Abstract
CD8+ T cells respond to short peptides bound to MHC class I molecules. Although most antigenic proteins contain many sequences that could bind to MHC class I, few of these peptides actually stimulate CD8+ T cell responses. Moreover, the T cell responses that are generated often follow a very reproducible hierarchy to different peptides for reasons that are poorly understood. We find that the loss of a single enzyme, endoplasmic reticulum aminopeptidase 1 (ERAP1), in the antigen-processing pathway results in a marked shift in the hierarchy of immunodominance in viral infections, even when the responding T cells have the same T cell receptor repertoire. In mice, ERAP1 is the major enzyme that trims precursor peptides in the endoplasmic reticulum and, in this process, can generate or destroy antigenic peptides. Consequently, when ERAP1 is lost, the immune response to some viral peptides is reduced, to others increased, and to yet others unchanged. Therefore, many epitopes must be initially generated as precursors that are normally trimmed by ERAP1 before binding to MHC class I, whereas others are normally degraded by ERAP1 to lengths that are too short to bind to MHC class I. Moreover, peptide trimming and the resulting abundance of peptide–MHC complexes are dominant factors in establishing immunodominance.
Keywords: antigen presentation, antigen processing, peptidases
Cells infected with a virus or otherwise producing foreign proteins can be recognized and eliminated by CD8+ T cells. CD8+ T cells recognize their targets through interactions between their T cell receptor (TCR) and MHC class I on the surface of the target cell. MHC class I is composed of a light chain (β2-microglobulin), a highly polymorphic heavy chain, and a small peptide that is produced by degradation of intracellular proteins.
MHC class I alleles bind to peptides with a particular set of anchor residues that fit into pockets in the MHC class I peptide-binding groove. Virus proteomes typically contain many peptides with anchor residues suitable for particular MHC class I alleles, yet immune responses are generally directed toward a very limited number of epitopes. This phenomenon, in which only a few epitopes are functionally immunogenic, is known as immunodominance and is, as yet, incompletely understood (reviewed in ref. 1). Although many explanations for immunodominance have been proposed (1–9), their relative importance is not well established.
The generation of most MHC class I epitopes requires proteolysis by proteasomes in the cytosol. Proteasomes can generate either the mature epitope that is ultimately presented by MHC class I molecules or precursors that are extended by several amino acids at the amino terminus. Such N-extended peptides are further processed by aminopeptidases to generate the final presented epitope. Although a number of peptidases have been proposed to play roles in MHC class I antigen presentation, few have been shown to play a major role in antigen presentation in intact cells. In cell lysates, aminopeptidases, including leucine aminopeptidase (LAP) (10), bleomycin hydrolase (BH) (11), and puromycin-sensitive aminopeptidase (PSA) (11) can convert various N-extended peptides into the mature epitopes. However, in intact cells and mice, LAP appears dispensable for antigen presentation (12), and the importance of BH and PSA in antigen presentation remains to be established. Thimet oligopeptidase in cell lysates has been shown to be important in destroying many peptides (13–15). It has recently been proposed that most antigenic peptides are generated as very long precursors (>15 residues) and that tripeptidyl peptidase II (TPPII) is essential for trimming these precursors for antigen presentation (16, 17). However, we find that, in vivo, proteasomes generate relatively few very long peptides, and silencing of TPPII has only a small effect on overall antigen presentation (I.A.Y., N. Bhutani, S.Z., A. L. Goldberg, and K.L.R., unpublished data).
Endoplasmic reticulum (ER) aminopeptidase 1 (ERAP1; ERAAP) is an IFN γ-inducible ER-localized aminopeptidase expressed in many tissues, although at widely different levels (18–20). ERAP1 was previously shown to influence the presentation of several peptides in cultured cells. Human cells lacking ERAP1 because of short interfering RNA-mediated knockdown showed an increase in the amount of peptide associated with MHC class I but also had markedly reduced presentation of specific peptides, such as the immunodominant H-2Kb-binding peptide from chicken ovalbumin, SIINFEKL (21, 22), whereas cells from mice lacking ERAP1 (ERAAP) had a generally reduced peptide supply (23, 24). Unlike other aminopeptidases, ERAP1 trims only peptides that are >8 or 9 aa (20, 25), the optimal length for binding to MHC class I, helping to explain why ERAP1 creates MHC class I epitopes in some cases (by trimming N-extended precursors to the mature epitope) and destroys them in others (by trimming mature epitopes that are nine or more residues to a size that is too small to bind to MHC class I).
To better understand the role and importance of ERAP1 in normal immune responses, we generated and analyzed a mutant mouse lacking ERAP1. We find that not only is ERAP1 important in generating MHC class I epitopes but also that its absence drastically alters immunodominance hierarchies, pointing to a central role for antigen processing and MHC class I–peptide complex abundance in establishing immunodominance.
Results
Generation of ERAP1 Knockout (KO) Mice.
C57/BL6 mice heterozygous for floxed ERAP1 (in which loxP sites were present in the introns flanking ERAP1 exons 5 and 6) were generated and crossed with cre-deleter mice (26). The resulting loss of exons 5 and 6 causes a frame-shift mutation that would lead to early termination of the exon4/exon7 fusion (see Fig. 6, which is published as supporting information on the PNAS web site); moreover, because exon 5 of ERAP1 contains the enzyme’s active site, the truncated protein (if expressed) would not be enzymatically active. The resulting heterozygous ERAP1+/− mice were bred to homozygosity. No gross effects of ERAP1 deficiency were noted. The mice were fertile, grew normally, and showed no obvious signs of pathology. Therefore, ERAP1 is not an essential gene for survival.
MHC Class I Levels in Fibroblasts from ERAP1 KO Mice Are Not Altered.
We prepared fibroblasts from day-14–17 embryos from ERAP1+/− crosses and typed the cell lines to identify ERAP1+/+ and ERAP1−/− lines. Several independent mouse embryonic fibroblast (MEF) cell lines were generated and analyzed for surface MHC class I expression. ERAP1+/+ and ERAP1−/− lines had similar levels of H-2Kb and H-2Db (Fig. 1A); however, constitutive MHC class I expression on these fibroblasts was very low, and, under these conditions, surface expression of MHC class I is probably not limited by peptide supply.
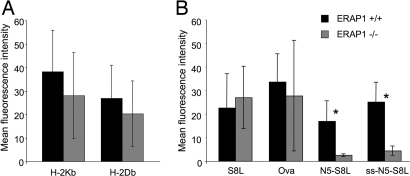
Fig. 1.
ERAP1−/− mouse embryonic fibroblast lines are defective for peptide trimming in the ER. Fibroblast cell lines were prepared from ERAP1+/+ (black bars) and ERAP1−/− (gray bars) embryonic mice. (A) Cells were stained with mAb directed against H-2Kb (mAb B8.24.3) or H-2Db (28.14.8S) and analyzed by flow cytometry. (B) MEF cells were transfected with plasmids expressing SIINFEKL as a cytosolic ubiquitin fusion (S8L), ovalbumin protein targeted to the cytosol (Ova), LEQLE-SIINFEKL as a cytosolic ubiquitin fusion (N5-S8L), or ALEQLE-SIINFEKL targeted to the ER by a signal sequence (ss.N5-S8L). After 48 h, the cells were stained with mAb 25.D1.16 (anti-H-2Kb-SIINFEKL) and analyzed by flow cytometry. Data (representative of five experiments) show the mean fluorescence intensity with background staining by an irrelevant antibody subtracted (averages of three independent cell lines; error bars represent 1 SD). Asterisks indicate statistically significant differences (P < 0.05, Student t test) between ERAP1+/+ and ERAP1−/− MEF cell lines.
Trimming of Peptide Precursors in the ER of Fibroblasts from ERAP1 KO Mice Is Reduced.
We transfected three ERAP1−/− and three ERAP1+/+ MEF lines with various precursors of SIINFEKL (the immunodominant H-2Kb-restricted epitope from chicken ovalbumin) using the mAb 25.D1.16 (27). First, we expressed SIINFEKL as a ubiquitin fusion in the cytosol. Ubiquitin C-terminal hydrolyases efficiently cleave the peptide bond between the ubiquitin moiety and SIINFEKL, thereby generating the mature epitope. There was no statistically significant difference between ERAP1-deficient and WT cells (Fig. 1B). This result is consistent with the fact that SIINFEKL requires no trimming for presentation and with earlier findings that ERAP1 does not destroy the mature 8-residue SIINFEKL peptide (20, 21, 25). These findings also demonstrate that, with the exception of peptide trimming in the ER, the MHC class I pathway is otherwise intact in ERAP1-deficient cells.
To analyze peptide trimming in the ER, we expressed in MEFs SIINFEKL precursors extended at the N terminus by 6 aa, targeted into the ER by a signal sequence (ss-ALEQLE-S8L). Presentation of H-2Kb-SIINFEKL from this precursor was reduced to levels barely above background staining (Fig. 1B). Therefore, in MEFs as in human cells (21), ERAP1 is critical for trimming peptides in the ER. Similarly, presentation of SIINFEKL from a cytosolic ubiquitin fusion containing the 13-residue precursor LEQLE-SIINFEKL was markedly reduced in ERAP1−/− MEFs. Therefore, this peptide is transported into the ER by transporter associated with antigen presentation (TAP) as an N-extended form, rather than being processed by cytosolic aminopeptidases to SIINFEKL, and trimming of this precursor to the mature epitope requires ERAP1.
In additional experiments, we analyzed the presentation of SIINFEKL from full-length ovalbumin. In all experiments (n > 6), presentation from this construct was highly variable between individual MEF lines and, on average, did not differ significantly between WT and ERAP1−/− cell lines. This result was surprising, because it differed from earlier findings in HeLa cells, where the presentation of SIINFEKL from the full-length protein was markedly reduced in the absence of ERAP1 (21). However, there may be effects of ERAP1 that are obscured by the large variation between different MEF cell lines.
MHC Class I Levels Are Reduced on Splenocytes from ERAP1 KO Mice.
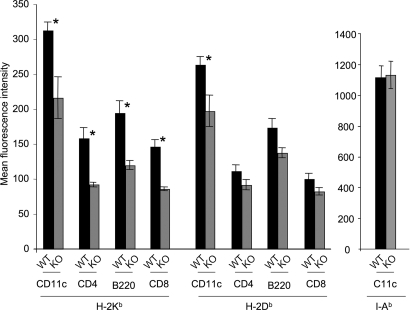
We stained spleen cells for the MHC class I molecules H-2Kb and H-2Db and, as a control, for the MHC class II I-Ab molecule. In contrast to the findings with MEFs, H-2Kb was reduced to ≈60–65% of control levels (P < 0.05, Student t test) on B220+ B cells, CD4+ and CD8+ T cells, and on CD11c cells (predominantly dendritic cells) from the spleen of mice lacking ERAP1 compared with their WT littermates (Fig. 2). Expression of H-2Db was moderately reduced on the same cell types, to ≈75–80% of control levels; however, this difference was statistically significant (P < 0.05, Student t test) only for CD11c+ cells. In contrast, there was no difference in the levels of I-Ab between WT and ERAP1 on any of the cell populations analyzed (Fig. 2). Therefore, the loss of ERAP1 selectively affects the surface expression of MHC class I molecules on splenocytes. Similarly, thymocytes (all subsets) from ERAP1−/− mice expressed significantly lower levels of both H-2Kb and H-2Db than did thymocytes from ERAP1+/+ mice (data not shown).
Fig. 2.
Splenocytes from ERAP1−/− mice express lower cell-surface MHC class I than those from ERAP1+/+ mice. Splenocytes from ERAP1+/+ mice (WT, black bars) or ERAP1−/− mice (KO, gray bars) were stained with mAb to H-2Kb or H-2Db or, as a control, to I-Ab and various antibodies to distinguish splenocyte subsets (CD11c, predominantly dendritic cells; CD8+and CD4, predominantly T lymphocyte subsets; B220, predominantly B cells) as indicated on the x axis. Shown are mean fluorescence intensities with background staining by an irrelevant antibody subtracted (averages of three mice; error bars represent 1 SD; representative of five experiments). Asterisks indicate statistically significant differences (P < 0.05, Student t test) between ERAP1+/+ and ERAP1−/− splenocytes.
CD8+ T Cell Responses to Viral Epitopes Are Altered in ERAP1 KO Mice.
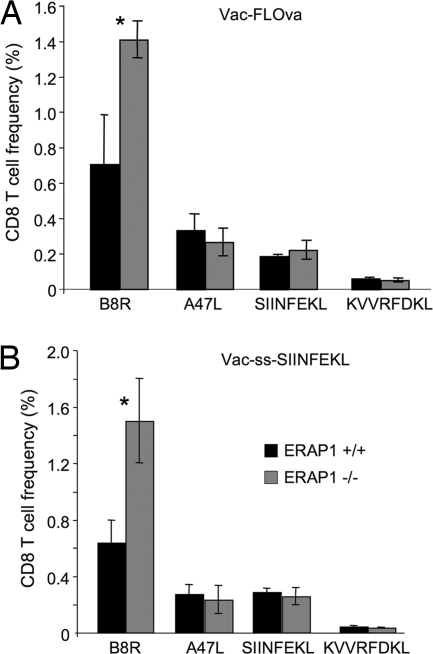
To determine whether, in the absence of ERAP1, responses to MHC class I-restricted peptides were altered, we infected ERAP1−/−, or age-matched WT C57BL/6 mice with recombinant vaccinia viruses expressing full-length ovalbumin (Vac-FLOVA) or SIINFEKL from ovalbumin preceded by a signal sequence (Vac-ES-Ova) and measured responses to two vaccinia-derived peptides (B8R20–27 and A47L138–146) (28) as well as SIINFEKL. The response to B8R was significantly increased in ERAP1−/− mice compared with WT mice, whereas the responses to A47L and SIINFEKL were not significantly different whether mice were infected with Vac-FLOVA or Vac-ES-OVA (Fig. 3). The response to the subdominant H-2Kb-restricted peptide from ovalbumin, KVVRFDKL (29), was not detectable in mice infected with Vac-ES-OVA (which do not express this epitope) and was barely above background in mice infected with Vac-FLOVA in the presence or absence of ERAP1 (Fig. 3).
Fig. 3.
CD8+ T cell responses to vaccinia virus are altered in ERAP1−/− mice. ERAP1+/+ (black bars) and ERAP1−/− (gray bars) mice were infected with recombinant vaccinia virus expressing full-length ovalbumin (Vac-FLOva) (A) or SIINFEKL targeted to the ER by a signal sequence (Vac-ss-SIINFEKL) (B). Eight days later, splenocytes were stimulated with peptides corresponding to vaccinia epitopes (B8R and A47L) or ovalbumin epitopes (SIINFEKL and KVVRFDKL), permeabilized, stained with anti-IFNγ antibody, and analyzed by flow cytometry. Shown is the percentage of CD8+ T cells producing IFNγ in response to each peptide (averages of three mice; error bars represent 1 SD; representative of at least three experiments). Asterisks indicate statistically significant differences (P < 0.05, Student t test) between ERAP1+/+ and ERAP1−/− mice.
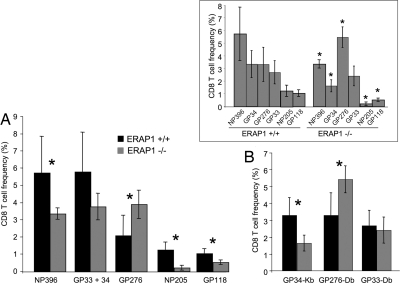
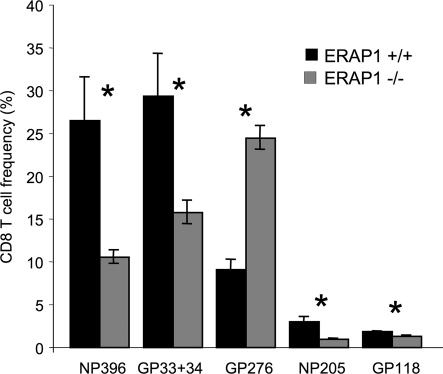
We also infected ERAP1−/− or WT mice with lymphocytic choriomeningitis virus (LCMV). Eight days later, we measured the frequency of CD8+ T cells specific for particular LCMV peptides. Of six LCMV H-2b-restricted epitopes tested, the frequencies of CD8+ T cells for four were reduced, for one unchanged, and for one somewhat increased (Fig. 4A and B). Specifically, there were only ≈1/6 to 2/3 as many CD8+ T cells specific for the H-2Kb-restricted peptides GP34, GP118, NP205, and the H-2Db-restricted peptide NP396, whereas those specific for the H-2Db-restricted epitope GP33 were unchanged. T cells specific for the H-2Db epitope GP276 were modestly increased, up to 1.5-fold, although this difference was statistically significant (P = 0.049, Student t test) in only one of three experiments.
Fig. 4.
CD8+ T cell responses to LCMV are altered in ERAP1−/− mice. ERAP1+/+ (black bars) or ERAP1−/− (gray bars) mice were infected with LCMV. Eight days later, splenocytes were stimulated with peptides corresponding to LCMV epitopes, permeabilized, and stained with anti-IFNγ antibody (A) or stained with tetramers consisting of H-2Kb or H-2Db complexed with the indicated LCMV epitopes and analyzed by flow cytometry (B). Shown is the percentage of CD8+ T cells reactive with each peptide (averages of three mice; error bars represent 1 SD; representative of at least three experiments). Asterisks indicate statistically significant differences (P < 0.05, Student t test) between ERAP1+/+ and ERAP1−/− mice. (Inset) Same data as in A and B, arranged to show the normal and altered immunodominance hierarchies.
Notably, all of the 8-mer epitopes (GP34, GP118, and NP205) showed reduced immunogenicity, whereas the longest peptide, the 11-mer GP276 showed a trend toward increased immunogenicity. This finding is consistent with the notion that ERAP1 is most important for generating shorter peptides, whereas longer peptides are more likely to be destroyed by ERAP1 trimming. ERAP1 trims ≈50% of peptides down to 9 mer and the other 50% to 8 mer (25). In addition to peptide size, internal sequences also affect the ability of ERAP1 to trim peptides, but this specificity is not yet well defined.
In WT mice, the magnitude of T cell responses to the different LCMV epitopes follows a stereotypical hierarchy of immunodominance: NP396 > GP34 ≈ GP276 > GP33 > NP205 > GP118 (30–32) (Fig. 4 Inset). This pattern is reproducible between individual mice, independent experiments, and different laboratories. It is, therefore, remarkable that the altered responses in the ERAP1-deficient mice markedly changes this pattern, establishing a new and reproducible hierarchy of immunodominance: GP276 > NP396 > GP33 > GP34 > GP118 > NP205 (Fig. 5Inset).
Fig. 5.
Altered CD8+ T cell responses in ERAP1−/− mice are not due to altered T cell repertoire. Splenocytes from LCMV-immune B6/SJL mice were adoptively transferred into ERAP1+/+ (black bars) or ERAP1−/− (gray bars) mice. One day after transfer, recipient mice were infected with LCMV, and the recall CD8+ T cell response generated by the donor cells was analyzed by intracellular IFNγ staining. Shown is the percentage of CD8+ CD45.1+ T cells producing IFNγ in response to each peptide (averages of three mice; error bars represent 1 SD; representative of two experiments). Asterisks indicate statistically significant differences (P < 0.05, Student t test) between ERAP1+/+ and ERAP1−/− mice.
ERAP1 Effects on CD8+ T Cells Do Not Depend on Differences in the T Cell Repertoire.
The effects of ERAP1 on the magnitude of T cell responses and the immunodominance hierarchy could be due to changes in peptide processing in peripheral cells or in the thymus, where different peptide presentation could lead to changes in TCR repertoire because of effects on positive or negative thymic selection. The number and frequency of T cell subsets were not altered in the thymus (see Table 1, which is published as supporting information on the PNAS web site) or spleen (see Table 2, which is published as supporting information on the PNAS web site) of ERAP1−/− vs. ERAP1+/+ mice, suggesting that thymic selection was not grossly affected by the absence of ERAP1. Nevertheless, it remained possible that the T cell repertoire could be altered in ways that led to the observed changes. To test this issue, we used an adoptive transfer approach to normalize the repertoire of responding T cells in the WT and ERAP1-deficient mice.
LCMV-immune splenocytes from WT B6/SJL mice (congenic for CD45.1) that had been infected with LCMV 5 months previously were transferred into ERAP1−/− or ERAP1+/+ mice, and the recipients were infected with LCMV to stimulate a recall response. After 7 days, donor CD45.1 splenocytes were analyzed by intracellular cytokine staining. In WT mice, the transferred memory cells showed the typical WT pattern of immunodominance. In contrast, the responses of the transferred memory cells in ERAP1−/− mice were very different from those in ERAP1+/+ mice (Fig. 5). These differences followed the same altered pattern of immunodominance that was observed in the primary response, except that, if anything, the magnitude of the differences between ERAP1+/+ and ERAP−/− mice was generally greater, all the more remarkable because the repertoire of the transferred memory cells was initially biased toward the WT pattern of immunodominance. Therefore, these results demonstrate that a major factor in establishing the pattern of immunodominance in the LCMV system is antigen presentation by host antigen-presenting cells (APCs).
Discussion
In studies using cultured cells, ERAP1 either enhanced MHC class I antigen presentation by trimming peptides to the proper size for MHC class I binding or reduced presentation by trimming peptides until they were shorter than MHC class I-binding requirements (21, 22). Recent studies of cells from ERAP1 (ERAAP) KO mice showed similar effects, although, in most cases, antigen presentation was reduced in the absence of ERAP1 (23, 24). Here, we confirm and extend these findings and show that, in vivo, ERAP1 plays an important role in immune response to viruses, either enhancing or reducing cytotoxic T lymphocyte (CTL) responses to particular viral epitopes and, thereby, helping establish immunodominance hierarchies. In addition to supporting a role for aminopeptidases in antigen presentation, these findings indicate that, in LCMV infections, immunodominance is, to a very large extent, determined by antigen processing and epitope density on APCs.
Consistent with previous studies (21, 22), we found that MEF cell lines from ERAP1-deficient mice presented SIINFEKL on H-2Kb much less efficiently than did WT MEFs, when transfected with precursors of SIINFEKL with 5- or 6-residue N-terminal extensions (Fig. 1B). Mature SIINFEKL was presented at similar levels in the WT and ERAP1−/− MEFs (Fig. 1B), demonstrating that all steps in the antigen-presentation pathway other than ER trimming were intact in the ERAP1−/− cells. These findings confirmed that ERAP1 is the major trimming enzyme in the ER lumen. Interestingly, ER-targeted SIINFEKL with a relatively short N-terminal extension (ALL-SIINFEKL, a 3-residue extension) was presented relatively well in ERAP1−/− cells, being reduced by ≈50% (but still easily detectable) compared with WT cells (data not shown), raising the possibility that another as-yet-unidentified ER aminopeptidase may contribute weakly to trimming of some ER peptides. In humans, another aminopeptidase that is closely related to ERAP1, termed L-RAP (31) or ERAP2 (32), is also expressed in some tissues, especially after IFNγ treatment and, in addition to acting directly on peptides in the ER, may also modify the activity of ERAP1, because the two enzymes form a complex (32). However, ERAP2 is not present in mice.
We found no significant difference in cell-surface MHC class I between ERAP1−/− and WT MEFs. However, it is likely that, in primary fibroblasts (which express much lower levels of MHC class I and other components of the MHC class I antigen-presentation pathway than do the transformed cells used in previous studies), peptide is not always limiting for surface expression of MHC class I. Surface levels of MHC class I were significantly reduced on ERAP1−/− B cells, T cells, and thymocytes. T cell and thymocyte subset ratios were not altered in KO vs. WT mice, suggesting that the reduction in MHC class I levels was not enough to affect positive or negative thymic selection. Importantly, MHC class I surface expression was also reduced on CD11c+ dendritic cells (DC), which are critical for immune surveillance and for activating T cell responses. T and B cells and DC express mainly immunoproteasomes, and, in HeLa-Kb cells in which immunoproteasomes were induced by chronic treatment with IFNγ, ERAP1 knockdown also led to reduced surface H-2Kb levels (21). The effect of ERAP1 KO in splenocytes was greater on H-2Kb than on H-2Db, consistent with the observation that ERAP1 acts only very slowly on peptides of eight or fewer residues but rapidly hydrolyzes ≈50% of nonamers to octamers (20, 25). Because H-2Kb mainly binds peptides that are 8 aa in length, ERAP1 will primarily function to generate these peptides, whereas, in contrast, ERAP1 can both generate and destroy many of the 9-mer peptides that bind to H-2Db (33). In any case, these data show an important role for ERAP1 in MHC class I antigen presentation in mouse primary cells.
One of the most important reasons to develop an ERAP1 KO mouse was to evaluate the importance of ERAP1 in the generation of immune responses in vivo. We infected ERAP1−/− or ERAP1+/+ mice with viruses that mainly infect either LCMVs or nonlymphoid tissue (recombinant vaccinia viruses) and measured the frequency of CD8+ T cells specific for various epitopes from these viruses. ERAP1 proved to be very important in establishing immune responses to viral epitopes: Six of the nine epitopes that gave a detectable response showed significantly different responses in the absence of ERAP1. In two cases, eliminating ERAP1 increased the response, whereas, in four, the response was markedly reduced in the absence of ERAP1. These observations show that, in vivo, as in vitro, ERAP1 plays an important role in modifying antigenic peptides.
It is noteworthy that the effects of ERAP1 are, to some extent, predictable based on its biochemical characterization (20, 25). ERAP1 only slowly trims peptides that have eight or fewer residues. Three of the peptides with altered responses are octamers that are presented by H-2Kb (LCMV GP34, NP205, and GP118); these octamers showed a reduced response. ERAP1 also trims about half of nonamers slowly and the other half rapidly. One of the two nonamers analyzed in the LCMV response (NP396, presented by H-2Db), showed a reduced response in the absence of ERAP1, whereas the other nonamer (GP33, presented on H-2Db) was not affected by the lack of ERAP1. ERAP1 rapidly trims peptides between 10 and 16 residues long; the CD8+ response to LCMV GP276, which is 11 residues long, was significantly enhanced in secondary responses and showed a similar trend in primary responses in the absence of ERAP1. Therefore, presentation of the shortest peptides, which are most likely to depend on ERAP1 for trimming, is the most reduced by its loss; whereas presentation of the longest peptide, which is most likely to be destroyed by ERAP1, was enhanced in its absence.
An exception to this trend occurred with the vaccinia virus epitope B8R, an octamer (TSYKFESV) that is presented by H-2Kb. In the absence of ERAP1, B8R induced about twice as many CTL as in its presence. This difference between LCMV and vaccinia peptides may be related to the primary sequence of the peptides or may be because the viruses infect different cell types. The frequency of CD8+ T cells responding to the B8R epitope changes when vaccinia is administered to different tissues (28), and it is tempting to speculate that this change may be related to different levels of ERAP1 in different tissues. In addition, consistent with observations on MEFs, the immune response to ovalbumin (SIINFEKL) was not affected when mice were infected with recombinant vaccinia virus expressing full-length ovalbumin or SIINFEKL targeted to the ER by a signal sequence (Fig. 3 A and B). The main point, however, is that ERAP1 clearly plays an important role in vivo in processing peptides for MHC class I antigen presentation.
One striking result of ERAP1’s broad effect on antigen presentation was its impact on immunodominance. The CTL response to LCMV, as with virtually all immune responses, is focused on a limited number of peptides, and, in mice with the same genetic background, the response to different peptides follows a strict and stable hierarchy, with (in C57BL/6 mice) the peptide NP396 inducing the most CTL and GP118 the fewest (31). LCMV is a remarkably potent inducer of CTL, stimulating a CTL response and maintaining its immunodominance hierarchy in the absence of CD28 costimulation (34) and in the absence of immunoproteasomes (35) and showing only slight changes in immunodominance even in the absence of CD4 help (36). In ERAP1−/− mice, however, the immunodominance hierarchy was disrupted: The response to several peptides was reduced by 1/2 to 2/3, whereas the response to other peptides was unchanged or even increased. Remarkably, even when responding cells were all from the WT mice that had previously been exposed to LCMV (and were, therefore, already skewed toward the WT pattern of immunodominance), the pattern of altered immunodominance emerged in ERAP1−/− mice.
These findings have important implications for the mechanisms responsible for immunodominance. The causes of immunodominance are as yet incompletely understood. Although high-affinity binding to MHC class I is important for responses and immunodominance (37–40), many peptides with high affinity for MHC class I are not immunodominant, and, conversely, several immunodominant peptides are of relatively low affinity (41–44). CD8+ T lymphocytes themselves have been proposed to play an important role in establishing immunodominance through several different mechanisms. CD8+ T cells responsive to immunodominant epitopes may be the most abundant ones in the T cell repertoire, either naturally (3, 44, 45) or as a result of cross-reactive immunity (4). Certain T cell clones may dominate responses by out-competing others for APCs (6, 7) by suppressing responses to subdominant epitopes (8, 9) or by more efficiently generating IFNγ (46). Other explanations for immunodominance include the induction of tolerance to certain epitopes (5), physical separation between epitopes (9), kinetics of protein expression (47), and agonist/antagonist peptide interactions with TCR (48). However, there is little definitive data demonstrating the relative importance of these explanations in vivo. The abundance of peptide–MHC complexes at the cell surface [which could be influenced by many factors, including the amount of peptide generation and destruction by proteasomes (49) and peptidases] may be another factor (50, 51), but quantitation of cell-surface MHC class I–peptide complexes suggests that peptide density may not be a major cause of immunodominance (52, 53) (reviewed in ref. 1); however, accurate quantitation of cell-surface peptide–MHC complexes is notoriously difficult. These various mechanisms are not mutually exclusive and may all participate to varying extents in different situations.
Perhaps the most comparable study of the importance of antigen presentation in establishing immunodominance is one that analyzed LMP2-deficient mice (54), in which immunodominance of a few of the many influenza-derived epitopes is affected. However, in this model, altered T cell repertoire was also important for some, although not all, of the alterations in immunodominance. In the ERAP1-deficient mouse model, we can rule out T cell-intrinsic factors, such as a biased T cell repertoire, because the same effects were seen even when the responding T cells from the same LCMV-immune WT host were transferred into ERAP1+/+ versus ERAP1−/− mice. In addition, the results from these adoptive transfer experiments indicate that T cell immunodomination or competition at the level of the APC does not underlie the major mechanism of immundominance for LCMV, and, because the source of the epitopes (i.e., protein expressed from the virus) is the same in WT and ERAP1−/− mice, antigen-intrinsic differences can be ruled out. The CTL transfer experiments map the effects of ERAP1 to host APCs, where ERAP1 is almost certainly mediating its effects through alterations in the generation and destruction of the individual viral epitopes.
These findings are particularly remarkable because LCMV stimulates an extraordinarily strong immune response. Similarly, we observed changes in CD8+ T cell responses to vaccinia virus, another strongly immunogenic virus. Thus, the changes in the immune response we observe are not subtle ones nor ones occurring under very limiting conditions. Together, these findings demonstrate that the major factor in establishing the immunodominance hierarchy to LCMV epitopes is almost certainly the abundance of peptide epitopes at the cell surface. By altering antigen processing and, therefore, affecting epitope abundance, the absence of ERAP1 reprogrammed the immunodominance hierarchy in the immune response to LCMV.
Materials and Methods
ERAP1 Mutant Mice.
Heterozygous mice with a conditional KO of ERAP1 were generated under contract by OzGene (Bentley, Australia). Briefly, LoxP sequences were inserted between exons 4 and 5 and between exons 6 and 7 (Fig. 6). A phosphoglycerine kinase (PGK) Neo cassette flanked by FLP recombinase target (FRT) sequences, inserted between exon 6 and the second loxP site, was used to confer resistance to C57BL/6 ES cells that had successfully integrated the targeting vector and was removed by treatment with Flippase (FLP) recombinase. This procedure produced ES cells with exons 5 and 6 of ERAP1 flanked by LoxP sites (Fig. 6). ES cells were microinjected, and chimeric mice were bred to generate heterozygous F1 mice. These floxed mice were crossed with Cre-deleter C57BL/6 mice (26), leading to the removal of exons 5 and 6 on one chromosome. Mice were bred to homozygosity.
Plasmids.
Plasmids used in these experiments were pTracerSRα (pTracerSV40; Invitrogen) in which the SV40 promoter has been replaced by the SRα promoter (55) pTracer-SRα-Cyto-OVA (expressing full-length ovalbumin with the N-terminal 50 aa containing the signal sequence removed, yielding a cytosolic protein that is rapidly degraded by proteasomes) (56), pUG1 (a control vector containing ubiquitin followed by an internal ribosome entry site and GFP, under the control of the CMV promoter), pUG-S8L (same as pUG1 but with SIINFEKL fused to the C terminus of ubiquitin, so that C-terminal ubiquitin hydrolases rapidly release SIINFEKL from ubiquitin) (57), pUG-N5-S8L (pUG expressing LEQLE-SIINFEKL fused to the C terminus of ubiquitin), and pUG-ss-AN5-S8L (pUG with the Ad-E3gp19K signal sequence preceding ALEQLE-SIINFEKL).
Cells and Flow Cytometry.
Six 9-week-old homozygous KO mice or their WT littermates were killed, and splenocytes or thymocytes were stained with anti-CD8, anti-CD4, anti-CD11c, anti-H-2Kb, anti-H-2Db, anti-B220, and anti-I-Ab in various combinations and analyzed by flow cytometry. Antibodies were obtained from Becton Dickinson and eBioscience, San Diego).
MEFs were prepared as described (12) from heterozygous females crossed to heterozygous males. After culture, DNA from each cell line was analyzed by genomic PCR, and homozygous WT and KO lines (three each) were stained with B8.24.3 (anti-H-2Kb) (58) or 28.14.8S (anti-H-2Db) (58) and analyzed by flow cytometry.
To analyze antigen presentation, MEFs were transfected by using FuGene6 (Roche) according to the manufacturer’s protocol, with plasmids expressing various SIINFEKL precursors stained with mAb 25.D1.16 (specific for the combination of SIINFEKL and H-2Kb) (27) and analyzed by flow cytometry. Transfected cells were identified by GFP expression. Transfection efficiency, based on GFP expression, ranged from ≈5% to 25%, depending on the vector.
Viral Infections and Detection of Epitope-Specific CD8+ T Cells.
Homozygous KO mice, age-matched WT littermates, or C57BL/6 mice (The Jackson Laboratory), were infected i.p. with 5 × 104 plaque-forming unit (PFU) of LCMV (strain Armstrong) or 5 × 106 PFU of vaccinia recombinants expressing full-length ovalbumin (Vac-FLOVA) (50) or A-SIINFEKL preceded by a signal sequence (Vac-ES-Ova) (50) provided by Jon Yewdell (National Institutes of Health, Bethesda). Seven or 8 days later, epitope-specific CD8+ T cells were detected by using intracellular cytokine staining or tetramers as described in refs. 4, 28, and 59.
Adoptive Transfer of LCMV-Immune Splenocytes.
To generate LCMV-immune mice, WT B6/SJL mice (CD45.1 from Taconic Farms) were infected with 5 × 104 PFU of LCMV and were used at 5–6 months after infection. Splenocytes from LCMV-immune B6/SJL mice were adoptively transferred into ERAP1−/− or ERAP1+/+ mice. One day after transfer, recipient mice were infected with 5 × 104 PFU of LCMV, and the recall CD8+ T cell response generated by the donor cells was analyzed by intracellular cytokine staining as described in ref. 4. Donor CD8+ T cells were identified by staining with a mAb specific for the congenic marker CD45.1 (A20; eBioscience).
Supplementary Material
Acknowledgments
We thank Keith Daniels for constructing MHC class I tetramers and Sharlene Hubbard for expert assistance with mice. This work was supported by National Institutes of Health grants (to K.L.R.).
Abbreviations
- APC
antigen-presenting cell
- CTL
cytotoxic T lymphocyte
- ER
endoplasmic reticulum
- ERAP1
ER aminopeptidase 1
- KO
knockout
- LCMV
lymphocytic choriomeningitis virus
- MEF
mouse embryonic fibroblast
- TCR
T cell receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Yewdell J. W., Bennink J. R. Annu. Rev. Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 2.Busch D. H., Pamer E. G. J. Immunol. 1998;160:4441–4448. [PubMed] [Google Scholar]
- 3.Sandberg J. K., Grufman P., Wolpert E. Z., Franksson L., Chambers B. J., Karre K. J. Immunol. 1998;160:3163–3169. [PubMed] [Google Scholar]
- 4.Brehm M. A., Pinto A. K., Daniels K. A., Schneck J. P., Welsh R. M., Selin L. K. Nat. Immunol. 2002;3:627–634. doi: 10.1038/ni806. [DOI] [PubMed] [Google Scholar]
- 5.Wherry E. J., Blattman J. N., Murali-Krishna K., van der Most R., Ahmed R. J. Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grufman P., Sandberg J. K., Wolpert E. Z., Karre K. J. Leukoc. Biol. 1999;66:268–271. doi: 10.1002/jlb.66.2.268. [DOI] [PubMed] [Google Scholar]
- 7.Grufman P., Wolpert E. Z., Sandberg J. K., Karre K. Eur. J. Immunol. 1999;29:2197–2204. doi: 10.1002/(SICI)1521-4141(199907)29:07<2197::AID-IMMU2197>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 8.Weidt G., Utermohlen O., Heukeshoven J., Lehmann-Grube F., Deppert W. J. Immunol. 1998;160:2923–2931. [PubMed] [Google Scholar]
- 9.Rodriguez F., Harkins S., Slifka M. K., Whitton J. L. J. Virol. 2002;76:4251–4259. doi: 10.1128/JVI.76.9.4251-4259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beninga J., Rock K. L., Goldberg A. L. J. Biol. Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 11.Stoltze L., Schirle M., Schwarz G., Schroter C., Thompson M. W., Hersh L. B., Kalbacher H., Stevanovic S., Rammensee H. G., Schild H. Nat. Immunol. 2000;1:413–418. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 12.Towne C. F., York I. A., Neijssen J., Karow M. L., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Neefjes J. J., Rock K. L. J. Immunol. 2005;175:6605–6614. doi: 10.4049/jimmunol.175.10.6605. [DOI] [PubMed] [Google Scholar]
- 13.Saric T., Graef C. I., Goldberg A. L. J. Biol. Chem. 2004;279:46723–46732. doi: 10.1074/jbc.M406537200. [DOI] [PubMed] [Google Scholar]
- 14.York I. A., Mo A. X., Lemerise K., Zeng W., Shen Y., Abraham C. R., Saric T., Goldberg A. L., Rock K. L. Immunity. 2003;18:429–440. doi: 10.1016/s1074-7613(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 15.Kim S. I., Pabon A., Swanson T. A., Glucksman M. J. Biochem. J. 2003;375:111–120. doi: 10.1042/BJ20030490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reits E., Neijssen J., Herberts C., Benckhuijsen W., Janssen L., Drijfhout J. W., Neefjes J. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- 17.Kloetzel P. M. Nat. Immunol. 2004;5:661–669. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- 18.Hattori A., Matsumoto H., Mizutani S., Tsujimoto M. J. Biochem. (Tokyo) 1999;125:931–938. doi: 10.1093/oxfordjournals.jbchem.a022371. [DOI] [PubMed] [Google Scholar]
- 19.Schomburg L., Kollmus H., Friedrichsen S., Bauer K. Eur. J. Biochem. 2000;267:3198–3207. doi: 10.1046/j.1432-1327.2000.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., Goldberg A. L. Nat. Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 21.York I. A., Chang S. C., Saric T., Keys J. A., Favreau J. M., Goldberg A. L., Rock K. L. Nat. Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 22.Serwold T., Gonzalez F., Kim J., Jacob R., Shastri N. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 23.Yan J., Parekh V. V., Mendez-Fernandez Y., Olivares-Villagomez D., Dragovic S., Hill T., Roopenian D. C., Joyce S., Van Kaer L. J. Exp. Med. 2006;203:647–659. doi: 10.1084/jem.20052271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer G. E., Gonzalez F., Champsaur M., Cado D., Shastri N. Nat. Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 25.Chang S. C., Momburg F., Bhutani N., Goldberg A. L. Proc. Natl. Acad. Sci. USA. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwenk F., Baron U., Rajewsky K. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porgador A., Yewdell J. W., Deng Y., Bennink J. R., Germain R. N. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 28.Tscharke D. C., Karupiah G., Zhou J., Palmore T., Irvine K. R., Haeryfar S. M., Williams S., Sidney J., Sette A., Bennink J. R., Yewdell J. W. J. Exp. Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W., Khilko S., Fecondo J., Margulies D. H., McCluskey J. J. Exp. Med. 1994;180:1471–1483. doi: 10.1084/jem.180.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Most R. G., Concepcion R. J., Oseroff C., Alexander J., Southwood S., Sidney J., Chesnut R. W., Ahmed R., Sette A. J. Virol. 1997;71:5110–5114. doi: 10.1128/jvi.71.7.5110-5114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Most R. G., Murali-Krishna K., Whitton J. L., Oseroff C., Alexander J., Southwood S., Sidney J., Chesnut R. W., Sette A., Ahmed R. Virology. 1998;240:158–167. doi: 10.1006/viro.1997.8934. [DOI] [PubMed] [Google Scholar]
- 32.Murali-Krishna K., Altman J. D., Suresh M., Sourdive D. J., Zajac A. J., Miller J. D., Slansky J., Ahmed R. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 33.Falk K., Rötzschke O., Stevanovic S., Jung G., Rammensee H.-J. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 34.Suresh M., Whitmire J. K., Harrington L. E., Larsen C. P., Pearson T. C., Altman J. D., Ahmed R. J. Immunol. 2001;167:5565–5573. doi: 10.4049/jimmunol.167.10.5565. [DOI] [PubMed] [Google Scholar]
- 35.Nussbaum A. K., Rodriguez-Carreno M. P., Benning N., Botten J., Whitton J. L. J. Immunol. 2005;175:1153–1160. doi: 10.4049/jimmunol.175.2.1153. [DOI] [PubMed] [Google Scholar]
- 36.Khanolkar A., Fuller M. J., Zajac A. J. J. Immunol. 2004;172:2834–2844. doi: 10.4049/jimmunol.172.5.2834. [DOI] [PubMed] [Google Scholar]
- 37.Sette A., Vitiello A., Reherman B., Fowler P., Nayersina R., Kast W. M., Melief C. J. M., Oseroff C., Yuan L., Ruppert J., et al. J. Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 38.van der Most R. G., Sette A., Oseroff C., Alexander J., Murali-Krishna K., Lau L. L., Southwood S., Sidney J., Chesnut R. W., Matloubian M., Ahmed R. J. Immunol. 1996;157:5543–5554. [PubMed] [Google Scholar]
- 39.Levitsky V., Zhang Q. J., Levitskaya J., Masucci M. G. J. Exp. Med. 1996;183:915–926. doi: 10.1084/jem.183.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Micheletti F., Bazzaro M., Canella A., Marastoni M., Traniello S., Gavioli R. Immunology. 1999;96:411–415. doi: 10.1046/j.1365-2567.1999.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pion S., Fontaine P., Desaulniers M., Jutras J., Filep J. G., Perreault C. Eur. J. Immunol. 1997;27:421–430. doi: 10.1002/eji.1830270212. [DOI] [PubMed] [Google Scholar]
- 42.Vitiello A., Yuan L., Chesnut R. W., Sidney J., Southwood S., Farness P., Jackson M. R., Peterson P. A., Sette A. J. Immunol. 1996;157:5555–5562. [PubMed] [Google Scholar]
- 43.Mullbacher A., Lobigs M., Yewdell J. W., Bennink J. R., Tha Hla R., Blanden R. V. Scand. J. Immunol. 1999;50:420–426. doi: 10.1046/j.1365-3083.1999.00619.x. [DOI] [PubMed] [Google Scholar]
- 44.Regner M., Mullbacher A., Blanden R. V., Lobigs M. Viral Immunol. 2001;14:135–149. doi: 10.1089/088282401750234510. [DOI] [PubMed] [Google Scholar]
- 45.Belz G. T., Stevenson P. G., Doherty P. C. J. Immunol. 2000;165:2404–2409. doi: 10.4049/jimmunol.165.5.2404. [DOI] [PubMed] [Google Scholar]
- 46.Liu F., Whitton J. L., Slifka M. K. J. Immunol. 2004;173:456–462. doi: 10.4049/jimmunol.173.1.456. [DOI] [PubMed] [Google Scholar]
- 47.Probst H. C., Tschannen K., Gallimore A., Martinic M., Basler M., Dumrese T., Jones E., van den Broek M. F. J. Immunol. 2003;171:5415–5422. doi: 10.4049/jimmunol.171.10.5415. [DOI] [PubMed] [Google Scholar]
- 48.Lau L. L., Jiang J., Shen H. J. Immunol. 2005;174:7970–7976. doi: 10.4049/jimmunol.174.12.7970. [DOI] [PubMed] [Google Scholar]
- 49.Basler M., Youhnovski N., Van Den Broek M., Przybylski M., Groettrup M. J. Immunol. 2004;173:3925–3934. doi: 10.4049/jimmunol.173.6.3925. [DOI] [PubMed] [Google Scholar]
- 50.Restifo N. P., Bacik I., Irvine K. R., Yewdell J. W., McCabe B. J., Anderson R. W., Eisenlohr L. C., Rosenberg S. A., Bennink J. R. J. Immunol. 1995;154:4414–4422. [PMC free article] [PubMed] [Google Scholar]
- 51.Gallimore A., Dumrese T., Hengartner H., Zinkernagel R. M., Rammensee H. G. J. Exp. Med. 1998;187:1647–1657. doi: 10.1084/jem.187.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Y., Smith K. D., Kurilla M. G., Lutz C. T. J. Immunol. 1997;159:1844–1852. [PubMed] [Google Scholar]
- 53.Crotzer V. L., Christian R. E., Brooks J. M., Shabanowitz J., Settlage R. E., Marto J. A., White F. M., Rickinson A. B., Hunt D. F., Engelhard V. H. J. Immunol. 2000;164:6120–6129. doi: 10.4049/jimmunol.164.12.6120. [DOI] [PubMed] [Google Scholar]
- 54.Chen W., Norbury C. C., Cho Y., Yewdell J. W., Bennink J. R. J. Exp. Med. 2001;193:1319–1326. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takebe Y., Seiki M., Fujisawa J. I., Hoy P., Yokota K., Arai K. I., Yoshida M., Arai N. Mol. Cell. Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen L., Rock K. L. Proc. Natl. Acad. Sci. USA. 2004;101:3035–3040. doi: 10.1073/pnas.0308345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bachmair A., Finley D., Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 58.Ozato K., Hansen T. H., Sachs D. H. J. Immunol. 1980;125:2473–2477. [PubMed] [Google Scholar]
- 59.Mylin L. M., Schell T. D., Roberts D., Epler M., Boesteanu A., Collins E. J., Frelinger J. A., Joyce S., Tevethia S. S. J. Virol. 2000;74:6922–6934. doi: 10.1128/jvi.74.15.6922-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.