Abstract
The NF-κB pathways have been implicated in tumorigenesis in several lymphoid malignancies, including non-Hodgkin's and Hodgkin's lymphomas. However, the antiapoptotic functions and the mechanism responsible for signaling through each NF-κB pathway remain to be elucidated. In the current study, lymphoma cell lines with constitutively active NF-κB were found to be resistant to inducers of the extrinsic and intrinsic apoptosis pathways. Resistance to cell death resulted from blocks early and late in the apoptosis cascade. Several NF-κB target genes were overexpressed in these cell lines, including Bcl-xL, Fas-associated death domain-like IL-1β-converting enzyme inhibitor protein, cellular inhibitor of apoptosis, and X inhibitor of apoptosis. Inhibition of the canonical or noncanonical NF-κB pathways with small interfering RNAs or adenovirus expressing a stable form of inhibitor of NF-κB (IκB) enhanced sensitivity to apoptosis inducers and resulted in lower levels of Bcl-xL or Fas-associated death domain-like IL-1β-converting enzyme inhibitor protein, cellular inhibitor of apoptosis, and X inhibitor of apoptosis. These findings demonstrate an important role of both NF-κB pathways in mediating resistance to apoptosis and distinctive antiapoptotic downstream target gene profiles responsible for this effect.
Keywords: Epstein–Barr virus, human T cell leukemia virus, Tax
Nuclear factor κB (NF-κB) was originally identified as a transcription factor that regulated expression of the κ Ig gene in B lymphocytes, but has subsequently been shown to be activated by different mitogenic stimuli, stress signals, or inflammatory cytokines in a broad range of cell types (1, 2). The NF-κB pathways include the “canonical” (classical) and “noncanonical” (alternative) pathways using NF-κB precursor proteins, p105 (NF-κB-1) and p100 (NF-κB-2), respectively (3, 4). These proteins are processed to the mature p50 NF-κB-1 and p52 NF-κB-2 proteins that heterodimerize with other members of the NF-κB family, p65 Rel-A or c-Rel, or Rel-B, respectively. Activation of the canonical pathway involves release of p50–p65 from IκB, as a result of phosphorylation by IκB kinase (IKK) and degradation of IκB by the proteasome. Activation of the noncanonical pathway involves cleavage of p100 and dimerization of the mature p52 product with Rel-B. In both cases, the mature dimeric NF-κB proteins translocate to the nucleus and activate genes involved in antiapoptotic function and genes involved in the modulation of immune and inflammatory response, cell proliferation, adhesion, and angiogenesis.
The antiapoptotic functions of the NF-κB family result from a direct interaction with tumor suppressor proteins, such as p53, or an imbalance in expression of proapoptotic and antiapoptotic proteins. The antiapoptotic function of NF-κB involves the intrinsic (mitochondrial) pathway or the extrinsic (receptor) pathway (5). Overexpression of members of the Bcl-2 family, such as A1 or Bfl1 and Bcl-xL, inhibit the activation of caspase 9 or proapoptotic Bcl-2 family members produced by inducers of the intrinsic pathway. Inducers of the intrinsic pathway include agents that cause DNA damage or oxidative stress. On the other hand, overexpression of the cellular inhibitors of apoptosis (CIAPs) and Fas-associated death domain-like IL-1β-converting enzyme inhibitor protein (FLIP) modulate the activation of caspase 8 produced by induction of TNF receptor family members. Finally, NF-κB regulates the activation of common “executioner” caspases, including caspases 3 and 7, which result from activation of both apoptotic pathways, by overexpression of the X chromosome-linked inhibitor of apoptosis (XIAP).
NF-κB is constitutively active in a variety of lymphomas, including Hodgkin's and non-Hodgkin's lymphomas, such as mantle cell, mediastinal diffuse large cell, and virally mediated lymphomas, including adult T cell leukemia lymphoma (ATLL), Burkitt lymphoma, and primary effusion lymphomas (6–9). Activation occurs by direct effects on NF-κB genes as a result of mutations, deletions, or rearrangements of NF-κB regulatory genes or indirect effects of viral oncogenes (1). One such viral oncogene, Epstein–Barr virus (EBV) latent membrane protein 1, activates NF-κB, acting as a homologue of constitutively active CD40 (10–12). Similarly, several human herpesvirus 8 proteins activate NF-κB in primary effusion lymphomas (13, 14). In ATLL, the Tax protein of human T cell leukemia virus (HTLV) activates NF-κB through interactions with the IKK-γ subunit or protein kinases that phosphorylate and activate the IKK-α or IKK–β subunits, by direct binding to IκB proteins resulting in their degradation, or by accelerating the cleavage of the NF-κB-1 or NF-κB-2 precursor proteins (15, 16).
A variety of different NF-κB pathway inhibitors have been developed and are undergoing clinical studies in lymphomas and other malignancies. These include agents that block IKK activity, IκB degradation, NF-κB nuclear targeting, or NF-κB target gene activity (6, 17, 18). The current work provides further understanding of the relative contribution of each NF-κB pathway and its downstream target genes on the activation of each apoptotic pathway, which is important for the design and assessment of novel targeted anticancer agents.
Results
Lymphoma Cell Lines with Constitutively Activated NF-κB Are Resistant to Inducers of Apoptosis.
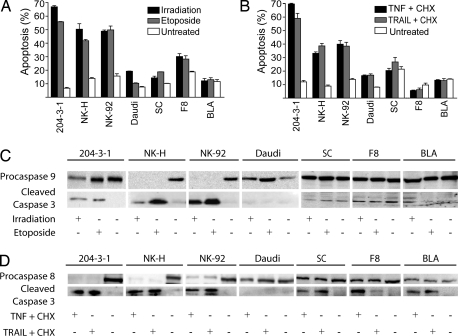
The NF-κB pathway is known to be constitutively active in Tax tumor-derived cells (SC, F8, and BLA) and EBV (+) Burkitt's lymphoma cell line (Daudi) (19, 20). Because NF-κB is associated with resistance to apoptosis, we evaluated the intrinsic and extrinsic apoptotic pathways in cells with constitutively active NF-κB activity (NF-κB-ca) and controls without NF-κB-ca, including a murine B cell leukemia cell line (204-3-1), primary human natural killer cells (NK-H), and a human natural killer cell line (NK-92). For this purpose, different inducers of apoptosis were used, including γ-irradiation and etoposide to activate the intrinsic pathway and TNF-α or TNF-related apoptosis-inducing ligand (TRAIL) to activate the extrinsic pathway. Apoptosis was measured by FACS with annexin V and propidium iodine (PI) staining. As seen in Fig. 1A, the level of apoptosis induced through the intrinsic pathway was 3- to 4-fold higher in control cells compared with those with an NF-κB-ca pathway (P < 0.001). Similar results were obtained when we evaluated the extrinsic apoptotic pathway (Fig. 1B; P < 0.001). Overall, cells with NF-κB-ca demonstrated resistance to the induction of apoptosis via the intrinsic or extrinsic apoptotic pathways.
Fig. 1.
Resistance to apoptosis in cells with constitutive activation of NF-κB. (A and B) Constitutive expression of NF-κB in lymphoma cells results in resistance to inducers of the intrinsic pathway (A) and extrinsic pathway (B) of apoptosis. Indicated cell lines with (Daudi, SC, F8, BLA) or without constitutive NF-κB expression (204-3-1, NK-H, NK-92) were treated, as described in Methodology with γ-irradiation or etoposide (A) or TNF-α or TRAIL with cycloheximide (CHX) (B) and then assayed 24 h later for the proportion of annexin V and PI-positive cells by FACS. (C and D) Alternatively, the same cells were assayed by immunoblot as described in Methodology for levels of procaspase 9 and caspase 3 (C) or procaspase 8 and caspase 3 (D). Identical protein loading was assured by using the same membranes for each of the blots.
To determine the step(s) perturbed in the apoptotic cascade in NF-κB-ca cell lines, we evaluated levels of different components of the intrinsic and extrinsic pathway. As seen in Fig. 1C, treatment with etoposide or γ-irradiation resulted in a significant decrease in procaspase 9 levels in control cell lines (NK-H and NK-92) and to a lesser extent in 204-3-1 cells, increased cleaved caspase 9 levels in 204-3-1 cells (Fig. 5A, which is published as supporting information on the PNAS web site), and an increase in cleaved caspase 3 levels in the control cell lines. In contrast, cells with NF-κB-ca demonstrated no apparent change in procaspase 9 or caspase 3 levels. For studies of the extrinsic apoptotic pathway, control cell lines demonstrated reduction in the levels of procaspase 8 and elevation of cleaved caspase 3 levels. This effect was reduced in cells with NF-κB-ca, although some cleaved caspase 3 was evident (Fig. 1 C and D). These findings demonstrate that cells with NF-κB-ca are more resistant to apoptosis through activation of the intrinsic or extrinsic pathways, by inhibiting multiple levels of these cascades.
Increased Levels of Bcl-xL, FLIP, and IAP Antiapoptotic Proteins in Cell Lines with an NF-κB-ca Transcription Factor Pathway.
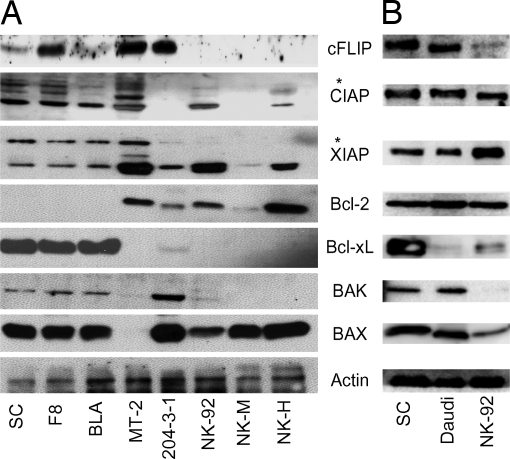
Previous studies showed that Daudi cells overexpress several proteins involved in regulation of apoptosis, including Bcl-xL, Bcl-2, Bad, Bax, Bak, and the IAP family members (21, 22). However, there has been less complete characterization of apoptosis regulators in Tax(+) tumor-derived cell lines (23). As seen in Fig. 2, Tax tumor-derived cell lines and Daudi cells have a similar expression profile for critical regulators of apoptosis. Compared with normal mouse NK cells (NK-M), Tax tumor-derived cell lines overexpress Bcl-xL, XIAP, CIAP1, CIAP2, FLIP, and survivin (Figs. 2 and 5B). Variable, but higher, levels of FLIP were seen in Tax transgenic cell lines than in NK-M, NK-92, and NK-H cells. Bcl-xL expression was also much lower in 204-3, NK-92, and NK-H cells compared with that of the Tax transgenic cell lines. The HTLV-1 immortalized lymphoid cell line, MT2, exhibited higher levels of FLIP, and to a lesser extent CIAP and XIAP, as compared with NK-92 and NK-H cells, but very low levels of Bcl-xL, Bak, and Bax. Daudi cells also overexpress FLIP and survivin, and to a lesser extent CIAP, compared with NK92 cells (Figs. 2 and 5B).
Fig. 2.
Expression of apoptosis-related proteins in lymphoma cell lines. Levels of FLIP, CIAP1, CIAP2, XIAP, Bcl-2, Bcl-xL, Bak, Bax, and actin are shown for Tax transgenic cell lines with constitutive NF-κB expression (SC, F8, BLA), HTLV-1 immortalized human lymphoid cells (MT2), and cells lacking constitutive NF-κB expression, including murine B (204–3-1) and NK cells (NK-M), and human NK cells (NK-92 and NK-H) (A) and Daudi cells (B). Immunoreactive bands of higher molecular weight (∗) were also identified in immunoblots with antibodies to CIAP and XIAP. The Bcl-2 antibodies used in the two blots were from Cell Signaling and BD Biosciences.
Comparison of the Canonical and Noncanonical NF-κB Pathways in Conferring Resistance to Apoptosis.
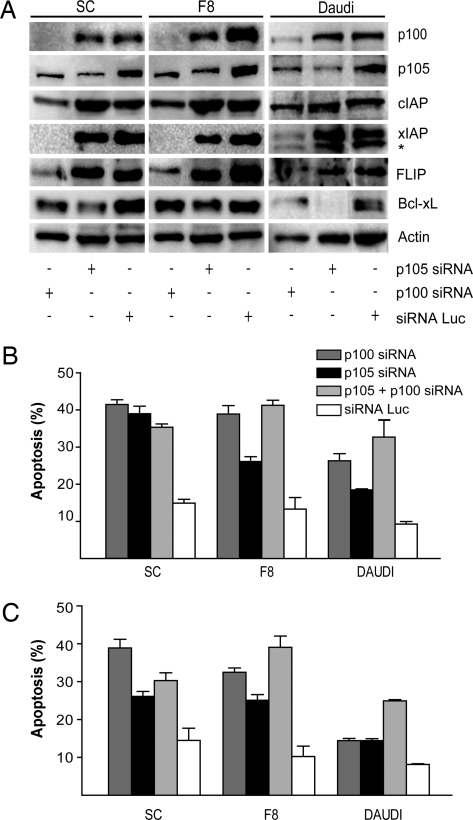
The association between alterations of NF-κB signaling and lymphomas has been suggested by the presence of mutations, deletions, or translocations in tumors of members of the NF-κB gene family, as well as by the presence of characteristic gene expression profiles (1, 24, 25). However, the relative contributions of the canonical and noncanonical NF-κB pathways in mediating resistance to apoptosis remain to be defined. To evaluate the role of both NF-κB pathways in the resistance to apoptosis, we generated stable Daudi and Tax tumor-derived cell lines expressing lentiviral vectors encoding short interfering RNA (siRNA) hairpins targeting p105, NF-κB1 (canonical pathway) and/or p100, NF-κB2 (noncanonical pathway), or luciferase, as a negative control. Western blot analysis of stable p100 and p105 siRNA-expressing cell lines demonstrated specific down-regulation of endogenous p100 and p105 expression, respectively (Figs. 3A and 5C). However, depression of p105 in p100 siRNA-expressing cells was also evident. The activity of the siRNAs was also confirmed by immunoflouroescence studies examining nuclear translocation of Rel-A and Rel-B (Fig. 6, which is published as supporting information on the PNAS web site) and EMSAs (Fig. 7, which is published as supporting information on the PNAS web site). There was no decrease in p105 or p100 expression in luciferase siRNA-expressing cells. The specificity of the targeted siRNA was also confirmed by a lack of down-regulation of actin expression. Inhibition of the canonical pathway with a siRNA against p105 resulted in depressed levels of Bcl-xL. Inhibition of the noncanonical pathway with a siRNA against p100 resulted in depressed levels of CIAP, XIAP, survivin, and Bcl-xL (Figs. 3A and 5D). These results demonstrate little or no interplay between the downstream targets of the canonical and noncanonical pathways and affirm the specificity of the siRNA expression methods used.
Fig. 3.
Repression of NF-κB-1 (p105) and NF-κB-2 (p100) with siRNA expression lentiviruses restores sensitivity to inducers of apoptosis. (A) Levels of p100, p105, CIAP, FLIP, Bcl-xL, XIAP, and actin were determined by immunoblot in Tax transgenic cell lines SC and F8, and Daudi cells expressing p100 and/or p105, or luciferase siRNAs, as described in Methodology. Nonspecific bands are indicated (∗). (B and C) The effects on apoptosis are shown for Tax transgenic cell lines SC and F8, and the Burkitt lymphoma cell line Daudi of luciferase, p100, p105, or a combination of p100 and p105 siRNAs on apoptosis induced by etoposide (B) or TNF-α treatment (C) as described in Methodology. Control levels of apoptosis and band intensity measurements are available in Supporting Text, which is published as supporting information on the PNAS web site.
Tumor cell lines expressing siRNA against p100 or p105 exhibited 2- to 3-fold increases in the extent of apoptosis after treatment with etoposide or TNF-α, as compared with cell lines expressing the luciferase siRNA (Fig. 3 B and C). Similar levels of apoptosis were seen in cells expressing both p100 and p105 siRNA vectors, as compared with those cells expressing either siRNA alone. In the absence of TNF-α or etoposide treatment, the siRNAs did not increase the level of apoptosis (data not shown). In contrast, expression of siRNAs against p100 or p105 did not affect the rate of apoptosis after treatment with etoposide of control NK-92 cells, as compared with NK-92 cells expressing siRNA against luciferase (data not shown).
Enhancement of Apoptosis by Expression of Dominant-Negative IκBα.
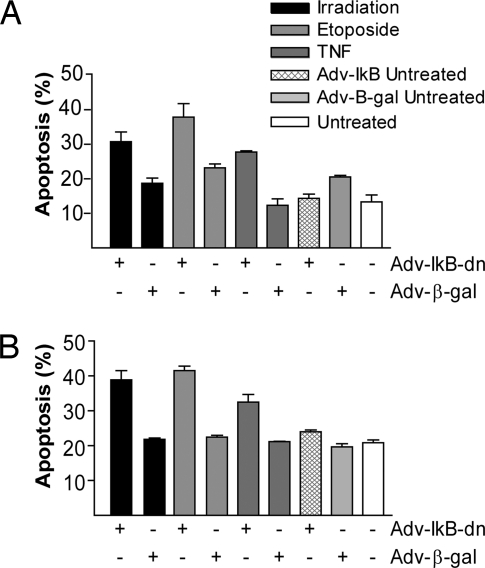
To confirm the results obtained from the p105 siRNA studies, we took a different approach for inhibition of the canonical NF-κB pathway, using a human adenovirus 5 expressing dominant-negative IκB (Adv-IκB-dn). For a negative control, tumor cell lines were infected with an adenovirus expressing β-gal (Adv-β-gal). The effect of Adv-IκB-dn on NF-κB activation was determined by luciferase assay using an NF-κB-Luc reporter plasmid and EMSA. After 24 h of infection, 60% inhibition of NF-κB activity was seen in Adv-IκB-dn-infected cells compared with Adv-β-gal-infected cells (Fig. 7), as well as down-regulation of Bcl-xL (Fig. 5 E and F).
Effects of Adv-IκB-dn on apoptosis were evaluated 8 h after infection with Adv-IκB-dn; SC and F8 cell lines were treated for 24 h with different activators of apoptosis. As seen in Fig. 4, infection with Adv-IκB-dn or Adv-β-gal did not have an apoptotic effect, but the addition of apoptosis activators increased the rate of apoptosis in Adv-IκB-dn-infected cells 1.5- to 2-fold compared with that of Adv-β-gal-infected cells. These results reinforce the previous findings that the canonical pathway plays a role in mediating resistance to apoptosis.
Fig. 4.
Inhibition of NF-κB activation with adenovirus expressing a stable form of IκB restores sensitivity to inducers of apoptosis. The effect of Adv-IκB-dn or Adv-β-gal on induction of apoptosis by γ-irradiation, etoposide, or TNF-α were examined in Tax transgenic SC (A) and F8 (B) cells, using annexin V and PI staining and FACS analysis.
Discussion
The constitutive activation of the NF-κB pathway has been implicated in tumorigenesis in a broad spectrum of aggressive lymphoid malignancies (9). Although advances have been made in understanding the molecular mechanisms through which the canonical and noncanonical NF-κB pathways facilitate the development of lymphomas, there remain significant gaps in our knowledge that must be overcome for the design and evaluation of novel targeted therapies. The present study makes several key observations regarding the link of NF-κB activation and lymphomagenesis. First, we demonstrated that an NF-κB-ca pathway rendered lymphoma cell lines resistant to the induction of apoptosis. Second, we found that overexpression of NF-κB downstream targets such as Bcl-xL, FLIP, and members of the IAP family (XIAP and CIAP) resulted in resistance to apoptosis. Third, our results indicated that both the canonical and noncanonical NF-κB pathways are important in resistance to apoptosis, which is mediated by different downstream targets. Inhibition of apoptosis through the canonical pathway was associated with Bcl-xL overexpression, whereas the noncanonical pathway was coupled with overexpression of a broader antiapoptotic profile including Bcl-xL, XIAP, CIAP, and FLIP. This result is consistent with the known differences in κB site affinity by different NF-κB proteins (26). Overall, our results suggest that both NF-κB pathways are indispensable in the mediation of tumorigenesis by reducing apoptosis through overexpression of different profiles of antiapoptotic proteins.
Many tumor cell lines and tumor tissues possess a constitutively active NF-κB pathway as a result of translocations, deletions, and mutations that uncouple members of the NF-κB family from their regulators (27–30). Characteristic gene signatures for lymphomas confirm the association of NF-κB with poor prognosis subtypes of diffuse large B cell lymphoma (activated B cell-like and mediastinal, DLBCL) (24, 25). Potential mechanisms to explain the association of NF-κB with poor-prognosis DLBCL include the promotion of cell proliferation, the prevention of apoptosis, and an increase in the angiogenic potential (31). However, previous studies have not explored in detail which specific NF-κB pathway is responsible for mediating these effects. Aberrant NF-κB activation also occurs in other hematopoietic malignancies, including Hodgkin's disease, Helicobacter pylori-associated mucosal-associated lymphoma tumors, mantle cell lymphomas, plasma cell malignancies, and acute myelogenous leukemia (32, 33).
Taking advantage of two distinct lymphoma models, we provide evidence that both NF-κB pathways regulate activation of apoptosis through different downstream targets. The first model was derived from the Tax transgenic mouse lymphoma model that we have previously described (19, 23, 34, 35). The second model was derived from an EBV (+) Burkitt's lymphoma tumor. Both models contain an NF-κB-ca pathway (19, 20). Our results demonstrate that lymphoma cell lines with NF-κB-ca exhibited very limited induction of apoptosis through either the intrinsic or extrinsic pathways, in comparison to cells lacking NF-κB activity. These results confirm and extend previous results (36–40). We also showed that the inhibition occurred early in the activation of both apoptotic cascades, because there was no apparent reduction in caspase 8 or 9 levels. Studies of NF-κB inhibitors helped explain these findings, demonstrating overexpression of Bcl-xL and XIAP to impede the intrinsic apoptosis pathway and CIAP and cFLIP to constrain the extrinsic apoptosis pathway. These critical regulatory proteins inhibit the effect of different activators of apoptosis, such as doxorubicin, etoposide, TNF-α, and TRAIL, among others. These findings reinforce the role of NF-κB as an antiapoptotic signal in lymphoid tissues (6).
Despite the limited levels of apoptosis, some cleaved caspase 3 is evident in Tax transgenic cell lines, suggesting an additional block to apoptosis at later steps in the pathway. This effect may be mediated by XIAP, CIAP, or survivin, which bind and inhibit caspase 3 and are overexpressed in the lymphoma cell lines with NF-κB-ca (41). Similarities and differences were noted in comparing the expression profiles of Tax transgenic cell lines with those of MT2 and Daudi cells. These differences may be caused by modulating effects of other HTLV-1 or EBV proteins expressed in MT2 or Daudi cells, respectively, and differences in the mechanism of NF-κB activation by Tax compared with EBV latent membrane protein 1.
The antiapoptotic function of each NF-κB pathway was tested by reducing the level of expression of p100 and p105 subunits with specific siRNAs. This methodology was successful in decreasing the formation of the processed subunits p52 and p50. The siRNA to p100 also had some effect on p105, consistent with cross-talk between these two pathways demonstrated in p100-deficient murine fibroblasts (42). This finding may be caused by the formation of p50-RelB heterodimers when p100 is depleted or later phases of NF-κB activity resulting from altered gene expression of NF-κB or IκB proteins. The value of this siRNA approach was further demonstrated by EMSA and immunofluorescent studies. Cells expressing these siRNAs had restored sensitivity to TNF or etoposide. The noncanonical pathway appeared to be more significant than the canonical pathway in mediating the effect of TNF-α in Tax tumor-derived cells. These results were supported by the strong down-regulation of FLIP and CIAP seen after the inhibition of the noncanonical pathway in these lymphoma models. Combined repression of p100 and p105 resulted in similar levels of apoptosis as when using either siRNA alone. This result is consistent with our findings that there is overlap in gene targets regulated by the canonical and noncanonical pathways.
Contrary to our results, Wang et al. (43) demonstrated in HT1080 and U2OS cells that p100 expression was essential for TNF-α-induced apoptosis. This apparent discrepancy may be explained in part by differences in the cell types used to perform the experiments and by the predominant cytoplasmic localization of the p100 subunits used, which could represent a reduction of p52, and therefore, the expression of NF-κB-related antiapoptotic proteins. However, both studies concluded that the antiapoptotic signal produced by the processed form of p100, p52, is stronger than the tumor suppressor effect produced by p100, suggesting that the apoptotic-inducing activity of the death domain of p100 seems to be negatively regulated by other regions or subunits of p100. Both studies hypothesize the existence of a balance between the tumor suppressor activity produced by the large subunits, p100 and p105, and the antiapoptotic functions produced by their processed subunits, p52 and p50. This hypothesis is supported by studies of mice with mutations in the nfkb1 or nfkb2 genes, resulting in inactive precursor proteins, but active cleaved products (44, 45). This “balance” may result from either the regulation in the protein levels of each subunit produced by proteasome processing or posttranslational modifications that could alter the tumor suppressor effect of the large subunits.
Finally, after demonstrating that both NF-κB pathways are relevant in the resistance to apoptosis, we extended our studies to search for explanations of these results. We discovered that each pathway mediates its antiapoptotic effect through the regulation of different downstream targets. The canonical pathway was predominantly involved in the regulatory process of the intrinsic pathway through Bcl-xL, and the noncanonical pathway targeted both apoptotic pathways through Bcl-xL, FLIP, XIAP, and CIAP. Our results suggest that the noncanonical pathway has a broader antiapoptotic effect, making it a more powerful target. Expression of siRNAs to XIAP sensitizes Tax transgenic cells to apoptotic stimuli to an even greater degree than expression of siRNAs to Bcl-xL (L.B.-M. and L.R., unpublished work).
Our studies suggest that NF-κB is essential for the resistance to apoptosis seen in lymphoma models. More importantly, we have identified an antiapoptotic role for the noncanonical pathway and showed that the NF-κB pathways have both redundant and independent antiapoptotic downstream targets. Overall, these results suggest that each NF-κB pathway has a specific function, as demonstrated by the broader spectrum of antiapoptotic proteins regulated by the noncanonical pathway compared with the canonical pathway and the lack of synergism that occurs when both pathways are targeted. Therefore, our study serves as a cornerstone for the development of future in vivo animal and human experiments in lymphomas focused on the identification of the characteristic gene signature pattern of each NF-κB pathway that will translate into biological subclassification of hematological malignancies and the use of specific inhibitors of IKK or downstream NF-κB targets.
Methodology
Cell Lines.
The tumor-derived F8 and SC large granular lymphocytic cell lines, maintained in Iscove's medium, have been described (35). The BLA cell line was derived from a Tax transgenic hepatic tumor. The HTLV-1-infected T cell line (MT-2), Abelson murine leukemia virus-transformed pre-B cell line containing WT p53 (204-3-1, provided by N. Rosenberg, Tufts University, Medford, MA), human natural killer diffuse large cell lymphoma line (NK-92), EBV-positive Burkitt's lymphoma cell line (Daudi), and primary human natural killer (NK-H) cells and NK1.1-selected normal murine cells (NK-M; provided by M. Cella and M. Colonna, Washington University) were maintained in RPMI medium 1640 (20–22). Media were supplemented with 10% FBS, 1% l-glutamine, 1 mM sodium pyruvate, and 50 μg/ml penicillin-streptomycin. Culture media for F8, SC, BLA, and NK-92 cell lines were supplemented with 250–500 units per ml of human IL-2.
Western Blot Analysis.
Cell lysates were prepared by using a mammalian cell lysis buffer [50 mM Tris·Cl, pH 8/5 mM EDTA/100 mM NaCl/0.5% Triton X-100 plus protease and phosphatase inhibitors (p8340 and p2850; Sigma-Aldrich)]. Twenty micrograms of lysate was separated by 10% SDS/PAGE and transferred to poly(vinylidene difluoride) membranes, blocked with 0.01% Tween 20 and 5% dry milk in PBS, and probed overnight with primary antibody. After washing with PBS, horseradish peroxidase-labeled secondary antibody was added (1:2,000 dilution, goat anti-rabbit IgG; Pierce). Proteins were visualized in an Alpha Innotech (San Leandro, CA) imager by using the supersignal west femto sensitivity substrate (Pierce).
Immunoblotting was performed by using antibodies directed against the following antigens: cleaved caspase 3 (9761), murine caspase 9 (9504), Bcl-2 (2876), Bcl-xL (2762), Bax (2772), XIAP (2042), and p100/p52 (4882) used in a 1:1000 dilution from Cell Signaling Technology (Beverly, MA), Bcl-2 (610538) from BD Biosciences, and FLIP (sc-8347, 1:500), CIAP 1/2 (sc-12410, 1:500), actin (sc-8432), p105/p50 (sc-7178, 1:1000), and hemagglutinin (sc-805) from Santa Cruz Biotechnology, and human caspase 9 (AAM-139) from Stressgen Biotechnologies (Victoria, Canada).
Plasmids.
siRNA hairpins against p100 and p105 and a control siRNA were expressed under the control of the U6 human promoter and were generated by using PLKopuro.1 and PLKoneo.1 (provided by S. Stewart, Washington University). Complementary siRNA oligos were annealed and cloned into vectors digested with AgeI and EcoRI and confirmed by sequence analysis (46). The sense siRNA oligonucleotide probes were as follows: murine and human p105, CCTTCCGCAAACTCAGCTTTA; murine p100, CCTGCATGTAACCAAGAAGAA; and human p100, GCTGCTAAATGCTGCTCAGAA. A plasmid expressing siRNA against luciferase was provided by S. Stewart (46). Recombinant lentiviruses were generated in 293T cells, for infection of SC, F8, and Daudi cell lines, selected in 2 μg/ml puromycin and 0.8 mg/ml G418.
Adenoviruses.
Adv-IκB-dn and Adv-β-gal were provided by A. Baldwin (University of North Carolina, Durham) and B. Vogelstein (The John Hopkins University, Baltimore), respectively (47), purified by using cesium chloride equilibrium centrifugation, and titered by using an Adeno-X Rapid Titer Kit (Clontech). Infection of 106 cells was performed for 2 h in serum-free medium, and then cultured in complete medium for 24 h. Optimal multiplicities of infection for each cell line were determined from the effects on an NF-κB luciferase reporter assay.
NF-κB Luciferase Assay.
For each assay, 106 cells were transfected by using Transit LTR1 reagent (Mirus, Madison, WI), with NF-κB luciferase reporter plasmid pGL2-NF-κB-Luc (provided by D. Piwnica-Worms, Washington University) (48), and infected 24 h later with Adv-IκB-dn or Adv-β-gal at the corresponding multiplicity of infection obtained from titration studies (data not shown). Results were analyzed from three independent experiments. Data are presented as means ± standard error, and comparisons were made by using the one-way ANOVA test.
Apoptosis Studies.
For apoptosis studies, 106 cells were treated with γ-irradiation (60 Gy), etoposide (150 nM; Sigma), or 10 ng/ml TNF (Sigma) or 100 ng/ml TRAIL (Biomol, Plymouth Meeting, PA) plus 10 μg/ml cycloheximide, and 24 h later 105 cells were stained with FITC-conjugated antibody against annexin V and PI (Molecular Probes). Apoptotic cells were measured with a FACScan flow cytometer (Becton Dickinson), quantitating annexin V+/PI+ cells. In experiments with adenovirus-infected cells, the treatment was performed 8 h after infection.
Supplementary Material
Acknowledgments
We thank Drs. S. Stewart, D. Piwnica-Worms, H. Piwnica-Worms, M. Cella, M. Colonna, B. Vogelstein, and A. Baldwin for gifts of plasmids, cells, expression adenoviruses, and advice and Mr. J. Harding and Ms. N. Campbell for advice and assistance. This work was supported by Public Health Service Grants CA10073, CA63417, and CA105218.
Abbreviations
- EBV
Epstein–Barr virus
- HTLV
human T cell leukemia virus
- NF-κB-ca
constitutively active NF-κB
- FLIP
Fas-associated death domain-like IL-1β-converting enzyme inhibitory protein
- CIAP
cellular inhibitor of apoptosis
- XIAP
X inhibitor of apoptosis
- TRAIL
TNF-related apoptosis-inducing ligand
- PI
propidium iodide
- IKK
IκB kinase
- siRNA
short interfering RNA
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rabson A. B., Weismann D. Clin. Cancer Res. 2005;11:2–6. [PubMed] [Google Scholar]
- 2.Xiao C., Ghosh S. Adv. Exp. Med. Biol. 2005;560:41–45. doi: 10.1007/0-387-24180-9_5. [DOI] [PubMed] [Google Scholar]
- 3.Pomerantz J. L., Baltimore D. Mol. Cell. 2002;10:693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 4.Karin M., Cao Y., Greten F. R., Li Z. W. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 5.Ghobrial I. M., Witzig T. E., Adjei A. A. CA Cancer J. Clin. 2005;55:178–194. doi: 10.3322/canjclin.55.3.178. [DOI] [PubMed] [Google Scholar]
- 6.Nakanishi C., Toi M. Nat. Rev. Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 7.Li Q., Withoff S., Verma I. M. Trends Immunol. 2005;26:318–325. doi: 10.1016/j.it.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Wu J. T., Kral J. G. J. Surg. Res. 2005;123:158–169. doi: 10.1016/j.jss.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Aggarwal B. B. Cancer Cell. 2004;6:203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Hiscott J., Kwon H., Genin P. J. Clin. Invest. 2001;107:143–151. doi: 10.1172/JCI11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luftig M., Yasui T., Soni V., Kang M.-S., Jacobson N., Cahir-McFarland E., Seed B., Kieff E. Proc. Natl. Acad. Sci. USA. 2004;101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luftig M., Prinarakis E., Yasui T., Tsichritzis T., Cahir-McFarland E., Inoue J.-I., Nakano H., Mak T. W., Yeh W.-C., Li X., et al. Proc. Natl. Acad. Sci. USA. 2003;100:15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guasparri I., Keller S. A., Cesarman E. J. Exp. Med. 2004;199:993–1003. doi: 10.1084/jem.20031467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Sgarbanti M., Arguello M., tenOever B. R., Battistini A., Lin R., Hiscott J. Oncogene. 2004;23:5770–5780. doi: 10.1038/sj.onc.1207707. [DOI] [PubMed] [Google Scholar]
- 15.Jeang K.-T. Cytokines Growth Factor Rev. 2001;12:207–217. doi: 10.1016/s1359-6101(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 16.Kfoury Y., Nasr R., Hermine O., deThe H., Bazarbachi A. Cell Death Differ. 2005;12(Suppl.):1–7. doi: 10.1038/sj.cdd.4401624. [DOI] [PubMed] [Google Scholar]
- 17.Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., et al. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 18.Schimmer A. D., Welsh K., Pinilla C., Wang Z., Krajewska M., Bonneau M. J., Pedersen I. M., Kitada S., Scott F. L., Bailly-Maitre B., et al. Cancer Cell. 2004;5:25–35. doi: 10.1016/s1535-6108(03)00332-5. [DOI] [PubMed] [Google Scholar]
- 19.Portis T., Harding J. C., Ratner L. Blood. 2001;98:1200–1208. doi: 10.1182/blood.v98.4.1200. [DOI] [PubMed] [Google Scholar]
- 20.Thompson M. P., Aggarwal B. B., Shishodia S., Estrov Z., Kurzrock R. Leukemia. 2003;17:2196–2201. doi: 10.1038/sj.leu.2403130. [DOI] [PubMed] [Google Scholar]
- 21.Alas S., Emmanouilides C., Bonavida B. Clin. Cancer Res. 2001;7:709–723. [PubMed] [Google Scholar]
- 22.Jazirehi A. R., Gan X. H., DeVos S., Emmanouilides C., Bonavida B. Mol. Cancer Ther. 2003;2:1183–1193. [PubMed] [Google Scholar]
- 23.Portis T., Harding J., Grossman W. J., Ratner L. J. Virol. 2001;75:2185–2193. doi: 10.1128/JVI.75.5.2185-2193.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenwald A., Wright G., Wiestner A., Chan W. C., Connors J. M., Campo E., Gascoyne R. D., Grogan T. M., Muller-Hermelink H. K., Smeland E. B. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 25.Monti S., Savage K. J., Kutok J. L., Feuerhake F., Kurtin P., Mihm M., Wu B., Pasqualucci L., Neuberg D., Aguiar R. C., et al. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 26.Natoli G., Saccani S., Bosisio D., Marazzi I. Nat. Immunol. 2005;6:439–444. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- 27.Houldsworth J., Olshen A. B., Cattoretti G., Donnelly G. B., Teruya-Feldstein J., Qin J., Palanisamy N., Shen Y., Dyomina K., Petlakh M., et al. Blood. 2004;103:1862–1868. doi: 10.1182/blood-2003-04-1359. [DOI] [PubMed] [Google Scholar]
- 28.Hosokawa Y. Apoptosis. 2005;10:25–34. doi: 10.1007/s10495-005-6059-6. [DOI] [PubMed] [Google Scholar]
- 29.Hatta Y., Arima N., Machino T., Itoh T., Hashimoto S., Takeuchi J., Sawada U., Hayakawa S., Yamamoto T., Horie T. Int. J. Mol. Med. 2003;11:239–242. [PubMed] [Google Scholar]
- 30.Emmerich F., Theurich S., Hummel M., Haeffkere A., Vry M. S., Dohner K., Bommert K., Stein H., Dorken B. J. Pathol. 2003;201:413–420. doi: 10.1002/path.1454. [DOI] [PubMed] [Google Scholar]
- 31.Davis R. E., Brown K. D., Siebenlist U., Staudt L. M. J. Exp. Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skinnider B. F., Mak T. W. Blood. 2002;99:4283–4297. doi: 10.1182/blood-2002-01-0099. [DOI] [PubMed] [Google Scholar]
- 33.Isaacson P. G., Duy M. Q. Nat. Rev. Cancer. 2004;4:644–653. doi: 10.1038/nrc1409. [DOI] [PubMed] [Google Scholar]
- 34.Grossman W. J., Kimata J. T., Wong F.-H., Zutter M., Ley T. J., Ratner L. Proc. Natl. Acad. Sci. USA. 1995;92:1057–1062. doi: 10.1073/pnas.92.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grossman W., Ratner L. Blood. 1997;90:783–794. [PubMed] [Google Scholar]
- 36.Meli M., D'Alessandro N., Tolomeo M., Rausa L., Notarbartolo M., Dusonchet L. Ann. N.Y. Acad. Sci. 2003;1010:232–236. doi: 10.1196/annals.1299.041. [DOI] [PubMed] [Google Scholar]
- 37.Mathas S., Lietz A., Janz M., Hinz M., Jundt F., Scheidereit C., Bommert K., Dorken B. Blood. 2003;102:1028–1034. doi: 10.1182/blood-2002-04-1154. [DOI] [PubMed] [Google Scholar]
- 38.Izban K. F., Ergin M., Qin J. Z., Martinez R. L., Pooley R. J., Saeed S. Hum. Pathol. 2000;31:1482–1490. doi: 10.1053/hupa.2000.20370. [DOI] [PubMed] [Google Scholar]
- 39.Giri D. K., Aggarwal B. B. J. Biol. Chem. 1998;273:14008–14014. doi: 10.1074/jbc.273.22.14008. [DOI] [PubMed] [Google Scholar]
- 40.Kawai H., Yamada Y., Tatsuka M., Niwa O., Yamamoto K., Suzuki F. Cancer Res. 1999;59:6038–6041. [PubMed] [Google Scholar]
- 41.Deveraux Q. L., Reed J. C. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 42.Lo J. C., Basak S., James E. S., Quiambo R., Kinsella M. C., Alegre M. L., Weih F., Franzoso G., Hoffmann A., Fu Y. X. Blood. 2005;107:1048–1055. doi: 10.1182/blood-2005-06-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Cui H., Schoering A., Ding J. L., Lane W. S., McGill G., Fisher D. E., Ding H.-F. Nat. Cell Biol. 2002;4:888–893. doi: 10.1038/ncb872. [DOI] [PubMed] [Google Scholar]
- 44.Ishikawa I., Carrasco D., Claudio E., Ryseck R.-P., Bravo R. J. Exp. Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishikawa H., Claudio E., Dambach D., Raventos-Suarez C., Ryan C., Bravo R. J. Exp. Med. 1998;187:985–996. doi: 10.1084/jem.187.7.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., et al. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C.-Y., Mayo M. W., Baldwin A. S. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 48.Jeong S. J., Radonovich M., Brady J. N., Pise-Masison C. A. Blood. 2004;104:1490–1497. doi: 10.1182/blood-2003-12-4174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.