016–1024. doi: 10.3732/ajb.90.7.1016. [DOI] [PubMed] [Google Scholar]
- 46.Fleming T. H., Nuñez R. A., Stemberg S. L. Oecologia. 1993;94:72–75. doi: 10.1007/BF00317304. [DOI] [PubMed] [Google Scholar]
- 47.Dávila-Aranda P., Arias-Montes S., Lira-Saade R., Villaseñor J. L., Valiente-Banuet A. In: Columnar Cacti and Their Mutualists. Fleming T. H., Valiente-Banuet A., editors. Tucson, Arizona: Univ. of Arizona Press; 2002. pp. 25–41. [Google Scholar]
- 48.Nyfeller R. Am. J. Bot. 2002;89:312–326. [Google Scholar]
- 49.Hodges S. A., Arnold M. L. Proc. R. Soc. London B.; 1995. pp. 343–348. [Google Scholar]
- 50.Young T. P., Augspurger C. K. Trends Ecol. Evol. 1991;6:285–289. doi: 10.1016/0169-5347(91)90006-J. [DOI] [PubMed] [Google Scholar]
- 51.Bogler D. J., B. S. B. Am. J. Bot. 1996;83:1225–1235. [Google Scholar]
- 52.Bogler D. J., Simpson B. B. Syst. Biol. 1995;20:191–205. [Google Scholar]
- 53.Thiede J. In: Illustrated Handbook of Succulent Plants, Monocotyledons. Eggli U., editor. Berlin: Springer; 2001. p. 354. [Google Scholar]
- 54.Taberlet P., Gielly L., Pautou G., Bouvet J. Plant Mol. Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 55.Eguiarte L. E., Duvall M. R., Learn G. H. J., Clegg M. T. Boletín de la Sociedad Botánica de México. 1994;54:35–60. [Google Scholar]
- 56.Hall T. A. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 57.Posada D., Crandall K. A. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 58.Kumar S., Tamura K., Nei M. mega: Molecular Evolutionary Genetics Analysis Software for Microcomputers. University Park, PA: Pennsylvania State Univ.; 1993. [DOI] [PubMed] [Google Scholar]
- 59.Swofford D. L. paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- 60.Takezaki N., Rzhetsky A., Nei M. Mol. Biol. Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 61.Sanderson M. J. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 62.Bremer K. Proc. Natl. Acad. Sci. USA. 2000;97:4707–4711. doi: 10.1073/pnas.080421597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pybus O. G., Harvey P. H. Proc. R. Soc. London Ser. B.; 2000. pp. 2267–2272. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.