Abstract
Telomerase plays a crucial role in telomere maintenance in vivo. To understand telomerase regulation, we have been characterizing components of the enzyme. To date several components of the mammalian telomerase holoenzyme have been identified: the essential RNA component (human telomerase RNA [hTR]), the catalytic subunit human telomerase reverse transcriptase (hTERT), and telomerase-associated protein 1. Here we describe the identification of two new proteins that interact with hTR: hStau and L22. Antisera against both proteins immunoprecipitated hTR, hTERT, and telomerase activity from cell extracts, suggesting that the proteins are associated with telomerase. Both proteins localized to the nucleolus and cytoplasm. Although these proteins are associated with telomerase, we found no evidence of their association with each other or with telomerase-associated protein 1. Both hStau and L22 are more abundant than TERT. This, together with their localization, suggests that they may be associated with other ribonucleoprotein complexes in cells. We propose that these two hTR-associated proteins may play a role in hTR processing, telomerase assembly, or localization in vivo.
INTRODUCTION
Telomerase is a specialized reverse transcriptase that is essential for telomere maintenance. The telomerase ribonucleoprotein (RNP) uses an internal RNA template to synthesize telomeric repeat sequences onto chromosome ends. Deletion of the essential RNA component of telomerase leads to progressive telomere shortening, chromosome instability, and cell death in both yeast and mouse cells (Singer and Gottschling, 1994; McEachern and Blackburn, 1996; Blasco et al., 1997; Lee et al., 1998).
The telomerase enzyme is made up of an essential core as well as several accessory proteins. The core telomerase enzyme consists of the RNA component (telomerase RNA [TR]) and the catalytic subunit (telomerase reverse transcriptase [TERT]). The structure of the RNA component is conserved (Romero and Blackburn, 1991; Lingner et al., 1994) in ciliates where the RNA is 150–200 nucleotides in length (Greider and Blackburn, 1989). In mammalian cells the RNA component is significantly larger, 390–450 nucleotides (Blasco et al., 1995; Feng et al., 1995), and the structure is highly conserved among vertebrates (Chen, Blasco, and Greider, unpublished results). The catalytic TERT component, first identified in the ciliate Euplotes (Lingner and Cech, 1996), has homologues in yeast (EST 2), human (hTERT), and mouse (mTERT) (Harrington et al., 1997b; Lingner et al., 1997b; Meyerson et al., 1997; Nakamura et al., 1997; Greenberg et al., 1998; Martin-Rivera et al., 1998). TERT contains sequence motifs similar to the conserved core of reverse transcriptases, and mutation of the conserved essential aspartate residues in the catalytic triad of reverse transcriptases eliminates telomerase activity (Lingner et al., 1997b; Weinrich et al., 1997). Minimal telomerase activity can be reconstituted in an in vitro transcription/translation extract using just the TERT and TR components (Weinrich et al., 1997; Beattie et al., 1998), suggesting that these components may be sufficient for catalysis.
In addition to the catalytic incorporation of nucleotides onto telomeric 3′ ends, telomerase has a number of additional properties including substrate recognition, processivity (Greider and Blackburn, 1987; Harrington and Greider, 1991; Collins and Greider, 1995), and primer cleavage (Collins and Greider, 1993; Melek et al., 1996). Furthermore, in mammalian cells telomerase activity is regulated in different cell types as well as in different proliferative states. It is possible that other proteins play important roles in these processes. Two proteins, p80 and p95 from Tetrahymena, copurify with telomerase (Collins et al., 1995) and are associated with the enzyme in vivo (Gandhi and Collins, 1998). The role of these two telomerase-associated proteins is not yet clear. A homologue of p80, telomerase-associated protein 1 (TEP1), is associated with telomerase in human cells (Harrington et al., 1997a; Nakayama et al., 1997). Purification of the Euplotes telomerase led to the identification of a telomerase-associated protein p43 in addition to TERT (p123) (Lingner and Cech, 1996). This p43 protein has been identified as a homologue of the La protein in mammalian cells, which binds the 3′ end of many RNA polymerase III transcripts (Lingner, Cech, and Wolin, personal communication). Recently, two chaperone proteins, p23 and Hsp90, were shown to associate with human telomerase and facilitate RNP assembly (Holt et al., 1999). In yeast at least three proteins, Est1p, Est3p, and Cdc13p, are associated with telomerase (Lin and Zakian, 1995; Steiner et al., 1996) (Lundblad, personal communication). Although they are dispensable for telomerase activity assayed in vitro (Lingner et al., 1997a), mutations in these proteins lead to telomere shortening in vivo and loss of cell viability (Lendvay et al., 1996). Thus telomerase-associated proteins may play essential yet undefined roles in telomerase biogenesis, assembly, and regulation.
Both telomere length and telomerase activity have been implicated in cellular senescence and cancer. During immortalization of mammalian cells in culture and in many human tumors, telomerase is activated (Kim et al., 1994). Artificial elongation of telomeres by ectopic hTERT expression in primary human cells leads to the bypass of cellular senescence, suggesting that telomerase expression and telomere length play a direct role in cellular immortalization (Bodnar et al., 1998; Vaziri and Benchimol, 1998). Recently it was demonstrated that expression of TERT along with SV40 T antigen and activated Ras are sufficient to transform primary human cells (Hahn et al., 1999).
To explore fully the role of telomerase both in normal cells and in tumor formation, it is important to characterize the components of this complex enzyme. Biochemical evidence indicates that human telomerase is >1000 kDa (Nakayama et al., 1997; Schnapp et al., 1998), suggesting that it contains numerous subunits in addition to hTERT and the RNA component. We set out to identify proteins that interact directly with the human telomerase RNA component. We describe here the identification of two RNA-binding proteins that interact with telomerase and may play a role in its assembly, transport, or regulation.
MATERIALS AND METHODS
Three-Hybrid Screen
The three-hybrid system (SenGupta et al., 1996) was modified and used to screen for human TR (hTR)-binding proteins. The new reporter strain L40coatng was used. This strain is similar to the L40coat strain described previously (SenGupta et al., 1996) but has a nonaggregating form of coat protein instead of the wild-type MS2 coat. Nucleotides 64–222 of hTR were fused to the 5′ end of the MS2 phage DNA to generate the pLS112 plasmid. To avoid transcriptional termination by the RNA polymerase III, we changed four nucleotides in hTR-MS2 RNA. Positions 82, 83, 102, and 103 of hTR were changed to A, A, A, and C, respectively, to disrupt a string of Us that was observed to cause transcriptional termination in initial experiments. The fusion RNA of the expected size was detected on Northern blots, indicating that it was transcribed and stable in yeast cells (our unpublished results). The pLS112 plasmid was transformed into the L40coatng strain along with cDNA–GAD fusion libraries from HeLa, Jurkat, or human testes (provided by Dr. G. Hannon and Dr. L. VanAelst, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, and Clontech, Palo Alto, CA). The positive clones that grew in the presence of 5 mM 3-amino triazol and stained positive for β-galactosidase were picked. These clones were tested for reporter gene activation in the absence of RNA plasmids, and only clones that activated the reporters in the presence of the hTR-MS2 RNA, but not MS2 alone, were further characterized. Positive clones from this screen were characterized further by DNA sequencing and expression in mammalian cells as described in the text.
Cloning Full-Length hStau
Rapid-amplification-of-cDNA-ends PCR was used to clone hStau. By the use of a tagged human testis cDNA library (Marathon ready human testis cDNA from Clontech) and gene-specific primers within the cloned region, both the 5′ and 3′ half of the gene were amplified by PCR according to the manufacturer's protocols using AP1- and hStau-specific primers 10-9 (CACCTCCAGCCTCTCTGGCAGGGGCTC) and 10-10 (GGCAAAGGAAAGACAAGACATGGCTGCG). Each half was subcloned into pCRScript SK+ (Stratagene, La Jolla, CA) and sequenced. The entire gene was then reconstructed by cloning both halves together into the pCRScript vector using a unique BamHI restriction site. The full-length cDNA was then completely sequenced. The first in-frame methionine was assigned as the first amino acid in the protein, although there is no direct evidence of translation starting at this point. An AAUAAA polyadenylation signal was found 1.3 kb downstream of the stop codon for the longest predicted ORF. By the use of this assignment, the calculated molecular weight of the hStau protein is 55 kDa. This agrees well with the size of the protein identified on Western blots (see Figure 2).
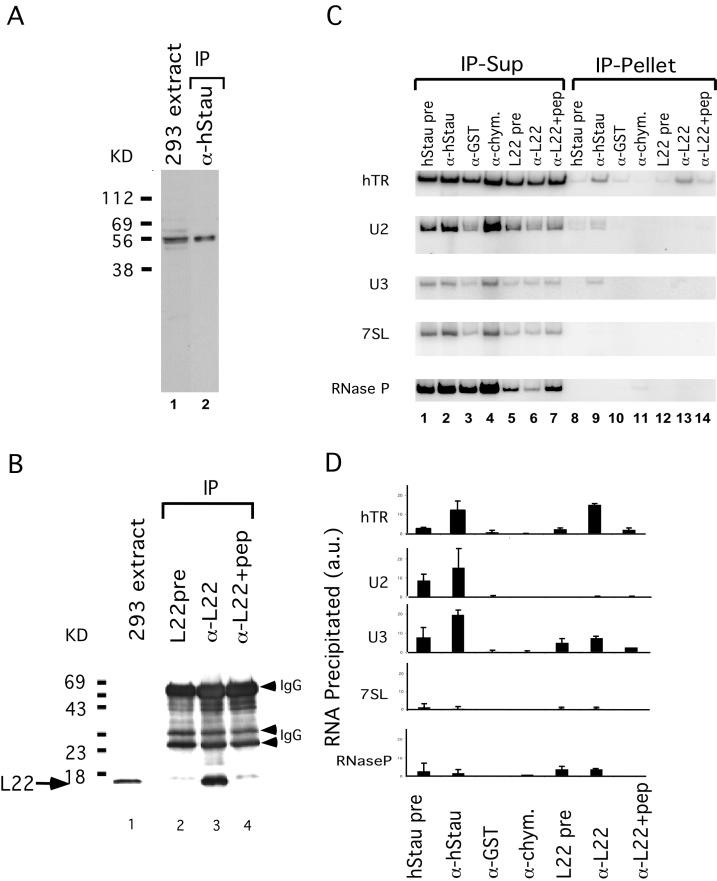
Figure 2.
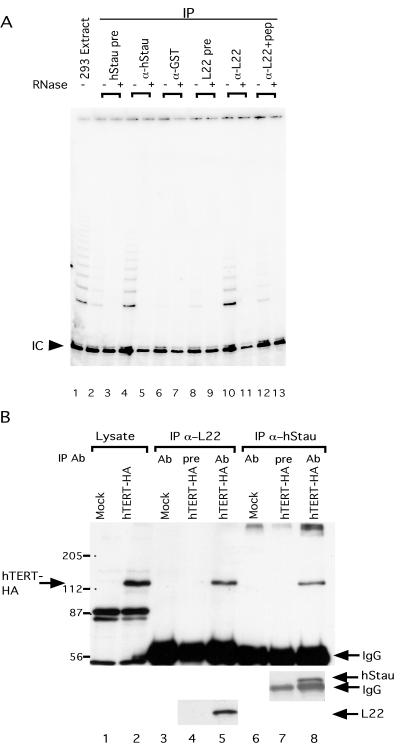
hStau and L22 interact with hTR. (A) hStau antibody specificity. Western blot analysis on a 293 cell extract (lane 1) and hStau immunoprecipitates (lane 2) using anti-hStau antibody shows the 55-kDa hStau protein. The relative mobility of molecular weight markers (in kilodaltons) is indicated on the left. (B) L22 antibody specificity. Western blot analysis on 293 cell extracts (lane 1) and L22 immunoprecipitates (lanes 2–4) is shown. Lane 2, precipitation using preimmune serum (pre); lane 3, precipitation using anti-L22 serum; and lane 4, preincubation of L22 peptide (pep) with anti-L22 serum before immunoprecipitation. The relative mobility of molecular weight markers (in kilodaltons) is indicated on the left. The arrow indicates the L22 band (15 kDa). The IgG heavy and light chains (arrowheads) are also indicated. (C) RT-PCR analysis of RNAs in the supernatant (Sup; lanes 1–7) and pellet (lanes 8–14) fractions of hStau and L22 immunoprecipitation reactions. Lanes 1 and 8, hStau precipitation using preimmune serum; lanes 2 and 9, precipitation using anti-hStau antibody; lanes 3 and 10, precipitation using anti-GST antibody; lanes 4 and 11, precipitation using anti-chymotrypsin (-chym.) antibody; lanes 5 and 12, precipitation using L22 preimmune serum; lanes 6 and 13, precipitation using anti-L22 antibody; and lanes 7 and 14, precipitation using anti-L22 antibody preincubated with L22 peptide. The RNAs amplified in the fractions are indicated on the left (hTR, U2, U3, 7SL, and RNase P). (D) Quantitation of RNAs precipitated in C. The RNA precipitated is expressed as a fraction of the total signal intensity obtained in both the pellet and supernatant. RNAs assayed are indicated on the left, and antibodies used are indicated on the bottom. L22+pep, L22 antibodies preincubated with the L22 peptide before immunoprecipitation. a.u., arbitrary units.
Generation of Antibodies
For L22, a synthetic peptide was generated that corresponds to the N-terminal region of the protein (CMAPVKKLVVKGG). The peptide was coupled to KLH and used to generate antisera in rabbits. To generate antibodies to hStau, we subcloned the partial cDNA that was obtained in the initial three-hybrid library into the bacterial expression vector to generate an N-terminal fusion of GST with the C-terminal 448 amino acids of hStau. The fusion protein GST-hStau was overexpressed in bacteria, purified over a glutathione column (Pharmacia, Piscataway, NJ), and used to generate antibodies in rabbits (Covance Research Products, Denver, PA).
Antisera from the rabbits were initially screened for protein or peptide binding using an ELISA assay. Positive antisera were then tested for specificity on Westerns and by immunoprecipitation. The antisera against L22 recognized a 15-kDa protein as expected both in cell lysates and in immunoprecipitations. Antibodies directed against hStau recognized a major 55-kDa protein in Westerns on extracts and immunoprecipitations.
Western, Immunoprecipitation, Immunodepletion, and Immunofluorescence
Western analysis, immunoprecipitation, and immunofluorescence were performed according to the procedures of Harlow and Lane (1988). Human 293 cell extracts were made using either Nonidet P-40 (up to 1%) buffer (Harlow and Lane, 1988) or hypotonic buffer (Counter et al., 1992) and used for Western and immunoprecipitation (IP) analysis. Some hStau immunoprecipitations were performed using hStau serum that was coupled to CNBr-activated Sepharose beads to avoid IgG bands that have a molecular weight similar to that of hStau in the subsequent Western analysis.
Immunodepletion was done by sequential immunoprecipitation. After four rounds of immunoprecipitation, the supernatant was Western blotted, and the hStau level was quantitated relative to the level in the starting extract.
In L22 or nucleolin immunofluorescence experiments, fixed cells were stained either with L22 or nucleolin (C23; Santa Cruz Biotechnology, Santa Cruz, CA) antiserum. After staining with Texas Red– or indocarbocyanine-conjugated secondary antibodies, cells were mounted with Vectashield mounting media (Vector Laboratories, Burlingame, CA) (with or without DAPI) and viewed using a Zeiss (Thornwood, NJ) fluorescence microscope.
Green Fluorescent Protein (GFP) Fusion Proteins
To determine the subcellular localization of the hStau protein, we fused hStau cDNA to GFP in pEGFP-C1 (Clontech), and the fusion protein was expressed in 293 cells. The expression of fusion protein was confirmed by Western blot analysis. Green fluorescence images were obtained using a Zeiss fluorescence microscope after 48 h of transfection.
Reverse Transcriptase (RT)-PCR
RT-PCR was used to quantitate RNAs in the supernatant and pellet of immunoprecipitation reactions. RNAs were prepared from both supernatant and pellet fractions by phenol and chloroform extraction and ethanol precipitation. The same amount of total RNA was then used for each reaction in a first-strand cDNA synthesis reaction using random hexamer primers and Superscript II reverse transcriptase (Life Technologies, Gaithersburg, MD). The cDNAs were then PCR amplified using hTR-, U2-, U3-, 7SL-, or RNase P-specific primers. The primers used are as follows: hTR, GCCTGGGAGGGGTGGTGGCCATTTTTTG and GTTTGCTCTAGAATGAACGGTGGAAG; U2, ATCGCTTCTCGGCCTTTT and GGGTGCACCGTTCCTGGGA; U3, GACTATACTTTCAGGGATCATTTC and CCACTCAGACCGCGTTCTCTC; 7SL, GTGCCTGTAGTCCCAGCTAC and GAGACGGGGTCTCGCTATG; and RNase P, GGAAGGTCTGAGACTAG and ATCTCCTGCCCAGTCTG. The number of PCR cycles was adjusted for different cDNA amplifications so that PCR was in the linear range. To control for genomic DNA contamination, cDNA synthesis reactions were also done in the absence of the RT. No signal was generated in the absence of RT. PCR products were separated on a 6% native polyacrylamide gel, dried, and exposed. Signal intensity was quantified on a STORM PhosphorImager system (Molecular Dynamics, Sunnyvale, CA).
Telomerase Assay
Cell extracts, immunoprecipitation supernatants, or pellet fractions were assayed in a two-step telomerase assay (telomeric repeat amplification protocol [TRAP]) similar to that described previously (Autexier et al., 1996). This two-step procedure uses a limited number of PCR cycles for amplification of the telomerase products so that the signal will be in the linear range. Thus relative signal intensities reflect relative activity in a semiquantitative manner. Negative controls that had either no extract or RNase-treated samples were used. An internal standard for product amplification in the PCR step of the assay was also included in each reaction (Kim and Wu, 1997).
RESULTS
Cloning Telomerase-associated Proteins
To identify proteins that bind to the hTR, we used a modified three-hybrid system (SenGupta et al., 1996). Nucleotides 64–222 of the 451 nucleotides of human telomerase RNA were fused to the MS2 phage RNA and expressed in yeast behind the RNase P promoter. This region of the RNA can form a highly conserved stem–loop structure (Chen, Blasco, and Greider, unpublished results) and is required for activity in in vitro reconstitution assays (Autexier et al., 1996; Beattie et al., 1998). Three different human cDNA–GAD fusion libraries (from HeLa, Jurkat, and human testes) were screened for proteins that interact with hTR (see MATERIALS AND METHODS). Positives that interacted with hTR-MS2, but not MS2 alone, were identified, and the partial cDNA clones were tagged with the hemagglutinin (HA) epitope. These tagged clones were used to transfect 293 cells to test whether they would interact with hTR in human cells. Two proteins were identified that showed specific association with hTR after immunoprecipitation with HA antibody (our unpublished results). They are ribosomal-associated protein L22, which was shown previously to bind Epstein-Barr virus (EBV)-encoded RNA (EBER) in EBV-infected cells (Toczyski et al., 1994), and a new protein that we initially called telomerase-associated protein 3 (TEP3). This protein was identified recently by others and given the name hStau (Marion et al., 1999; Wickham et al., 1999).
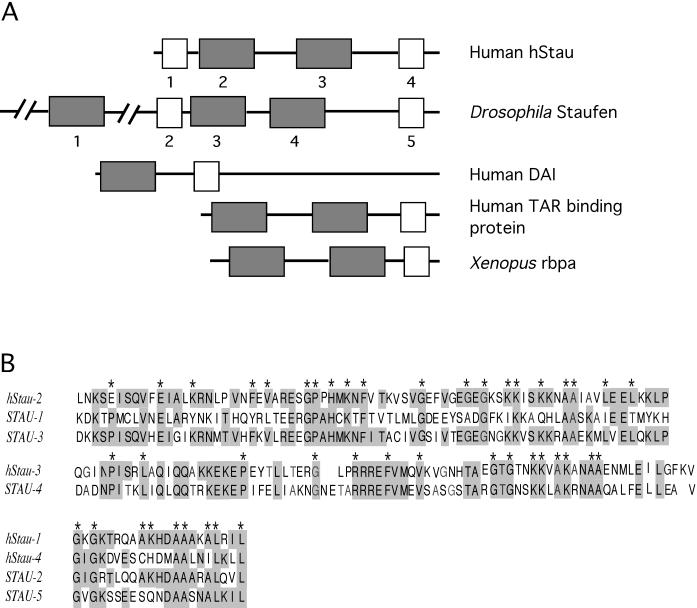
We cloned the full-length cDNA of hStau using rapid-amplification-of-cDNA-ends PCR from testis cDNA. The cDNA encodes a 496-amino-acid open reading frame with a 1.3-kb-long 3′-untranslated region. Our clone corresponds to the most abundant isoform of the several hStau isoforms described by Wickham et al. (1999). A motif search revealed several regions of the protein that contain significant homology to double-stranded RNA-binding domains that were originally identified in the Drosophila Staufen protein (Figure 1). hStau protein, like other double-stranded RNA-binding domain proteins, has two full-length and two short RNA-binding domains. The organization of the domains in hStau is similar to that of Staufen although hStau is shorter in length overall (Figure 1A). Furthermore the sequence similarity is limited to the RNA-binding domains; the fact that other regions of the proteins are not conserved suggests that although hStau uses Staufen-related motifs to bind to RNA, it may have a different function than does Staufen.
Figure 1.
Structural alignment and sequence comparison of domains in hStau and other double-stranded RNA (dsRNA)-binding proteins. (A) Structure of hStau and other proteins that contain double-stranded RNA-binding domains. Full-length dsRNA-binding domains are indicated by gray boxes, and short domains are indicated by white boxes. The numbers under the domains in hStau and Staufen correspond to the sequences listed in B. (B) Sequence homology between double-stranded RNA-binding domains of human Staufen and Drosophila Staufen (STAU). Top and Middle, alignment of the full-length domains. Bottom, alignment of the short domains. Identical residues are shaded; * symbols indicate the residues that are highly conserved in most of the double-stranded RNA-binding domains containing proteins (St. Johnston et al., 1992) (note that not all sequences are shown). Sequences used in the alignment are as follows: hStau-1, aa 59–79; hStau-2, aa 100–172; hStau-3, aa 202–275; hStau-4, aa 452–472; STAU-1, aa 308–380; STAU-2, aa 490–559; STAU-3, aa 575–647; STAU-4, aa 708–782; and STAU-5, aa 948-1020.
Interaction with hTR In Vivo
To examine further the interaction of L22 and hStau with hTR, we generated antibodies to both proteins and tested the ability of the antibodies to precipitate hTR. For L22, anti-peptide antibodies were made, whereas for hStau, antibodies to a recombinant GST fusion protein were generated (see MATERIALS AND METHODS). IP and Western blotting showed that these antibodies were specific for the appropriate proteins (Figure 2, A and B). Anti-hStau antibody recognized a major 55-kDa protein on Western blots of both cell lysates and immunoprecipitation reactions, consistent with its predicted molecular weight (see Figures 2A and 6A). Other isoforms of the protein were also detected, and they are likely to represent the other isoforms of the hStau protein containing different N termini (Wickham et al., 1999). The L22 antibody precipitated a 15-kDa protein that was not seen using preimmune serum or when the peptide was preincubated with the antibody before the IP (Figure 2B).
Figure 6.
Interaction between telomerase-associated proteins. (A) hStau and L22 do not interact with each other. A 293 cell lysate was immunoprecipitated with hStau preimmune serum (lane 2), hStau antibody (lane 3), L22 preimmune serum (lane 4), or L22 antibody (lane 5). The pellet fractions were analyzed by Western hybridization probed with either hStau antibody (top) or L22 antibody (bottom). (B) hStau and myc-TEP1 do not interact. 293 cells were either mock transfected (lanes 1, 3, and 4) or transfected with the myc-TEP1 construct (lanes 2, 5, and 6). Cell lysates were either run directly on a Western (lanes 1 and 2) or immunoprecipitated with either hStau preimmune serum (lanes 3 and 5) or hStau antibody (lanes 4 and 6). The Western was probed with anti-myc antibody (top) or hStau antibody (bottom). (C) L22 and myc-TEP1 do not interact. 293 cells were either mock transfected (lanes 1 and 3) or transfected with the myc-TEP1 construct (lanes 2, 4, and 5), and cell lysates were either run directly on a Western (lanes 1 and 2) or immunoprecipitated with either hStau preimmune serum (lane 4) or L22 antibody (lanes 3 and 5). The Western was probed with anti-myc antibody (top) or L22 antibody (bottom). The relative mobility of molecular weight markers (in kilodaltons) is indicated on the left in A–C.
To examine whether L22 and hStau interact with hTR in vivo, immunoprecipitations were performed on 293 cell lysates. The antibodies against L22 and hStau described above were used as well as several unrelated antibodies as controls. Using PCR, we measured the amount of hTR in the supernatant and pellet fraction of the immunoprecipitation reactions (Figure 2C). hTR was enriched in the pellets of the anti-hStau and anti-L22 immunoprecipitation reactions relative to reactions using the preimmune sera or two unrelated antibodies, anti-GST and anti-chymotrypsin. In addition when the anti-L22 antibody was preincubated with L22 peptide, a lower fraction of hTR was precipitated, indicating that the immunoprecipitation is relatively specific (Figure 2C). To quantify the level of enrichment, we repeated this experiment several times, and the average enrichments are shown in Figure 2D. As a second control for specificity of the interaction with hTR, we tested whether four abundant small RNAs (U2, U3, 7SL, and RNase P RNA) were also precipitated by our antibodies (Figure 2, C and D). Most of these RNAs were not detected at significant levels in the immunoprecipitation pellets, although some U2 and U3 RNA was detected in the pellet fraction of the anti-hStau precipitation. The precipitation of U2 and U3 could be caused by the high copy number of these RNAs (2–5 × 105 per cell) (Baserga and Steitz, 1993) compared with the hTR that is only present at 500 copies per cell (Avilion, 1995). Alternatively, hStau may have some affinity for U2 and U3 RNA.
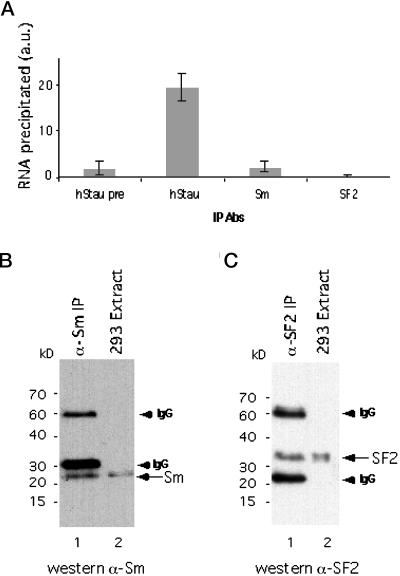
As a final control for specificity, we tested whether hTR was precipitated by antibodies directed against other abundant RNA-binding proteins, such as Sm (Baserga and Steitz, 1993) and SF2 (Krainer et al., 1991) proteins. Anti-Sm and anti-SF2 antibodies precipitated these two proteins, respectively (Figure 3, B and C). The level of hTR brought down by these antibodies was significantly less than that brought down by antibodies to hStau (Figure 3A). Together the immunoprecipitation experiments suggest that hStau and L22 specifically associate with hTR in vivo.
Figure 3.
Anti-Sm and anti-SF2 antibodies do not precipitate hTR. (A) Quantitation of hTR precipitated as described in Figure 2D. Antibodies used are indicated on the bottom. (B) Western blot analysis on 293 cell extracts (lane 2) and an Sm immunoprecipitate (lane 1). The arrow indicates Sm proteins (28–29 kDa) recognized by mAb Y12. IgG bands (arrowheads) are also indicated on the right. (C) Western blot analysis on 293 cell extracts (lane 2) and SF2 immunoprecipitates (lane 1). The arrow indicates the SF2 protein (33 kDa). IgG bands (arrowheads) are also indicated on the right.
To estimate how much hTR is associated with hStau or L22, we immunodepleted hStau or L22 from human 293 cell extracts and assayed for the presence of hTR by RT-PCR. Upon removal of ∼90% of the hStau protein from extracts, there was a 15–50% reduction of hTR (our unpublished results), whereas for L22, only ∼10% of hTR was associated with the protein (our unpublished results). These results suggest that, although hTR is associated with hStau and L22, not all of the hTR in the cell is associated with hStau or L22.
hStau and L22 Interact with Telomerase
To determine whether hStau or L22 is associated with telomerase, we assayed telomerase activity in anti-hStau and anti-L22 immunoprecipitates. Immunoprecipitation reactions were performed using either anti-hStau, anti-L22, preimmune serum, or an unrelated anti-GST antibody. The pellet fractions were assayed for telomerase activity by the TRAP assay. Telomerase activity was specifically immunoprecipitated with both the anti-hStau and anti-L22 antisera. Little activity was seen in immunoprecipitations with preimmune sera, and none was seen with the anti-GST antibody (Figure 4A). Preincubation with the L22 peptide significantly reduced the level of telomerase activity precipitated in the anti-L22 pellet fractions. Thus both hStau and L22 are associated with telomerase activity.
Figure 4.
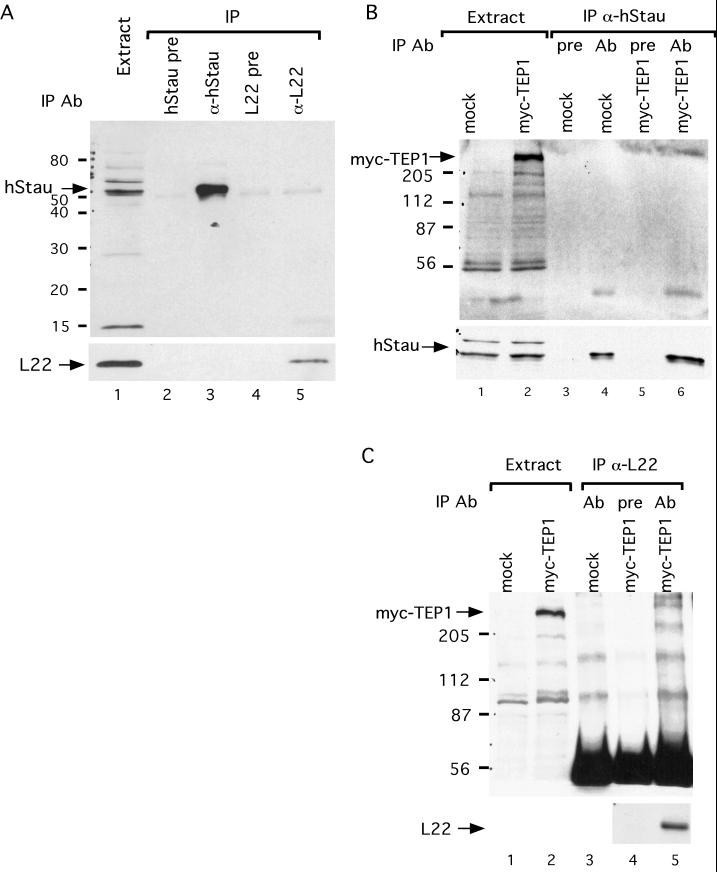
hStau and L22 interact with telomerase and hTERT. (A) hStau and L22 associate with telomerase activity in 293 cells. Telomerase assays (modified TRAP, see MATERIALS AND METHODS) were performed on a 293 extract (assayed at 5 μg of protein; lane1) and immunoprecipitation pellets (lanes 2–13) from immunoprecipitations with various antibodies. The antibodies used are indicated at the top of the gel. Lanes 2 and 3, precipitation using hStau preimmune serum; lanes 4 and 5, precipitation using anti-hStau antibody; lanes 6 and 7, precipitation using anti-GST antibody; lanes 8 and 9, precipitation using L22 preimmune serum; lanes 10 and 11, precipitation using anti-L22 antibody; and lanes 12 and 13, precipitation using anti-L22 antibody preincubated with L22 peptides. Samples in lanes 3, 5, 7, 9, 11, and 13 were pretreated with RNase (+) before the telomerase reactions. (B) hStau and L22 interact with hTERT. Western blot analysis using anti-HA (top), anti-hStau (middle), or anti-L22 (bottom) antibody on cell lysates (lanes 1 and 2) or various immunoprecipitation pellets (lanes 3–8) is shown. Extracts used are a mock-transfected 293 extract (lanes 1, 3, and 6) and an hTERT-HA–transfected cell extract (lanes 2, 4, 5, 7, and 8). The antibodies used in immunoprecipitation reactions were the following: lanes 3 and 5, precipitation using anti-L22; lane 4, precipitation using L22 preimmune serum; lanes 6 and 8, precipitation using anti-hStau; and lane 7, precipitation using hStau preimmune serum. The relative mobility of molecular weight markers (in kilodaltons) is indicated on the left. The identity of various bands is indicated by arrows.
Telomerase activity requires the catalytic subunit hTERT (Nakamura et al., 1997). We tested whether the hTERT protein was present in the immunoprecipitation reactions with antibodies to hStau and L22, but we failed to detect endogenous hTERT on Westerns (our unpublished results). This result is similar to that found using antibodies to the telomerase-associated protein TEP1 (Harrington et al., 1997b) and is likely caused by the low levels of endogenous hTERT in cells and/or by the low affinity of the hTERT antibody. Thus to examine the association of hStau and L22 with hTERT, we transfected 293 cells with a cDNA that expresses hTERT tagged with the HA epitope (Counter et al., 1998). Cell extracts were made, and antibodies directed against hStau and L22 were used in immunoprecipitation reactions. Western analysis of the pellet fraction using anti-HA antibodies showed that hTERT was precipitated by both anti-hStau and anti-L22 antisera but not by preimmune sera (Figure 4B). These results indicate that hStau and L22 protein associate with hTERT.
Subcellular Localization of hStau
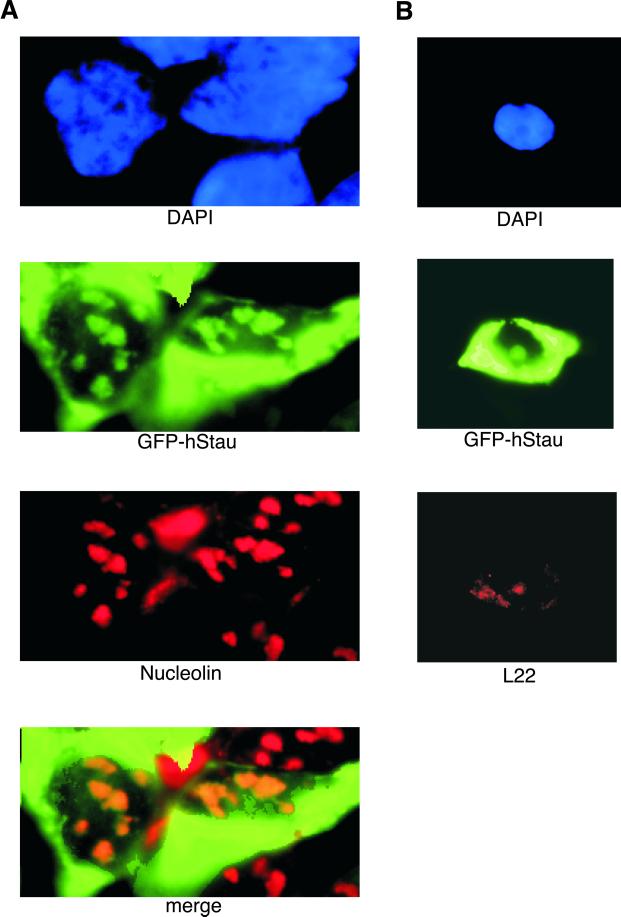
To determine the subcellular localization of the hStau protein, we fused hStau to GFP and transiently expressed the fusion protein in 293 cells. hStau localized to both nucleoli and cytoplasm (Figure 5). We assayed the location of nucleolin and L22 as a control for nucleolar staining. The hStau nucleolar localization was coincident with the staining by an anti-nucleolin or anti-L22 antibody (Figure 5). L22 has been shown previously to localize to both nucleoli and cytoplasm (Toczyski et al., 1994). Recent reports have suggested that the hStau protein localizes to the rough endoplasmic reticulum (Marion et al., 1999; Wickham et al., 1999), although nucleolar staining was also reported by one group (Marion et al., 1999). The hStau fluorescence observed was specific because it was not observed either when cells were mock treated or when GFP alone was expressed. Thus hStau is present in nucleoli where the RNA component hTR also localizes (Mitchell et al., 1999; Narayanan et al., 1999).
Figure 5.
Subcellular localization of the hStau and the L22 protein. (A) Green fluorescence of GFP-hStau and nucleolin immunofluorescence of 293 cells expressing the GFP-hStau fusion. Cells were stained with DAPI (blue, top), green fluorescence (green, second from top), and anti-nucleolin (red, second from bottom), and images were merged (bottom). (B) Green fluorescence of GFP-hStau and L22 immunofluorescence of 293 cells expressing the GFP-hStau fusion. Cells were stained with DAPI (blue, top) and L22 antiserum (red, bottom) and processed for a green fluorescence image (green, middle).
Interactions among Telomerase-associated Proteins
TEP1 was identified as a homologue of the Tetrahymena p80 telomerase component and was shown to interact with hTERT in vivo (Harrington et al., 1997a). To test whether hStau or L22 interact with each other or with TEP1, we performed a series of coimmunoprecipitation reactions. Antibodies directed against hStau immunoprecipitated hStau, but L22 was not detectable in the pellets. Similarly, antibodies directed against L22 brought down L22, but hStau was not detectable in the pellets (Figure 6A). A faint band migrating slightly faster than hStau was seen in both the hStau preimmune and L22 immunoprecipitation reactions. Because this band is seen at similar levels in the preimmune sera and the specific L22 immunoprecipitation, it likely represents a nonspecific association. These results suggest that hStau and L22 do not interact with each other.
To examine the interaction of hStau and L22 with TEP1, we used 293 cells transfected with expression plasmids for myc-tagged TEP1. In these transfected cells, anti-hStau antibody immunoprecipitated hStau, but TEP1 was not detected by Western analysis probing for the myc-epitope tag (Figure 6B). Similarly, anti-L22 antibody immunoprecipitated L22 but not myc-tagged TEP1 from myc-TEP1–expressed cells (Figure 6C). However, as described previously (Harrington et al., 1997b), myc-TEP1 protein was coprecipitated when overexpressed hTERT was immunoprecipitated (our unpublished results), indicating that TEP1 protein associates with telomerase. Thus, these results suggest that TEP1, hStau, and L22 are not associated with each other, yet they are individually associated with hTR and hTERT.
DISCUSSION
We have identified two new hTR-binding proteins, L22 and hStau. Although both of these proteins are associated with hTR in vivo, they appear not to associate with each other. The nucleolar localization suggests that these proteins might be involved with various stages of RNA processing or RNP assembly.
hStau Is an RNA-binding Protein
hStau is a double-stranded RNA-binding protein that associates with hTR in vivo. hStau belongs to a family of proteins that contains multiple double-stranded RNA-binding domains (St. Johnston et al., 1992). The structural organization of the domains as well as sequences within these domains is most similar to that of the Drosophila Staufen protein (St. Johnston et al., 1991). However sequences outside of the dsRNA-binding region are not conserved between hStau and Staufen, suggesting that although the RNA-binding function may be conserved, other functions may differ between these proteins. In Drosophila, the Staufen protein specifically binds a structure in the bicoid and oskar mRNA and is necessary for the proper localization of these RNAs to the anterior and posterior of the oocyte, respectively (St. Johnston et al., 1991). Staufen protein also binds to prospero mRNA and contributes to its localization to neuroblasts during development (Broadus et al., 1998). In vitro, Staufen binds directly to bicoid mRNA (St. Johnston et al., 1992). Although the binding of Staufen in vivo is specific, a 76-amino-acid recombinant fragment containing the double-stranded RNA-binding domain of Staufen will bind to many RNAs in vitro that contain extensive secondary structure but not unstructured RNAs (St. Johnston et al., 1992). Upon injection of various RNAs into Drosophila embryos, the Staufen protein binds the bicoid 3′-untranslated region strongly but has little affinity for other structured RNAs like VA1 or poly (rI/rC) (Ferrandon et al., 1994). Recently hStau was shown to bind bicoid RNA in vitro (Marion et al., 1999; Wickham et al., 1999). We found that in vivo hStau binds hTR and to some extent U2 and U3 RNA. In contrast there was no detectable association of hStau with 7SL or RNase P RNAs in vivo (Figure 2C). This in vivo binding specificity may be achieved in part via the specific conformation of the RNA and/or the association with other proteins in an RNP complex.
L22 Is an RNA-binding Protein
L22, which associates with hTR, is an RNA-binding protein. L22 was described previously as a ribosomal-associated protein. However, although it is localized in the nucleolus and purifies with ribosomes, little is known about its role in ribosome function (Lavergne et al., 1987; Toczyski et al., 1994). The binding site for L22 was determined previously by RNA selection to include a stem–loop structure with three conserved nucleotides at the top of the stem (Dobbelstein and Shenk, 1995). A similar stem–loop structure containing the consensus L22-binding site is present in the hTR fragment that was used in the three-hybrid screen (Chen, Blasco, and Greider, unpublished results). This suggests that L22 may bind directly to hTR via this stem–loop structure. L22 also binds EBER1 in EBV-infected human B lymphocytes (Toczyski et al., 1994). Upon infection with EBV, L22 translocates to the nucleoplasm and associates with EBER1 (Toczyski et al., 1994). It thus will be interesting to determine whether the interaction between L22 and hTR is altered by EBV infection.
The Role of RNA-binding Proteins in Telomerase Complexes
RNA-binding proteins may be involved in the maturation, localization, and assembly of RNPs. hStau and L22 may mediate these functions for the telomerase RNP. L22 was reported previously to localize to nucleoli (Toczyski et al., 1994). We found that hStau also localizes to nucleoli. Interestingly, recent studies have shown that hTR contains a conserved box H/ACA sno-RNA motif and localizes to nucleoli (Mitchell et al., 1999; Narayanan et al., 1999). Because many RNA-processing activities as well as RNP assembly are performed in the nucleolus (Pederson, 1998), L22 and hStau may play a role in hTR localization, processing, and telomerase RNP assembly. The p43 protein from Euplotes, which copurifies with TERT, is also an RNA-binding protein similar to the La antigen (Lingner, Cech, and Wolin, personal communication). The La antigen is involved in the stability of RNA polymerase III transcripts as well as their maturation and assembly into functional RNPs (Wolin and Matera, 1999). A variety of different RNA-binding proteins may associate with hTR at different stages of its biogenesis and contribute to the transport, localization, and assembly of telomerase.
Although hStau and L22 are individually associated with hTERT, they fail to interact with each other. This suggests that they may be associated with telomerase at distinct stages of biogenesis. In agreement with this, we found that only ∼15–50% of hTR is associated with hStau at steady state in vivo. hTR is thus likely to be present in other complexes in addition to those with hStau and L22.
Multiprotein Complexes and RNP Biogenesis
Another common feature shared among TEP1, hStau, and L22 is that they participate in other cellular RNP complexes in addition to telomerase association. TEP1 is associated with the vault RNP (Kickhoefer et al., 1999). Vaults are evolutionary-conserved, large cytoplasmic RNPs with a possible role in nucleocytoplasmic transport (Chugani et al., 1993; Kickhoefer et al., 1996). hStau mRNA is expressed at similar levels in a wide variety of human tissues; the protein level however varies significantly in different tissues, suggesting possible translational regulation (our unpublished results). The hStau protein is localized to the rough endoplasmic reticulum and the nucleolus (Marion et al., 1999; Wickham et al., 1999) (and this report). L22 is localized to the nucleolus and copurifies with ribosomes, in addition to its binding to EBER RNAs in EBV-positive cells (Toczyski et al., 1994). Such sharing of components has been seen in several multiprotein complexes. For example, p21 interacts with cyclin-dependent kinase (CDK) (Xiong et al., 1993) to inhibit its activity and also binds PCNA to inhibit PCNA-dependent DNA replication (Warbrick et al., 1995; Gulbis et al., 1996). The p21-binding site for CDK and PCNA appears to be separable (Chen et al., 1995). Similarly, the coactivator p300/CBP (histone acetyltransferase) is found in a large number of transcription complexes, including the nuclear receptors (Korzus et al., 1998) cAMP response element–binding protein (Chrivia et al., 1993; Kwok et al., 1994) and STAT-1 (Darnell, 1997). The high abundance of TEP1, L22, and hStau is consistent with their playing a role in a variety of multisubunit complexes and their role in RNA processing and RNP assembly.
ACKNOWLEDGMENTS
We thank Drs. Lea Harrington and Murray Robinson for providing expression constructs for human TEP1 and antibodies against TEP1 and hTERT. We thank Dr. Robert Weinberg for providing the expression construct for hTERT-HA, Dr. Maria Blasco for antibodies against hTERT, and Dr. Adrian Krainer for the Sm and SF2 antibodies. We thank the labs of Drs. S. Fields and M. Wickens for sharing three-hybrid information and reagents. We also thank Drs. J. Boeke and R. Green and the members of the Greider lab for critical reading of the manuscript. This work was supported by National Institutes of Health grants AG-09383 to C.W.G. and GM-28220 to R.S. S.L. was supported by the National Cancer Institute postdoctoral fellowship CA-68736.
REFERENCES
- Autexier C, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- Avilion AA. Program in Cellular and Developmental Biology. Stony Brook, NY: State University of New York at Stony Brook; 1995. Characterization and expression of human telomerase. [Google Scholar]
- Baserga SJ, Steitz JA. The diverse world of small ribonucleoproteins. In: Gesteland RF, Atkins JF, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 359–381. [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- Blasco MA, Funk W, Villeponteau B, Greider CW. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA [see comments] Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells [see comments] Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- Chen J, Jackson PK, Kirschner MW, Dutta A. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature. 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Chugani DC, Rome LH, Kedersha NL. Evidence that vault ribonucleoprotein particles localize to the nuclear pore complex. J Cell Sci. 1993;106:23–29. doi: 10.1242/jcs.106.1.23. [DOI] [PubMed] [Google Scholar]
- Collins K, Greider CW. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- Collins K, Greider CW. Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J. 1995;14:5422–5432. doi: 10.1002/j.1460-2075.1995.tb00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Kobayashi R, Greider CW. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Dobbelstein M, Shenk T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J Virol. 1995;69:8027–8034. doi: 10.1128/jvi.69.12.8027-8034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Elphick L, Nusslein-Volhard C, St. Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumor cells with defined genetic elements [see comments] Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein [see comments] Science. 1997a;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- Harrington L, Zhou W, McPhail T, Oulton R, Yeung DS, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997b;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- Holt SE, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Stephen AG, Harrington L, Robinson MO, Rome LH. Vaults and telomerase share a common subunit, TEP1. J Biol Chem. 1999;274:32712–32717. doi: 10.1074/jbc.274.46.32712. [DOI] [PubMed] [Google Scholar]
- Kickhoefer VA, Vasu SK, Rome LH. Vaults are the answer, what is the question. Trends Cell Biol. 1996;6:174–178. doi: 10.1016/0962-8924(96)10014-3. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer [see comments] Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kim NW, Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP) Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB [see comments] Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lavergne JP, Conquet F, Reboud JP, Reboud AM. Role of acidic phosphoproteins in the partial reconstitution of the active 60 S ribosomal subunit. FEBS Lett. 1987;216:83–88. doi: 10.1016/0014-5793(87)80761-5. [DOI] [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, II, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997a;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase [see comments] Science. 1997b;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Marion RM, Fortes P, Beloso A, Dotti C, Ortin J. A human sequence homologue of staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Rivera L, Herrera E, Albar JP, Blasco MA. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc Natl Acad Sci USA. 1998;95:10471–10476. doi: 10.1073/pnas.95.18.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- Melek M, Greene EC, Shippen DE. Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human [see comments] Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- Narayanan A, Lukowiak A, Jady BE, Dragon F, Kiss T, Terns RM, Terns MP. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 1999;18:5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Schnapp G, Rodi HP, Rettig WJ, Schnapp A, Damm K. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 1998;26:3311–3313. doi: 10.1093/nar/26.13.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase [see comments] Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Steiner BR, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- St. Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski DP, Matera AG, Ward DC, Steitz JA. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci USA. 1994;91:3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- Weinrich SL, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Wickham L, Duchane T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Matera AG. The trials and travels of tRNA. Genes Dev. 1999;13:1–10. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases [see comments] Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]