Abstract
IL-15 is normally presented in vivo as a cell-associated cytokine bound to IL-15Rα. We show here that the biological activity of soluble IL-15 is much improved after interaction with recombinant soluble IL-15Rα; after injection, soluble IL-15/IL-15Rα complexes rapidly induce strong and selective expansion of memory-phenotype CD8+ cells and natural killer cells. These findings imply that binding of IL-15Rα to IL-15 may create a conformational change that potentiates IL-15 recognition by the βγc receptor on T cells. The enhancing effect of IL-15Rα binding may explain why IL-15 normally functions as a cell-associated cytokine. Significantly, the results with IL-2, a soluble cytokine, are quite different; thus, IL-2 function is markedly inhibited by binding to soluble IL-2Rα.
Keywords: cytokines, T cells, soluble receptors, CD122, natural killer cells
In mice, certain cells, namely memory-phenotype (MP) CD8+ T cells and natural killer (NK) cells, are highly sensitive to IL-15 (1–9). MP CD8+ cells display high levels of CD44 and, like NK cells, also show high expression of CD122 (IL-2Rβ), a component of the receptor for both IL-15 and IL-2 (6). For resting cells, responsiveness to these two cytokines is controlled by a two-chain receptor, βγc, consisting of the β chain (CD122) plus the common γ chain, γc, which controls intracellular signaling.
IL-15 is normally not secreted in soluble form (8–10) but is held on the cell surface bound to a unique receptor, IL-15Rα, especially on dendritic cells (11–16). Cell-bound IL-15 then is presented in trans to T cells and NK cells and is recognized by the βγc receptor on these cells; such recognition maintains cell survival and intermittent proliferation.
IL-15Rα plays a mandatory role in presenting endogenous IL-15. Thus, like IL-15–/– mice (1), IL-15Rα–/– mice lack CD122hi CD8+ cells and NK cells (17), presumably because the IL-15 synthesized in IL-15Rα–/– mice fails to leave the cytoplasm. Nevertheless, βγc+ cells can proliferate in response to a soluble recombinant form of IL-15 in the absence of IL-15Rα (18). Moreover, under certain conditions, IL-15Rα can be inhibitory. Thus, injecting mice with a soluble (s) recombinant form of IL-15Rα is reported to suppress NK cell proliferation (10) and certain T dependent immune responses in vivo (19–22), and adding sIL-15Rα in vitro can block the response of cell lines to IL-15 (20–25). Despite these findings, there are other reports that sIL-15Rα (26), and also a soluble sushi domain of IL-15Rα (27), can enhance IL-15 responses of human cell lines.
In this paper, we investigated whether sIL-15Rα can alter the response of normal mouse T cells to IL-15. As discussed below, IL-15 responses of CD8+ T cells and NK cells are improved considerably by association with sIL-15Rα, both in vitro and in vivo.
Results
Stimulation by IL-15/IL-15Rα Complexes in Vitro.
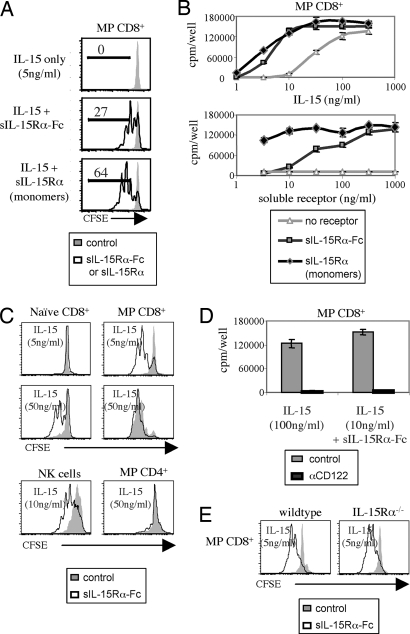
To examine whether the stimulatory function of soluble IL-15 is altered by binding to sIL-15Rα, purified MP CD44hi CD122hi CD8+ cells were cultured in vitro with mouse IL-15 ± mouse sIL-15Rα covalently linked to the Fc portion of human IgG1 (sIL-15Rα-Fc). For IL-15 alone, half-maximal responses required ≈30 ng/ml and responses were negligible with <10 ng/ml (Fig. 1A and B). Here, the notable finding was that supplementing a low concentration of IL-15, e.g., 5 ng/ml, with sIL-15Rα-Fc led to strong proliferative responses of MP CD8+ cells as measured either by carboxyfluorescein diacetate succinimidyl ester (CFSE) dilution (Fig. 1A) or by [3H]thymidine incorporation (Fig. 1B). No proliferation occurred with sIL-15Rα-Fc alone (Fig. 1B), and the addition of sIL-15Rα-Fc failed to alter the response of MP CD8+ cells to a different cytokine, IL-2 (data not shown). With IL-15, sIL-15Rα-Fc did not appear to act by enhancing the half-life of IL-15 in vitro (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Soluble IL-15Rα augments IL-15-mediated lymphocyte proliferation in vitro. (A) Purified MP (CD44hi) CD8+ T cells from IL-7 tg mice were labeled with CFSE and cultured at 5 × 104 cells per well with 5 ng/ml of IL-15. As indicated, 1 μg/ml of either sIL-15Rα-Fc (dimers) or sIL-15Rα (monomers) was added to the cultures. CFSE dilution was assessed on day 4. Representative data are shown. (B) Purified MP CD8+ T cells were cultured with either titrated amounts of IL-15 plus a fixed concentration of soluble receptor (1 μg/ml) (Upper) or titrated amounts of soluble receptor plus a fixed concentration of IL-15 (10 ng/ml) (Lower). The data show mean levels of [3H]thymidine incorporation (±SD) for triplicate cultures on day 3. (C) Purified naïve (CD44lo) CD8+ T cells, MP CD8+ T cells, NK cells, or MP CD4+ T cells were cultured with IL-15 as indicated. Soluble IL-15Rα-Fc was added at 1 μg/ml. CFSE dilution was assessed on day 3. (D) Same as in B except 10 μg/ml of anti-CD122 antibody was added as indicated. (E) MP CD8+ T cells from wild-type IL-7 tg (Ly5.2) and IL-15Rα–/–/IL-7 tg (Ly5.1) mice were mixed together, labeled with CFSE, and cultured as indicated. CFSE dilution on Ly5.1– (wild type) and Ly5.1+ (IL-15Rα–/–) cells was measured on day 3.
With a limiting concentration of cytokine, IL-15 responses were improved generally by 6- to 9-fold by the addition of sIL-15Rα-Fc. Adding sIL-15Rα-Fc also considerably improved the IL-15 response of CD122hi NK cells (Fig. 1C) but was relatively ineffective on MP (CD44hi) CD4+ cells, which express intermediate levels of CD122 (Fig. 1C). Unexpectedly, sIL-15Rα-Fc plus IL-15 led to significant proliferation of typical naïve CD44lo CD122lo CD8+ cells, although only with high concentrations of IL-15 (Fig. 1C).
For MP CD8+ cells, responses to both soluble IL-15 alone and IL-15 plus sIL-15Rα-Fc were mediated solely through βγc receptors. Thus, responses were abolished by addition of CD122 mAb (Fig. 1D) and were as high with MP CD8+ cells from IL-15Rα–/– mice as with normal MP CD8+ cells (Fig. 1E).
Being a dimeric molecule, sIL-15Rα-Fc might enhance IL-15 activity by presenting this cytokine in a cross-linked form. However, enzyme-cleaved monomeric fragments of sIL-15Rα (free of Fc) were no less potent in augmenting IL-15 responses than dimeric molecules (Fig. 1 A and B). Indeed, under limiting conditions, responses were appreciably higher with the receptor monomers than with the dimers (Fig. 1B). Why the receptor monomers were more effective than the dimers is unclear, although for steric reasons, the monomer/IL-15 complexes may bind more effectively to the βγc receptor.
The above data refer to mouse IL-15 and mouse soluble IL-15Rα. Quite similar data applied to human IL-15/IL-15Rα. Thus, the response of mouse MP CD8+ cells to either human or mouse IL-15 was enhanced considerably by the addition of human sIL-15Rα-Fc (Fig. 7, which is published as supporting information on the PNAS web site). The addition of human IL-15Rα monomers was even more effective (data not shown). Note that for βγc responses, human IL-15 is considerably weaker than mouse IL-15 (25).
For the in vivo experiments discussed below, we used mouse sIL-15Rα-Fc. Insufficient amounts of the monomers were available for in vivo studies.
In Vivo Responses.
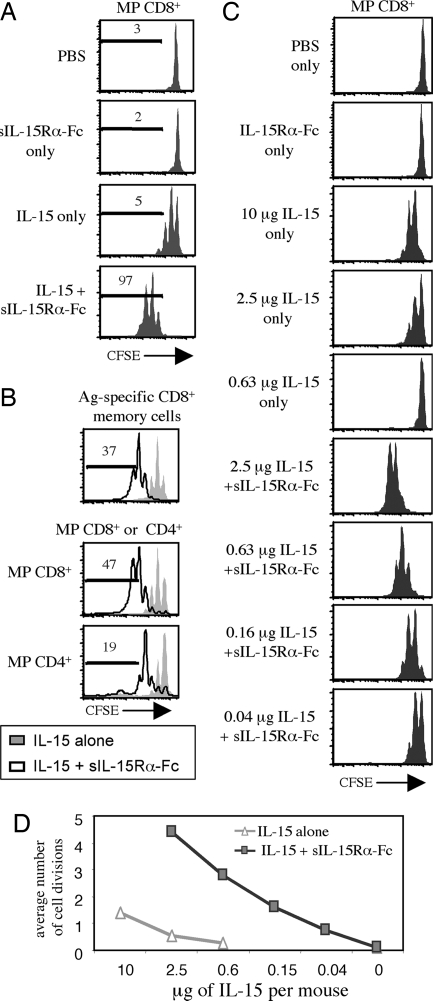
Confirming previous findings (2, 5), injecting mice i.p. with IL-15 after i.v. injection of CFSE-labeled MP CD8+ cells caused ≈50% of the donor cells to divide one to two times (Fig. 2A). With coinjection of sIL-15Rα-Fc, however, virtually all of the donor cells divided and >95% of the cells divided three times or more (compared with <5% for IL-15 injected alone); by contrast, injection of sIL-15Rα-Fc alone had no effect on proliferation. The capacity of sIL-15Rα-Fc to enhance responses of MP CD8+ cells to IL-15 also applied to antigen-specific memory CD8+ cells, i.e., to antigen-primed P14 TCR Tg CD8+ cells (Fig. 2B Upper). There was also enhancement of the IL-15 response of MP CD4+ cells (Fig. 2B Lower).
Fig. 2.
Soluble IL-15Rα augments IL-15-mediated donor lymphocyte proliferation in vivo. (A) CFSE-labeled T cells were transferred i.v. into C57BL/6 (B6) recipients. On days 1 and 2 after transfer, the recipients were given i.p. injections of PBS, sIL-15Rα-Fc alone (7 μg), IL-15 alone (1.5 μg), or sIL-15Rα-Fc plus IL-15 (7 μg and 1.5 μg, respectively, which represents a 1:2 molar ratio). CFSE dilution of the donor cells was measured in spleen on day 4. Representative data for gated MP CD8+ cells are shown. (B) As in A except that the cells transferred were from lymphocytic choriomeningitis virus-immune mice (Upper) versus normal mice (Lower). (C) CFSE-labeled MP CD8+ T cells were transferred to normal B6 hosts; one day later, the hosts were injected with the indicated dose of IL-15 with or without sIL-15Rα-Fc; the dose of sIL-15Rα-Fc varied such that a 2:1 molar ratio of IL-15 to sIL-15Rα-Fc was injected. CFSE profiles for donor MP CD8+ cells in spleen at 2 days after injection are shown. (D) Compilation of data from C. For A–C, data shown are representative of two mice per group and are also representative of two independent experiments.
For MP CD8+ cells, IL-15 titration experiments showed that in vivo responses to IL-15 were increased ≈50-fold when limiting doses of IL-15 were coinjected with sIL-15Rα-Fc (Fig. 2 C and D). Part of this increase in biological activity appears to reflect enhanced in vivo lifespan of IL-15 when bound to sIL-15Rα-Fc, because IL-15/sIL-15Rα-Fc complexes displayed prolonged biological activity compared with free IL-15 (Fig. 8, which is published as supporting information on the PNAS web site).
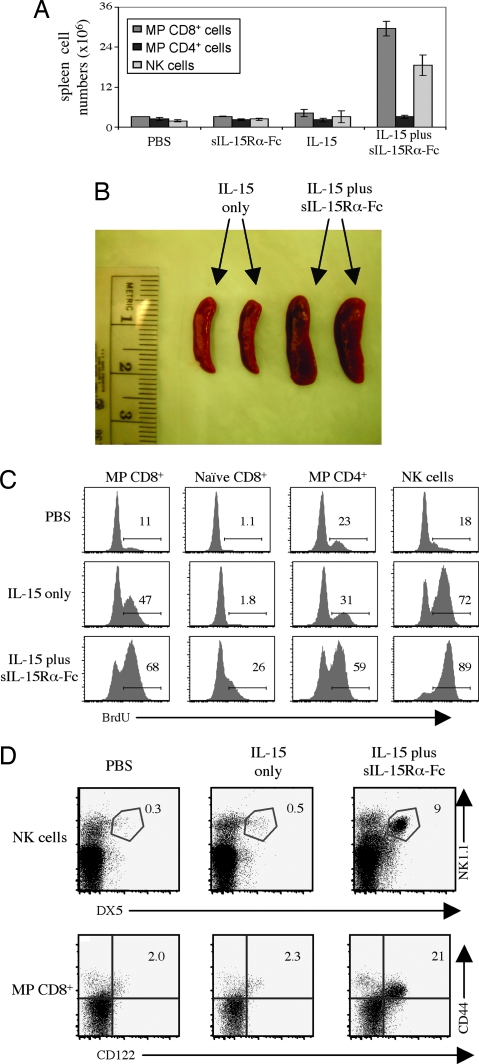
The above data apply to CFSE-labeled donor cells. For host cells, injection of IL-15 or sIL-15Rα-Fc alone had little effect on cell numbers. By contrast, two injections of IL-15 plus sIL-15Rα-Fc caused a marked increase in total numbers of host MP CD8+ cells and NK cells by day 3 after initial injection, and the spleen was obviously enlarged (Fig. 3A and B). Likewise, proliferation as measured by BrdU incorporation was much higher with injection of IL-15 plus sIL-15Rα-Fc than with IL-15 alone (Fig. 3C).
Fig. 3.
Soluble IL-15Rα augments IL-15-mediated host lymphocyte proliferation. (A) Normal B6 mice were injected i.v. on days 1 and 2 with PBS, sIL-15Rα-Fc alone, IL-15 alone, or sIL-15Rα-Fc/IL-15 as described for Fig. 2A. Total numbers of CD8+ MP T cells, CD4+ MP T cells, and NK cells recovered from spleen on day 3 are shown. (B) Spleens from A were photographed as indicated. (C) Mice were treated as in A, except that the mice also were given an i.v. injection of BrdU on day 1 and placed on BrdU in the drinking water until killing. Shown is BrdU staining for MP CD8+, naïve CD8+, MP CD4+, and NK cells. (D) IL-15Rα–/– mice were injected i.v. on days 1, 3, 5, and 7, with PBS, IL-15 (0.6 μg), or IL-15 (0.6 μg)/sIL-15Rα-Fc (3 μg). The data show staining of spleen cells on day 9. For B–D, representative data are shown. All data are representative of at least two independent experiments.
IL-15Rα–/– mice lack CD122hi MP CD8+ cells and NK cells (17), presumably because the absence of IL-15Rα precludes presentation of endogenous IL-15. As shown in Fig. 3D, injecting IL-15Rα–/– mice with a mixture of IL-15 and sIL-15Rα-Fc rapidly restored numbers of host NK1.1+ DX5+ NK cells and CD122hi MP CD8+ cells; at the dose used, IL-15 alone was ineffective.
Failure of sIL-15Rα-Fc to Block Presentation of Endogenous IL-15.
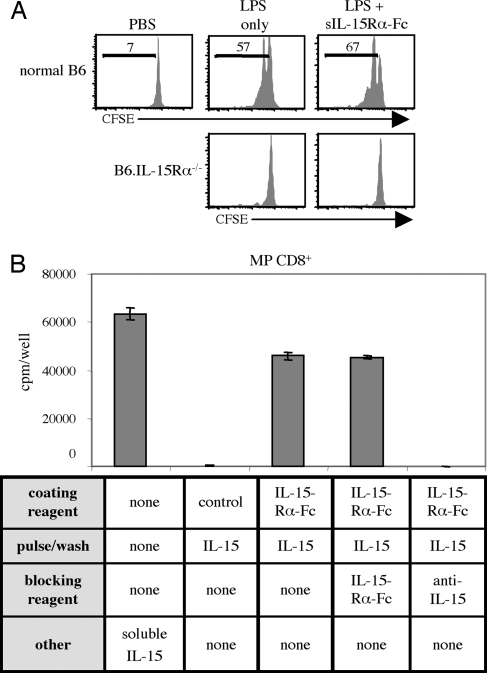
Injecting mice with LPS is known to cause a brief increase in endogenous IL-15 (and IL-15Rα) synthesis by non-T cells in vivo, with a consequent transient increase in the proliferation rate of IL-15-responsive CD122hi MP CD8+ cells (28, 29). Such LPS-induced bystander proliferation is illustrated in Fig. 4A, where most donor MP CD8+ cells underwent one to two cell divisions by day 3 after exposure to LPS in normal B6 hosts, which contrasted with the lack of proliferation in IL-15Rα–/– hosts. Significantly, injecting sIL-15Rα-Fc after LPS injection failed to reduce proliferation, even with daily injections of sIL-15Rα-Fc (10 μg per injection). Hence, injection of sIL-15Rα-Fc was unable to block T cell contact with endogenous IL-15 bound to endogenous IL-15Rα. Also, for IL-15Rα−/− hosts, sIL-15Rα-Fc was clearly unable to compensate for the lack of endogenous IL-15Rα, presumably because the latter is essential for conveying IL-15 to the cell surface.
Fig. 4.
Proliferation to IL-15 immobilized by IL-15Rα cannot be blocked by soluble IL-15Rα-Fc. (A) CFSE-labeled T cells were injected i.v. into Thy1-congenic B6 or IL-15Rα−/− hosts. One day later, mice were injected i.p. with PBS or 500 ng of LPS. As indicated, mice were also treated i.p. with 10 μg of sIL-15Rα-Fc daily beginning the day of LPS injection. Three days after LPS injection, spleens were harvested, and CFSE dilution of MP CD8+ T cells was assessed. (B) Ninety-six-well plates were precoated overnight with 10 μg/ml of sIL-15Rα-Fc. Plates were then washed and incubated with 1 μg/ml IL-15 for 1 h at 37°C. Thereafter, plates were washed, and 5 × 104 MP CD8+ T cells were added together with (i) 10 μg/ml sIL-15Rα-Fc, (ii) 10 μg/ml of polyclonal anti-IL-15 antibody, or (iii) control media; as an additional control, free IL-15 (32 ng/ml) was added to some wells. The data show mean levels of [3H]thymidine incorporation (SD) for triplicate cultures on day 3.
Similar findings applied to an in vitro system where MP CD8+ cells were cultured in wells that were first coated with sIL-15Rα-Fc and then pulsed with IL-15, followed by thorough washing to remove unbound cytokine (Fig. 4B). Thus, proliferative responses elicited by the bound IL-15Rα-Fc/IL-15 complexes could not be inhibited by the addition of soluble (unbound) IL-15Rα-Fc as a blocking reagent. By contrast, addition of a polyclonal antibody against IL-15 abolished proliferation.
Based on the above findings, the IL-15 molecule has only a single binding site for interaction with IL-15Rα. Once this site is occluded, by binding either to endogenous IL-15Rα on cells in vivo or to IL-15Rα attached to plastic in vitro, interaction with exogenous sIL-15Rα-Fc fails to occur, and there is no interference with presentation of IL-15 to T cells. This scenario does not explain why sIL-15Rα can block the response of cell lines to IL-15 (20–25). Here it may be relevant that these studies used human or simian IL-15, and not mouse IL-15 as in our study, which raises the possibility of distinct species differences in IL-15. In favor of this idea, we found that, as for MP CD8+ cells, the response of mouse CTLL cells to mouse IL-15 was enhanced by mouse sIL-15Rα-Fc (Fig. 9A, which is published as supporting information on the PNAS web site). By contrast, confirming the findings of others (20–23), the high response of CTLL cells to human IL-15 (25) was strongly inhibited by mouse sIL-15Rα-Fc (Fig. 9B). CTLL responses to IL-2 as a control were not affected by adding sIL-15Rα-Fc (Fig. 9C).
The above findings do not explain the reports that murine sIL-15Rα constructs are inhibitory for NK cell proliferation (10) and antigen-driven T cell responses in vivo (19–22). This discrepancy has yet to be resolved, although it is of interest that antigen-specific proliferative responses of naïve OT-1 TCR Tg CD8+ cells to specific peptide in vivo were not blocked by coinjection of sIL-15Rα-Fc, and the responses were considerably enhanced when a mixture of sIL-15R and IL-15 was injected (Fig. 9D). As yet we have not used the very large doses of sIL-15Rα required to block in vivo responses, i.e., 400 μg per injection for NK cell proliferation (10). Also, it is possibly relevant that the studies showing inhibition by sIL-15Rα in vivo used constructs grown in bacteria, whereas our constructs were grown in mammalian cells.
Stimulation by IL-2 Plus IL-2Rα.
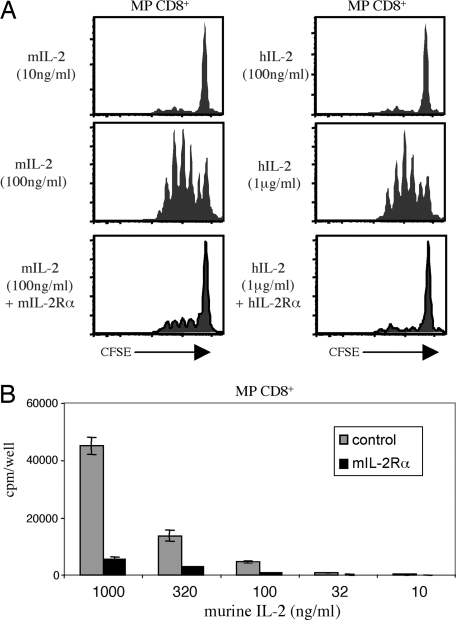
The observation that the biological activity of IL-15 was enhanced by binding to soluble IL-15Rα raised the question whether comparable findings would apply to IL-2 and IL-2Rα (CD25). As shown in Fig. 5, this possibility was clearly not the case. Thus, proliferative responses of MP CD8+ cells to mouse IL-2 in vitro were inhibited markedly by the addition of soluble mouse IL-2Rα (Fig. 5A Left and B). Similar inhibition applied to MP CD8+ cells (mouse) responding to human IL-2 and soluble human IL-2Rα (Fig. 5A Right).
Fig. 5.
Soluble IL-2Rα inhibits IL-2-mediated proliferation. (A) Purified CFSE-labeled MP CD8+ T cells were cultured with either murine IL-2 or human IL-2 at the concentration shown. As indicated, 2.5 μg/ml of either soluble murine IL-2Rα or soluble human IL-2Rα was added to the cultures. CFSE dilution was assessed on day 3. Representative data are shown. (B) Purified MP CD8+ T cells were cultured with titrated amounts of murine IL-2 with or without soluble mIL-2Rα (2.5 μg/ml). The data show mean levels of [3H]thymidine incorporation (±SD) for triplicate cultures on day 3.
Thus, whereas soluble IL-15Rα potentiated the function of IL-15, soluble IL-2Rα blocked the function of IL-2.
Discussion
The main conclusion from the above experiments is that soluble complexes of IL-15 and IL-15Rα are much more stimulatory than soluble IL-15 alone, both in vivo and in vitro. Without structural studies on IL-15/IL-15Rα interaction, one can only speculate on why and how this interaction potentiates IL-15 function. There are several possibilities.
First, binding of IL-15Rα to IL-15 might impair IL-15 internalization by T cells and, thereby, strengthen signaling through the βγc receptor. This idea is in line with reports that internalization of certain cytokines, e.g., IL-2, serves to attenuate receptor signaling (30). However, we do not favor this possibility for two reasons. First, if the strong stimulation by IL-15/sIL-15Rα complexes reflected reduced IL-15 internalization, one would expect to see a parallel reduction in internalization of CD122, the receptor for IL-15. As measured by down-regulation from the cell surface, however, the opposite applies, i.e., greater down-regulation of CD122 with IL-15/sIL-15Rα complexes than with IL-15 alone (data not shown). The second argument against IL-15/sIL-15Rα complexes preventing IL-15 internalization is that, if this possibility were the case, we should have seen similar findings with IL-2/sIL-2Rα, which was not so. Thus, IL-2/sIL-2Rα complexes were much less stimulatory than soluble IL-2 alone, which clearly contrasted with IL-15/sIL-15Rα complexes being more stimulatory than IL-15 alone.
A second possibility for how sIL-15Rα potentiates IL-15 activity is that sIL-15Rα might prevent degradation of IL-15. This notion deserves consideration because the enhancing effect of sIL-15Rα-Fc on IL-15 function was more pronounced in vivo than in vitro. Here, it is notable that binding of certain cytokines to antibodies or soluble receptors can extend cytokine survival in vivo (31–36). Consistent with this view, binding to sIL-15Rα-Fc did extend the half-life of IL-15 in vivo. However, additional mechanisms appear to be involved because sIL-15Rα-Fc improved the biological activity of IL-15 in vitro without affecting the cytokine half-life.
In light of the above findings, a third possibility needs to be considered, namely that IL-15Rα improves the function of IL-15 by inducing a conformational change in IL-15: this change augments interaction with the βγc receptor, thus changing IL-15 from an agonist to a superagonist. This model is in line with the affinity of IL-15/IL-15Rα interaction being far higher than for IL-2/IL-2Rα interaction (3, 8) and explains why, unlike IL-2, IL-15 functions so well as a cell-associated cytokine. Testing this idea directly will obviously require structural studies. In this respect, it is notable that the interaction between IL-15 and IL-15Rα involves a unique network of ionic interactions not found in other cytokine/cytokine receptor complexes (37). Whether this unique interaction results in a conformational change in IL-15 has yet to be determined.
There is accumulating evidence that IL-15 has beneficial effects on T cell survival and memory generation and also has potential for restoring the T cell pool after irradiation and other forms of cytoreduction (4–9, 18, 38, 39). As shown here, the biological activity of IL-15 as a therapeutic reagent could be considerably enhanced by administering preformed soluble IL-15/IL-15Rα complexes.
Materials and Methods
Mice.
C57BL/6 (B6), B6.Ly5.1, B6.Thy1.1, and OT-1 mice were purchased from The Jackson Laboratory. IL-15Rα−/− mice (17) were a generous gift of Averil Ma (University of California, San Francisco), and IL-7 transgenic (tg) mice (40) were a generous gift of J. Andersson (Basel Institute, Basel, Switzerland). P14 TCR tg mice were kindly provided by J. Lindsay Whitton (The Scripps Research Institute). IL-15Rα−/−, IL-7 tg, P14, and OT-1 TCR tg mice were all maintained on a B6 background and, for some experiments, crossed to either B6.Ly5.1 or B6.Thy1.1 mice. IL-15Rα−/− mice were crossed to IL-7 tg mice to generate IL-7 tg/IL-15Rα−/− mice. As we have previously described with IL-7 tg/IL-15−/− mice (41), IL-7 tg/IL-15Rα−/− mice have similar large numbers of CD122hi MP CD8+ T cells as IL-7 tg mice.
Recombinant Proteins.
Murine sIL-15Rα-Fc, human sIL-15Rα-Fc, and human IL-2Rα were purchased from R & D Systems. Monomeric sIL-15Rα and mouse IL-2Rα were purchased from R & D Systems as prerelease reagents. Monomeric sIL-15Rα was generated by the manufacturer by enzyme digestion of the dimeric sIL-15Rα-Fc, which resulted in the release of the Fc region. We verified complete digestion by Western blot with anti-IL-15Rα polyclonal antibodies (AF551, BAF551, and BAF847, R & D Systems) (data not shown). Recombinant cytokines (including mouse IL-15, human IL-15, mouse IL-2, human IL-2, mouse IL-4, and mouse GM-CSF) were purchased from eBioscience (San Diego) and/or R & D Systems.
Isolation of T Cells and CFSE Labeling.
To obtain adequate numbers of cells, in most experiments, MP CD8+ cells were prepared from IL-7 tg mice. By all parameters tested, MP CD8+ cells from IL-7 tg mice are identical to cells from normal mice. Moreover, the main findings reported here for IL-15/sIL-15Rα-Fc complexes also were observed with cells prepared from normal mice, both in vivo and in vitro. MP CD8+ T cells used for either in vitro or adoptive transfer experiments were isolated from lymph node (LN) and spleen and purified by cell sorting. In brief, single-cell suspensions were enriched first for CD3+ T cells by using a mouse T cell enrichment columns (MTCC-25, R & D Systems). Enriched T cells were labeled with antibodies and purified by cell sorting for CD8+CD44hi T cells. In some experiments, we used a similar protocol and isolated CD8+CD44lo, CD4+CD44hi, NK1.1+/DX5+ cells. Cell sorting was performed by using a BD FACSAria (BD Biosciences). Purity of sorted cells was routinely tested and >98%. In some experiments, total T cells or OT-1 cells were used as donor lymphocytes. For these experiments, cells from spleen and LN were purified by using a mouse T cell enrichment column (MTCC-25). For experiments using CFSE-labeled cells, T cells were labeled with 1.5 μm CFSE (Molecular Probes) according to the manufacturer’s directions.
Generation of Antigen-Specific CD8+ T Cells.
Lymphocytic choriomeningitis virus-specific P14 TCR tg CD8+ T cells (Thy1.2/Ly5.2) were adoptively transferred into IL-7 tg recipient mice (Thy1.1/Ly5.2) and infected with 2 × 105 plaque-forming units of lymphocytic choriomeningitis virus Armstrong strain. Two months later, T cells were purified by using a mouse T cell enrichment column (MTCC-25), labeled with CFSE, and adoptively transferred into B6.Ly5.1-recipient mice (Thy1.2/Ly5.1). P14 CD8+ T cells, which represented 15–20% of the donor CD8+ T cell population, were identified by triple staining for Thy1.2, Ly5.2, and CD8.
In Vitro Assays.
All cultures were performed in RPMI medium 1640 supplemented with 10% FCS, glutamine, 2-mercaptoethanol, nonessential amino acids, and antibiotics. FACS-purified T cells and NK cells were isolated as described above. CTLL (CTLL-2) cells were obtained from American Type Culture Collection (Manassas, VA), and cultured in RPMI medium 1640 supplemented with murine IL-2. For experiments with FACS-purified lymphocytes, 5 × 104 cells in 200 μl were plated per well in 96-well plates. Cytokine and/or soluble receptor were added at concentrations described in the figures. For CD122 blocking experiments, we used purified anti-CD122 antibody [TM-β1 (NA/LE); BD Pharmingen]. For experiments to block plate-bound IL-15, polyclonal anti-IL-15 antibody (AF447; R & D Systems) was used. Experiments with CTLL cells were plated as with FACS-purified lymphocytes except using 2 × 104 cells per well. For proliferation experiments with [3H]thymidine, 1 μCi/ml (1 Ci = 37 GBq) was added as indicated in the figure legends. Cells were cultured in triplicate wells.
In Vivo Assays.
For experiments assessing proliferation of adoptively transferred cells, T cells were isolated and labeled with CFSE (as described above), and then injected i.v. into Ly5 or Thy1 congenic-recipient mice. In experiments to measure proliferation of host cells, mice were injected i.p. with BrdU (2 mg) and then maintained on BrdU drinking water (0.8 mg/ml) by using methodology described in ref. 2. For injections of cytokine and soluble receptor, IL-15 and sIL-15Rα-Fc were incubated together for 20 min at 37°C. Samples were then diluted at least 10-fold in PBS to a volume of 500 μl before injection into mice. In control conditions, cytokine or receptor alone also was incubated for 20 min at 37°C. LPS (ALX-581–008; Alexis Biochemicals, San Diego) were injected i.p. in PBS. For vaccination experiments, dendritic cells were prepared as described by culture of bone marrow cells with GM-CSF and IL-4 (38). Dendritic cells were pulsed for 2 h with SIINFEKL peptide at 37°C, washed, and injected i.v.
Flow Cytometric Analysis.
Cells were analyzed by flow cytometric analysis by using standard protocols. Briefly, cells were washed in FACS buffer containing 1% FCS and 2 mM EDTA and stained with combinations of the antibodies: CD8-PerCP-Cy5.5, -APC, or -APC-Cy7 (53-6.7; eBioscience and BD Pharmingen); CD49b-PE and -APC (DX5; eBioscience); NK1.1-FITC and -PE (PK136; BD Pharmingen); CD3-PE, -PerCP-Cy5.5, -PE-Cy7, or -APC (145–2C11; eBioscience and BD Pharmingen); CD3-Pacific Blue (500A2; BD Pharmingen); CD4-PE, PE-Cy7, or -APC (RM4-5, eBioscience and BD Pharmingen); Ly5.1-FITC, -PE, -PE-Cy7, and -APC (A20, eBioscience and BD Pharmingen); Ly5.2-FITC, -PE, -PerCP-Cy5.5, and -APC (104; eBioscience and BD Pharmingen); Thy1.1-FITC, PE, -PE-Cy7, and -APC (HIS51; eBioscience); Thy1.2-FITC, PE, and -APC (53–2.1; eBioscience); CD44-FITC-APC and -Alexa Fluor 405 (IM7, eBioscience and Caltag, Burlingame, CA); CD122-PE (TM-β1; BD Pharmingen); B220-PerCP-Cy5.5 (RA3–6B2; BD Pharmingen); and TCR Vα2-PE (B20.1, BD Pharmingen). BrdU intracellular staining was performed with reagents from FITC or APC BrdU flow kits (559619 and 552598; BD Pharmingen) according to the manufacturer’s directions. Flow cytometric samples were analyzed by using a BD LSR II digital flow cytometer (BD Biosciences). Data were analyzed by using flowjo (Tree Star, San Carlos, CA).
Supplementary Material
Acknowledgments
We thank Ms. B. Marchand for typing the manuscript. The excellent support of the Flow Cytometry and Antibody Core Facility was invaluable. This work was supported by National Institutes of Health Grants CA038355, AI046710, AI045809, AI007244, AG020186, and AG001743. O.B. has been supported by the Swiss National Science Foundation and is the recipient of a Fellowship from the Novartis Foundation. J.F.P. is supported by a C. J. Martin Fellowship from the Australian National Health and Medical Research Council. Experiments involving the use of animals were approved by the Institutional Animal Care and Use Committee at The Scripps Research Institute. This work is publication no. 17918-IMM from The Scripps Research Institute.
Abbreviations
- CFSE
carboxyfluorescein diacetate succinimidyl ester
- MP
memory-phenotype
- NK
natural killer
- s
soluble.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kennedy M. K., Glaccum M., Brown S. N., Butz E. A., Viney J. L., Embers M., Matsuki N., Charrier K., Sedger L., Willis C. R., et al. J. Exp. Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Judge A., Zhang X., Fujii H., Surh C. D., Sprent J. J. Exp. Med. 2002;196:935–946. doi: 10.1084/jem.20020772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehniger T. A., Caligiuri M. A. Blood. 2001;97:14–32. doi: 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- 4.Becker T. C., Wherry E. J., Boone D., Murali-Krishna K., Antia R., Ma A., Ahmed R. J. Exp. Med. 2002;195:1541–1548. doi: 10.1084/jem.20020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Sun S., Hwang I., Tough D. F., Sprent J. Immunity. 1998;8:591–599. doi: 10.1016/s1074-7613(00)80564-6. [DOI] [PubMed] [Google Scholar]
- 6.Waldmann T. A. J. Clin. Immunol. 2002;22:51–56. doi: 10.1023/a:1014416616687. [DOI] [PubMed] [Google Scholar]
- 7.Zeng R., Spolski R., Finkelstein S. E., Oh S., Kovanen P. E., Hinrichs C. S., Pise-Masison C. A., Radonovich M. F., Brady J. N., Restifo N. P., et al. J. Exp. Med. 2005;201:139–148. doi: 10.1084/jem.20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Belle T., Grooten J. Arch. Immunol. Ther. Exp. (Warsz.) 2005;53:115–126. [PubMed] [Google Scholar]
- 9.Schluns K. S., Stoklasek T., Lefrancois L. Int. J. Biochem. Cell Biol. 2005;37:1567–1571. doi: 10.1016/j.biocel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen K. B., Salazar-Mather T. P., Dalod M. Y., Van Deusen J. B., Wei X. Q., Liew F. Y., Caligiuri M. A., Durbin J. E., Biron C. A. J. Immunol. 2002;169:4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- 11.Dubois S., Mariner J., Waldmann T. A., Tagaya Y. Immunity. 2002;17:537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- 12.Burkett P. R., Koka R., Chien M., Chai S., Boone D. L., Ma A. J. Exp. Med. 2004;200:825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkett P. R., Koka R., Chien M., Chai S., Chan F., Ma A., Boone D. L. Proc. Natl. Acad. Sci. USA. 2003;100:4724–4729. doi: 10.1073/pnas.0737048100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schluns K. S., Klonowski K. D., Lefrancois L. Blood. 2004;103:988–994. doi: 10.1182/blood-2003-08-2814. [DOI] [PubMed] [Google Scholar]
- 15.Zaft T., Sapoznikov A., Krauthgamer R., Littman D. R., Jung S. J. Immunol. 2005;175:6428–6435. doi: 10.4049/jimmunol.175.10.6428. [DOI] [PubMed] [Google Scholar]
- 16.Sandau M. M., Schluns K. S., Lefrancois L., Jameson S. C. J. Immunol. 2004;173:6537–6541. doi: 10.4049/jimmunol.173.11.6537. [DOI] [PubMed] [Google Scholar]
- 17.Lodolce J. P., Boone D. L., Chai S., Swain R. E., Dassopoulos T., Trettin S., Ma A. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 18.Lodolce J. P., Burkett P. R., Boone D. L., Chien M., Ma A. J. Exp. Med. 2001;194:1187–1194. doi: 10.1084/jem.194.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruckert R., Brandt K., Bulanova E., Mirghomizadeh F., Paus R., Bulfone-Paus S. Eur. J. Immunol. 2003;33:3493–3503. doi: 10.1002/eji.200324545. [DOI] [PubMed] [Google Scholar]
- 20.Ruckert R., Brandt K., Braun A., Hoymann H. G., Herz U., Budagian V., Durkop H., Renz H., Bulfone-Paus S. J. Immunol. 2005;174:5507–5515. doi: 10.4049/jimmunol.174.9.5507. [DOI] [PubMed] [Google Scholar]
- 21.Wei X., Orchardson M., Gracie J. A., Leung B. P., Gao B., Guan H., Niedbala W., Paterson G. K., McInnes I. B., Liew F. Y. J. Immunol. 2001;167:277–282. doi: 10.4049/jimmunol.167.1.277. [DOI] [PubMed] [Google Scholar]
- 22.Ruchatz H., Leung B. P., Wei X. Q., McInnes I. B., Liew F. Y. J. Immunol. 1998;160:5654–5660. [PubMed] [Google Scholar]
- 23.Budagian V., Bulanova E., Orinska Z., Ludwig A., Rose-John S., Saftig P., Borden E. C., Bulfone-Paus S. J. Biol. Chem. 2004;279:40368–40375. doi: 10.1074/jbc.M404125200. [DOI] [PubMed] [Google Scholar]
- 24.Mortier E., Bernard J., Plet A., Jacques Y. J. Immunol. 2004;173:1681–1688. doi: 10.4049/jimmunol.173.3.1681. [DOI] [PubMed] [Google Scholar]
- 25.Eisenman J., Ahdieh M., Beers C., Brasel K., Kennedy M. K., Le T., Bonnert T. P., Paxton R. J., Park L. S. Cytokine. 2002;20:121–129. doi: 10.1006/cyto.2002.1989. [DOI] [PubMed] [Google Scholar]
- 26.Giron-Michel J., Giuliani M., Fogli M., Brouty-Boye D., Ferrini S., Baychelier F., Eid P., Lebousse-Kerdiles C., Durali D., Biassoni R., et al. Blood. 2005;106:2302–2310. doi: 10.1182/blood-2005-01-0064. [DOI] [PubMed] [Google Scholar]
- 27.Mortier E., Quemener A., Vusio P., Lorenzen I., Boublik Y., Grotzinger J., Plet A., Jacques Y. J. Biol. Chem. 2005;281:1612–1619. doi: 10.1074/jbc.M508624200. [DOI] [PubMed] [Google Scholar]
- 28.Mattei F., Schiavoni G., Belardelli F., Tough D. F. J. Immunol. 2001;167:1179–1187. doi: 10.4049/jimmunol.167.3.1179. [DOI] [PubMed] [Google Scholar]
- 29.Tough D. F., Sun S., Sprent J. J. Exp. Med. 1997;185:2089–2094. doi: 10.1084/jem.185.12.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang D. Z., Wu Z., Ciardelli T. L. J. Biol. Chem. 1996;271:13349–13355. doi: 10.1074/jbc.271.23.13349. [DOI] [PubMed] [Google Scholar]
- 31.Finkelman F. D., Madden K. B., Morris S. C., Holmes J. M., Boiani N., Katona I. M., Maliszewski C. R. J. Immunol. 1993;151:1235–1244. [PubMed] [Google Scholar]
- 32.Ma Y., Hurst H. E., Fernandez-Botran R. J. Pharmacol. Exp. Ther. 1996;279:340–350. [PubMed] [Google Scholar]
- 33.Peters M., Jacobs S., Ehlers M., Vollmer P., Mullberg J., Wolf E., Brem G., Meyer zum Buschenfelde K. H., Rose-John S. J. Exp. Med. 1996;183:1399–1406. doi: 10.1084/jem.183.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenblum M. G., Unger B. W., Gutterman J. U., Hersh E. M., David G. S., Frincke J. M. Cancer Res. 1985;45:2421–2424. [PubMed] [Google Scholar]
- 35.Peleg-Shulman T., Roisman L. C., Zupkovitz G., Schreiber G. J. Biol. Chem. 2004;279:18046–18053. doi: 10.1074/jbc.M400033200. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi H., Tagaya Y., Han E. S., Kim I. S., Le N., Paik C. H., Pastan I., Nelson D. L., Waldmann T. A., Carrasquillo J. A. Cytokine. 1999;11:1065–1075. doi: 10.1006/cyto.1999.0509. [DOI] [PubMed] [Google Scholar]
- 37.Lorenzen I., Dingley A. J., Jacques Y., Grotzinerg J. J. Biol. Chem. 2006;281:6642–6647. doi: 10.1074/jbc.M513118200. [DOI] [PubMed] [Google Scholar]
- 38.Rubinstein M. P., Kadima A. N., Salem M. L., Nguyen C. L., Gillanders W. E., Cole D. J. J. Immunol. 2002;169:4928–4935. doi: 10.4049/jimmunol.169.9.4928. [DOI] [PubMed] [Google Scholar]
- 39.Diab A., Cohen A. D., Alpdogan O., Perales M. A. Cytotherapy. 2005;7:23–35. doi: 10.1080/14653240510018037. [DOI] [PubMed] [Google Scholar]
- 40.Mertsching E., Burdet C., Ceredig R. Int. Immunol. 1995;7:401–414. doi: 10.1093/intimm/7.3.401. [DOI] [PubMed] [Google Scholar]
- 41.Kieper W. C., Tan J. T., Bondi-Boyd B., Gapin L., Sprent J., Ceredig R., Surh C. D. J. Exp. Med. 2002;195:1533–1539. doi: 10.1084/jem.20020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.